Abstract
Kinesin is a homodimeric motor with two catalytic heads joined to a stalk via short neck linkers (NLs). We measured the torsional properties of single recombinant molecules by tracking the thermal angular motions of fluorescently labeled beads bound to the C terminus of the stalk. When kinesin heads were immobilized on microtubules (MTs) under varied nucleotide conditions, we observed bounded or unbounded angular diffusion, depending on whether one or both heads were attached to the MT. Free rotation implies that NLs act as swivels. From data on constrained diffusion, we conclude that the coiled-coil stalk domains are ≈30-fold stiffer than its flexible “hinge” regions. Surprisingly, while tracking processive kinesin motion at low ATP concentrations, we observed occasional abrupt reversals in the directional orientations of the stalk. Our results impose constraints on kinesin walking models and suggest a role for rotational freedom in cargo transport.
Keywords: microtubule, molecular motor, rotation, single molecule, yaw with one degree accuracy
Conventional kinesin (kinesin-1) is a motor protein that transports cargo along microtubules (MTs) in cells, converting the chemical energy of nucleotide hydrolysis into mechanical work (1). The structure of kinesin consists of twin catalytic motor domains (heads) that bind to the MT substrate with nucleotide-dependent affinity, joined by neck linkers (NLs) to the N terminus of an extended, α-helical coiled-coil (CC) stalk (Fig. 1A). Considerable recent progress has been made in understanding the mechanochemistry of the kinesin motor domains, which enable individual molecules to move processively, undertaking hundreds of successive 8-nm steps while hydrolyzing one ATP molecule per step (2–4). However, little is known about the structure of the stalk or its role in the currently favored model for kinesin movement, the so-called “asymmetric hand-over-hand” stepping mechanism (5–7). In this mechanism, the heads of kinesin advance in strict alternation, exchanging leading, and trailing roles at each step, analogous to a person walking (1). An asymmetric mechanism requires that the “left” and “right” steps be slightly different; however, in contrast with a symmetric hand-over-hand walk, it does not require continuous rotation of the kinesin stalk. Consistent with this model, a landmark experiment by the Gelles group (8) that tracked the angular motions of MTs driven by single kinesin motors affixed to a surface did not find evidence for large-scale rotations.
Fig. 1.
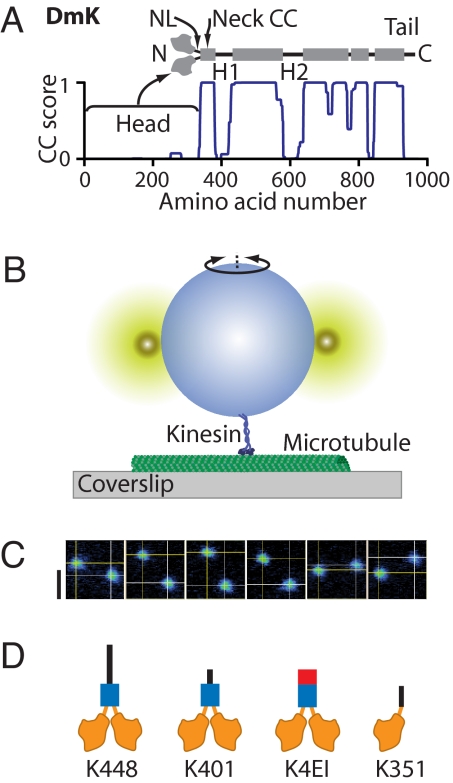
Kinesin structure, torsional assay, and constructs studied. (A) Drosophila kinesin (DmK) sequence, structure, and structural predictions. The kinesin dimer consists of two N-terminal motor domains (heads) attached by NLs to a largely CC stalk, which is interrupted by two large hinge regions (H1 and H2) and several smaller ones. The COILS score (blue line) (21) predicts CC propensity based on sequence similarity to known CCs. (B) Schematic representation showing the experimental geometry of the rotation assay (not to scale). A polystyrene bead, labeled by two smaller fluorescent particles, is attached to the terminus of the stalk of a kinesin molecule bound to a MT immobilized on a glass surface. Angular thermal motions of the tethered bead about the vertical axis are recorded. (C) Six false-color CCD camera frames show the thermal motions of fluorescent markers on a tethered bead; lines are drawn through the computed centroid coordinates (yellow). Time between frames, 1 s. (Scale bar, 1 μm.) (D) Diagram of the kinesin constructs used in this study: K448, K401, and K351. Predicted CC regions are colored blue, hinge regions are black, and motor domains and NLs are orange. K4EI has H1 replaced by an in-phase stable CC (red), resulting in a continuous CC stalk.
Although no net rotations of the kinesin stalk are expected during asymmetric hand-over-hand motion, significant momentary angular changes are nevertheless possible. If kinesin were a nearly rigid molecule, ±180° reorientations of the stalk would be anticipated whenever the two heads exchanged positions during stepping (1, 9). It has been speculated that flexible elements within the kinesin structure may relieve any such momentary torsional strain, facilitating forward motion. Although previous single-molecule studies found that full-length kinesin stalks exhibited both flexural (10) and rotational (11) compliance, the locations and elastic behavior of any compliant domains were unresolved. Two sources of flexibility have been hypothesized: (i) the so-called “hinge” domains, short regions of unknown structure interspersed in the predominantly CC stalk (H1 and H2; Fig. 1A); and (ii) the NL domains, ≈13-residue peptide chains that connect the heads to the stalk, which have been found to undergo nucleotide-dependent, order-to-disorder transitions (12–14). Besides playing a role in the stepping mechanism of individual kinesin molecules, compliance may facilitate cooperation among multiple motors ferrying the same cellular cargo (15).
Here, we explore the torsional properties of single kinesin molecules using an assay that tracks fluorescent markers attached to beads that report on the angular orientation of the stalk. The technique can determine the azimuthal bead angles with degree-level resolution at rates up to 100 Hz over intervals of thousands of seconds (yaw with one degree accuracy, YODA). To answer the question of whether individual kinesin heads can swivel, we tested motor heads that bound to MTs under various nucleotide conditions and found unrestricted stalk rotation. The locations of torsionally compliant domains within the kinesin stalk were also determined, by measuring recombinant constructs carrying progressively longer regions of the stalk and quantifying the rotational flexibilities of the CC and hinge regions. Last, the issue of stalk rotation during processive motion was assessed, and we report the unanticipated observation of occasional, abrupt reversals in the azimuthal orientation of the stalk when kinesin moves unidirectionally and processively along MTs.
Results
YODA.
We attached kinesin motors via antibody linkages to 1.3-μm-diameter polystyrene beads labeled with two smaller fluorescent “marker” particles on their surfaces, then allowed these motors to bind MTs (Fig. 1B). The motor density was kept sufficiently low that no bead was likely to become attached by more than a single kinesin molecule. A freely diffusing kinesin-bead complex was captured from solution with an optical trap and placed near a MT immobilized on a coverglass surface, visualized by Nomarksi differential interference contrast (DIC) illumination. On kinesin binding (induced by the presence or absence of nucleotides, see below), the optical trap was turned off, leaving the bead tethered to the MT via a single kinesin molecule and free to rotate with thermal motion. The microscope was switched to epifluorescence mode, and the positions of the fluorescent marker particles on the larger bead were imaged by an EM-CCD camera (Fig. 1C). Offline processing of the video array was used to generate records of the azimuthal bead angle (Movie S1).
Monomeric and Dimeric Kinesin Exhibit Unbounded and Bounded Rotational Diffusion, Respectively.
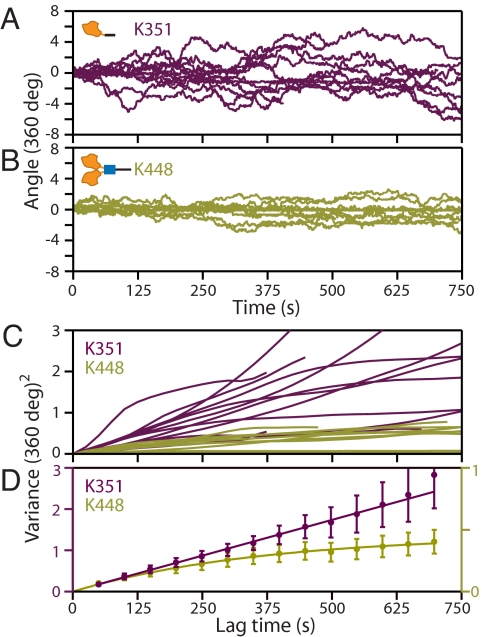
To test the sensitivity of our assay, we performed control experiments comparing the torsional behavior of two recombinant molecules: a single-headed, monomeric construct (K351) (16), and a double-headed, dimeric construct (K448) (5), consisting of the first 351 and 448 N-terminal residues of the kinesin heavy chain, respectively (Fig. 1D). The nonhydrolyzable ATP analog adenylyl β,γ-imidotriphosphate (AMP-PNP) was used to bind kinesin to MTs, placing motor dimers in a long-lasting, two-heads-bound (2-HB) state (17). As a further precaution, kinesin-MT linkages were stabilized by covalent cross-linking (XL; see Materials and Methods), ensuring that no transient unbinding could occur on the timescale of the experiments. The angular records showed that beads attached via the stalks of monomeric kinesin molecules frequently completed multiple, 360° rotations (Fig. 2A), whereas the angular excursions of dimeric molecules were far more limited during the periods of observation (up to 750 s) (Fig. 2B). We computed the running variance, 〈θ2(τ)〉, as a function of lag time, τ, in records (Fig. 2C). Ensemble-averaged variances displayed two distinct trends (Fig. 2D): unconstrained diffusion for the monomer (variance increases linearly) and bounded diffusion for the dimer (variance asymptotes to a final value). Data were well fit by 〈θ2(τ)〉 = 2Dτ for the monomer, and by 〈θ2(τ)〉 = (kBT/κθ)[1 − exp(−τ/τ0)] for the dimer (corresponding to Brownian motion in a flat or harmonic potential), where D is the effective angular diffusion coefficient of a free bead, κθ is the torsional stiffness of the kinesin molecule, kBT is thermal energy, and τ0 is a relaxation time of the complex. Fitting yielded DK351-XL-PNP = (6.8 ± 0.5) × 10−2 rad2·s−1, and κK448-XL-PNP = 0.23 ± 0.01 pN·nm·rad−1. These results indicate that monomeric kinesin undertakes free rotation (swiveling), but that dimeric kinesin exhibits bounded rotation, topologically constrained by its 2-HB state.
Fig. 2.
Control experiments with kinesin monomers and dimers. (A) Angle versus time records for the K351 monomer (purple), displaying multiple 360° revolutions (n = 12). (B) Angle versus time records for the K448 dimer (olive), which produced much more limited rotation (n = 10). (C) Computed variance of the individual angular records as a function of the lag time. (D) Ensemble-averaged variances (shown with SEs) reveal distinct trends: unbounded diffusion for kinesin monomers (filled purple circles), fit by a straight line (purple line), and bounded diffusion for kinesin dimers (filled olive circles), fit by an asymptotic expression (olive curve; see main text). Motors were chemically cross-linked to MTs in the presence of 2 mM AMP-PNP.
Unbounded and Bounded Diffusion with Kinesin Dimers.
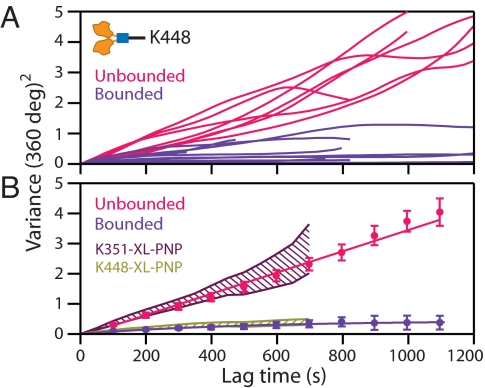
To investigate whether the NL of the dimer would swivel, we tested for the free rotation of dimeric K448 attached in a 1-HB state. According to one recent FRET-based study (18), kinesin remains predominantly in a 1-HB state at limiting ATP concentrations. Therefore, we cross-linked K448 molecules to MTs in the presence of 2 μM ATP, hypothesizing that a significant number of motors would be trapped with a single head bound to the MT when transiently exposed to the cross-linking reagent. Consistent with this inference, the records of angular variance displayed two distinct subpopulations: one that increased linearly with time, corresponding to unbounded diffusion (head swiveling), and another that converged to a fixed variance (within ≈200 s), corresponding to motors cross-linked by both heads (Fig. 3A). The subpopulation assignments were made by computing the time derivatives of records with respect to a fixed reference point: constant or decaying derivatives corresponded to unconstrained or constrained variance, respectively (Fig. S1). The ensemble averages of the two subpopulations (Fig. 3B) were statistically distinct (t test; t > 4, P < 0.002 for τ > 400 s), and fitting yielded DK488-XL-Low ATP = (6.8 ± 0.3) × 10−2 rad2·s−1 (unbounded), and κK448-XL-Low ATP = 0.24 ± 0.02 pN·nm·rad−1 (bounded). Also, these values, which we attributed to 1- and 2-HB states, were statistically identical to the corresponding quantities obtained with the monomeric and dimeric controls, respectively (Fig. 3B). The unconstrained rotation of the dimer directly shows that the NL of a functional kinesin molecule can act as a swivel. As a further control, K448 was attached to MTs in the absence of exogenous nucleotides. Once again, two distinct subpopulations were observed, consistent with the values of the monomeric and dimeric controls (Fig. S2 and SI Materials and Methods). Fitting the ensemble averages of the variances gave DK448-rigor = (5.1 ± 0.2) × 10−2 rad2·s−1, and κK448-rigor = 0.43 ± 0.03 pN·nm·rad−1 for the two populations.
Fig. 3.
Angular data for K448, cross-linked to MTs in the presence of 2 μM ATP, reveals an admixture of unbounded and bounded diffusion. (A) The variance records of individual dimers displayed either unbounded (Nu = 7; red) and bounded (Nb = 14; purple) rotational diffusion, which persisted over 1,200 s. (B) Ensemble variance averages for two groups identified in A as a function of lag time (same color scheme; shown with SEs). The computed ensemble variances for the unbounded K448 traces (red points and line) coincide with the control data for monomeric K351 attached by a single head (purple hatched area), whereas the bounded K448 variances (purple points and curve) coincide with the control data for dimeric K448 attached by both its heads (olive hatched area).
Torsional Properties of the Kinesin Stalk.
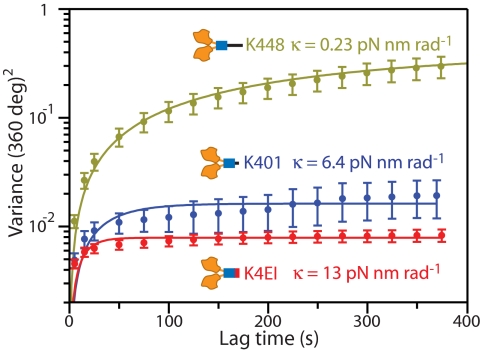
Attaching dimeric motors in a 2-HB state allowed us to explore the torsional compliance of the hinge domains of the stalk. We measured the stiffness of two kinesin constructs with modifications to hinge (H)1 (Fig. 1A). K401 has the identical CC-forming segment as K448, but carries 47 fewer residues in the H1 region at its C terminus. K4EI has the identical length as K401, but the H1 region of its stalk was engineered to carry a continuous CC, based on four tandem repeats of the “stable coil” motif, EIEALKA (Fig. 1D) (19). Motors were attached to MTs in a 2-HB state using AMP-PNP (2 mM). We observed bounded angular diffusion in all records for both constructs; fits to the ensemble variances yielded κK401-PNP = 6.4 ± 0.4 pN·nm·rad−1 and κK4EI-PNP = 13 ± 0.4 pN·nm·rad−1, increases of 28- and 57-fold, respectively, over κK448-XL-PNP (Fig. 4). These high torsional stiffnesses confirm the notion that the elastic compliance of the kinesin stalk resides chiefly in the H1 domain (15), and provide a basis for quantitative analysis of the elastic properties of CC and unstructured domains (see Discussion).
Fig. 4.
Angular variance and associated stiffnesses of dimeric kinesin constructs with different stalk compositions. Data are shown with SEs; curves were fit to the asymptotic expression in the text. Constructs K401 (n = 14, blue points and curve) and K4EI (n = 17, red points and curve) were attached to MTs in a 2-HB state using 2 mM AMP-PNP. K448 data are also shown (olive points and curve; data from Fig. 2D). The ensemble-averaged data show increasing torsional stiffness (small asymptotic variance) from K448 to K401 to K4EI.
Kinesin Rotates Occasionally While Stepping.
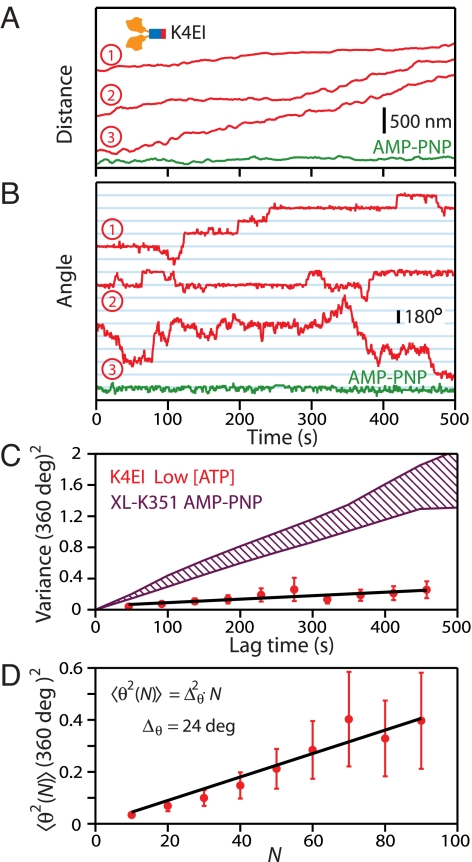
The high stiffness of the K4EI construct leads to greatly reduced angular noise, and therefore afforded an opportunity to revisit the conclusion that the kinesin stalk does not rotate appreciably during stepping (8), by tracking the angular motions of individual K4EI motors as these advanced processively under limiting ATP concentrations. The K4EI stiffness [κK4EI-PNP = 13 pN·nm·rad−1; 〈σθ〉 < 33°] corresponds to a relaxation time of τ0 = 0.5 s for a 1.3-μm bead. To observe rotation concurrently with translation, experiments were performed at [ATP] = 200–800 nM, yielding velocities of ≈4 nm/s (Fig. 5A), leading to an average time between steps that is 4-fold longer than the time resolution of the assay. Under these conditions, to our surprise, records displayed occasional, abrupt stalk rotations of ±180° superposed on a background of otherwise constant angles (Fig. 5B; Movie S2). Despite the complete reversals in the angular orientation of the attached beads, which persisted for up to hundreds of seconds, kinesin motors continued to step processively in their original directions. The average angular variance, 〈θ2(τ)〉, increased approximately proportional to the observation time; a linear fit yielded an effective diffusion constant of DK4EI-Low ATP = (9 ± 2) × 10−3 deg2·s−1 (Fig. 5C). To quantify the frequency of the angular reversals, we computed the ensemble variance, 〈θ2(N)〉, as a function of N, the number of steps (distance traveled divided by the 8.2-nm step size) (Fig. 5D). The linear relationship between variance and step number was consistent with an unbiased random walk of slope Δθ2 = 575 deg2. Therefore, based on an elemental angular step of ±180°, the stalk reversal probability per step is P = (Δθ/180°)2 ≈ 2%.
Fig. 5.
The stalk of kinesin can reverse its angular orientation during processive stepping. (A) Video-tracking records of three K4EI molecules moving in the presence of 500 nM ATP (red traces), contrasted with a motor immobilized by 2 mM AMP-PNP (green trace). (B) Angular records of the stalk orientation (red) for the same three traces shown in A displayed abrupt changes in angle, mostly through ±180°. No such transitions were observed under AMP-PNP (green). (C) The ensemble-averaged angular variance as a function of lag time (n = 23; red circles with SEs). A linear fit to data (black line) yielded an effective angular diffusion coefficient ≈8-fold lower than the 1-HB control (purple hatched area; data of Fig. 2D). (D) The ensemble-averaged angular variance (red circles, shown with SEs) correlates with the number of steps taken by motors. The line-fit (black line) is consistent with a random walk that rotates ±180° with a probability of ≈2% per 8-nm step taken.
Discussion
Our measurements of the torsional properties of kinesin were facilitated by a fluorescent bead-based assay that allows long observation times at high resolution (SI Materials and Methods). Under stalled conditions, the contrasting data from monomers (in a 1-HB state, by definition) and dimers (attached in a 2-HB state) show that the assay can clearly distinguish a swivel from a torsional spring on the time scale of the assay. Unbounded behavior was anticipated for K351, due to permissible rotation about one or more of the single α-carbon bonds (Cα–C and N–Cα) (20) found in the ≈20 residues of unstructured peptide emerging from the head domain, which include the 13 amino acids of the NL. For dimers attached by both heads, topological constraints lead to no bonds about which the stalk may rotate freely, yielding bounded diffusion. For dimeric constructs of K448, either bound in rigor or cross-linked to the MT in the presence of ATP, the identification of a subpopulation in a 1-HB state provides evidence for NL swiveling during kinesin-driven movement.
The torsional stiffness of motors bound in the 2-HB conformation enable us to compare the flexibilities of various stalk subdomains: the neck-CC (start position Ala-345 in Drosophila, subtending ≈35 residues) and the H1 domain (start position Gly-379, subtending ≈55 residues in the full-length protein). Three constructs with different amounts of CC or H were scored: K448 (NCC = 39, NH = 73), K401 (NCC = 39, NH = 26), and K4EI (NCC = 67, NH = 0), where NCC and NH are the predicted numbers of CC and H residues (21), respectively. For simplicity, we modeled the stalk compliance as two rods in series. For an isotropic rod of radius a and length l, the torsional stiffness is κ = Yπa4/6l, where Y is Young's modulus (22). The net torsional stiffness is related through κ−1 = κCC−1 + κH−1 = (6l/πa4)(NCC/YCC + NH/YH). Stalk length was computed by assuming 0.15 nm per residue (20), and the cross-sectional area of a hydrated CC (23) has been estimated as 2 nm2, yielding a ≈ 0.8 nm. We obtained independent estimates of stiffness for the three constructs under various conditions: K448 (κK448-XL-PNP = 0.23 pN·nm·rad−1, κK448-XL-Low ATP = 0.24 pN·nm·rad−1, κK448-rigor = 0.43 pN·nm·rad−1), K401 (κK401-PNP = 6.4 pN·nm·rad−1), and K4EI (κK4EI-PNP = 13 pN·nm·rad−1). A least-squares routine was used to fit these values, giving YCC = 0.6 GPa, and YH = 0.02 GPa [χν2 = 1.5; ν = 3; P(χν2) = 0.21]. Our value for CC stiffness is comparable with an earlier estimate of 2 GPa based on the apparent persistence length (23), which carried an uncertainty of a factor of 2 or more. The modulus ratio of CC to noncoiled domains is YCC/YH ≈ 30, implying that the flexibility of the overall stalk is dominated by the torsional flexibility of the H1 region. These measurements should also contribute to the understanding of the mechanical properties of biological protein fibers (24).
Two previous studies have examined the rotational flexibility of single kinesin molecules using a gliding-filament assay, where thermally driven rotations of MTs propelled by single motors bound to a coverglass surface were scored. In the first study, full-length native kinesin (11) was found to twist through several complete turns, implying considerable compliance (κKNative = 0.12 ± 0.02 pN·nm·rad−1). By contrast, a second study, employing a short, recombinant construct (K448) fused to biotin carboxyl carrier protein (BCCP) (8), reported little thermal rotation, less than half a turn (κK448-BCCP = 20 ± 9 pN·nm·rad−1). This disparity in stiffness cannot be explained trivially by the ≈5-fold difference in stalk length, because the torsional rigidity of a uniform rod is proportional to its length, yet the measured values differ by a factor of ≈170. Our data on constructs of different lengths and compositions suggest a possible reconciliation of these results by attributing the flexibility of kinesin primarily to its hinge elements. The full-length molecule kinesin contains all such elements, but the K448-BCCP construct is presumably stiffer, because it lacks some of these. The presence of the 87-residue BCCP domain (8) precludes direct comparison with our shorter K448-His construct. Here, our stiffest construct (K4EI) supplies a lower bound to the attachment stiffness, corresponding to the His-tag in series with the kinesin-bead antibody linkage, whereas a comparable quantity is not available for the BCCP tag. However, we note that the torsional stiffness reported for K448-BCCP is still approximately an order of magnitude greater than K448-His (20 versus 0.2–0.4 pN·nm·rad−1), so it seems possible that the comparatively bulky C-terminal BCCP domain may interfere with the adjacent H1 structure. Last, one recent study using synthetic kinesin polypeptides suggested that the H1 subdomain may be able to switch spontaneously between folded and unfolded states (15). In our data, we observed seconds-long transient angular excursions superposed on the general angular drift behavior, which may reflect the proposed hinge switching in the intact molecule (Fig. S3). Future experiments using recombinant variants of the H1 domain should be able to address this conjecture.
The striking discovery that beads occasionally reversed their angular orientations during processive motion (at a rate of ≈2% per step) was not detected previously (8), possibly due to surface-proximity effects (SI Materials and Methods). The events cannot be trivially explained by assuming that both heads unbind and rapidly rebind the MT (see SI Materials and Methods). Stalk reversals were observed under limiting ATP conditions, where the kinesin reaction cycle spends most of its time in the so-called “ATP waiting state,” with one nucleotide-free head firmly attached to the MT. The disposition of the partner head in this waiting state has been the subject of considerable controversy (25, 26). Single-molecule fluorescence results from Sosa and coworkers (27), and Vale and coworkers (18), along with independent optical trapping results from our own lab (28), indicate that the partner head is unbound from the MT, whereas Cross and coworkers (29) have proposed that it remains unbound but parked against the bound head. However, two studies by Yildiz et al. (7, 30) reached the opposite conclusion, and argued that both heads are bound. A 2-HB waiting state would topologically constrain stalk rotation, and so would a 1-HB state where the two heads are tightly associated. However, our records for K4EI stepping clearly exhibited the occasional stalk rotation, but at ≈8-fold lower angular diffusion rates than for the monomeric control (a cross-linked 1-HB state). This type of behavior is consistent with certain possibilities, depending on the nature of the waiting state. If the waiting state is 2-HB, it follows that the kinesin stalk cannot rotate during the ATP-waiting event, but manages to do so only briefly, during the course of stepping, when the NL of the bound head has an opportunity to swivel. If the waiting state is 1-HB, then it is possible that NL of the bound head is somehow prevented from freely swiveling while waiting for ATP to bind, but becomes free to do so only during the course of stepping, exactly as in 2-HB case. Alternatively, the NL of the 1-HB state may be able to swivel during the waiting state, but much less freely than the control monomer, such that rotations only tend to take place during ≈2% of reaction cycles.
Together, our findings on stalk rotation support a modified stepping model where kinesin generally advances by an asymmetric, hand-over-hand walk (1), but with occasional stepping imperfections that lead to the heads swapping roles. A mechanism that leads to a infrequent, but persistent stalk reversal, and which is consistent with both NL swivel and hand-over-hand motion is illustrated in Fig. 6. To execute an asymmetric hand-over-hand walk without producing concomitant stalk reorientation, the kinesin heads must alternately step forward on opposite sides of a vertical plane defined by the MT and kinesin stalk, in such a way as to generate a left step followed by a right step, rather than repeating any of these steps. In other words, one kinesin head must consistently take left steps while its partner head consistently takes right steps; these distinct motions compensate for one another in a rotational sense and prevent any net twist from building up, consistent with a model advanced by Hoenger et al. (31). Reorientation of the kinesin stalk by ±180° requires two separate things to occur. First, a given head must switch roles: a left-type stepping head must instead undertake a right step, or vice versa. This switch leads to two left (or right) steps being executed in a row, producing a change of phase in the overall stepping pattern. Second, the NL on the head switching phase must be able to swivel to accommodate the reorientation of the head in such a way as to keep it facing forward. At this point, the stalk is now reoriented by either ±180° (depending on which head switched phase) and roles of the two otherwise identical heads are henceforth reversed: The head which formerly took only left steps now takes right steps, and vice versa. The data for K4EI (Fig. 5) show that the sign of the stalk reorientation angle (±180°) is entirely random, implying that the two stepping kinesin heads have an unbiased propensity to break the phase of alternate stepping during hand-over-hand motion. Our model incorporates the presence of swiveling joints within the kinesin structure, a feature first invoked by the “rotary engine” mechanism originally proposed by Howard (12), involving a symmetric hand-over-hand walk where the stalk rotated at each step. However, our data do not support 180° stalk rotations with every step, but only infrequently.
Fig. 6.
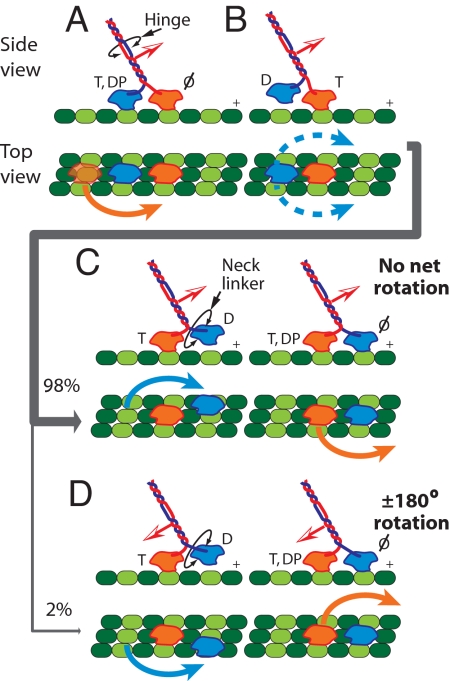
A model for asymmetric, hand-over-hand stepping that accommodates kinesin stalk reversals. (A) During normal processive motion, kinesin must pass through a 2-HB state, where the stalk (red and blue; orientation is indicated by the red and white arrow) cannot freely rotate, but remains torsionally flexible due to the hinge regions. The nucleotide occupancies of the two heads (colored orange and blue here to distinguish them, but otherwise identical in sequence) are shown, T, ATP; DP, ADP·Pi; D, ADP; Ø, no nucleotide. The MT protofilaments are depicted in green, with the plus-end of the MT to the right. (B) In the next ATP cycle, a 1-HB state develops, and the trailing head (blue) is poised to step. (C) During normal stepping, the blue head passes its partner to the left (blue arrow). Before rebinding, the NL of the blue head swivels to accommodate this motion (black circular arrow). (D) However, during a minor fraction of steps, the blue head passes instead to the right of its partner (blue arrow), generating two right steps in a row. After neck swiveling to accommodate the rebinding event, the blue head now occupies the leading position, but the orientation of the stalk is now reversed (rearward-facing red and white arrow).
The behavior of kinesin is in sharp contrast with a distantly related processive motor that is also known to move hand-over-hand, myosin-V, which has recently been reported to undergo random, ±90° stalk rotations per step (32), indicating that the heads indiscriminately take left and right steps. The source of these rotations was attributed to accumulation and relief of torsional strain within the otherwise rigid dimer. For kinesin, its swiveling NLs and flexible hinges domains appear to be able to relieve momentary strains that develop during stepping. Also, as we have seen, kinesin can even accommodate complete reorientations of its stalk, and thereby, relieve any level of twist that might develop while transporting cargo attached to its tail while maintaining both directionality and processivity. In this fashion, rotational freedom appears likely to play an important role in kinesin by allowing different molecular elements to achieve independent, stereospecific orientations.
Materials and Methods
Protein Preparation.
All recombinant constructs were derived from the sequence of the Drosophila melanogaster kinesin heavy chain (DmKHC), and carried a six-histidine tag (plus one to four additional residues) at the C terminus, which was used for affinity purification and bead attachment. DmK401 and DmK448 have been described previously (5). DmK378–4EI (pAF21) was generated by modifying an existing plasmid coding for DmK401-His (pCA1) (5), to substitute residues 379–401 with four tandem repeats coding for the heptad sequence EIEALKA (19). Care was taken to ensure the phase continuity of the existing CC, based on placing hydrophobic residues at the a and d positions of the heptad repeat. A plasmid for DmK351 was produced by using similar techniques (pNG). Motors were expressed in Escherichia coli as previously described (5), then purified to homogeneity by affinity chromatography. All dimeric constructs were tested in optical trapping motility assays under force-clamped conditions (5). Monomeric DmK351 was tested by scoring motion in a bead assay under multiple motor conditions.
Assays.
The 1.26-μm-diameter polystyrene beads coated with streptavidin (Spherotech) were incubated with 210-nm-diameter fluorescent, biotin-labeled beads (yellow-green; Molecular Probes) in phosphate buffer, then washed using PEMBSA buffer (80 mM Pipes, pH 6.9/1 mM EGTA/4 mM MgCl2/2 mg/mL BSA). Next, 5 μL of anti-histidine biotin-conjugated antibody (Qiagen) was added to 100 μL of 1 pM labeled beads, incubated at 22 °C for 1 h, then washed with PEMBSA. For AMP-PNP experiments, kinesin-bead complexes were produced by overnight incubations at 4 °C of protein with labeled beads, in the presence of assay buffer (80 mM Pipes, pH 6.9/50 mM potassium acetate/4 mM MgCl2/2 mM DTT/1 mM EGTA/10 μM Taxol/2 mg/mL BSA) with 2 μM AMP-PNP.
To assay kinesin motility, a microscope flow cell was prepared with Taxol-stabilized MTs immobilized on a polylysine-coated coverglass (33). A “motility” sample of kinesin-bead complexes was prepared by adding 2 mM ATP and 1% (vol/vol) of an oxygen-scavenging mixture (235 μg/mL glucose oxidase/42 μg/mL catalase/4.6 mg/mL glucose), then introduced in the flow chamber. Kinesin concentrations were adjusted such that <50% of beads moved (or bound, in the case of K351) when placed against a MT using an optical trap (estimated probability of two or more motors bound simultaneously to the MT: ≈1%) (34). The trapping laser power was kept <5 mW (measured at the objective rear pupil) to allow thermal and kinesin-driven motion of the beads. To collect rotation data, a “measurement” sample was prepared based on the motility sample, this time adding 2 mM AMP-PNP, instead of ATP, to bead complexes. Using the optical trap, freely diffusing beads, carrying two 200-nm fluorescent particles bound at nearly diametrically opposite positions, were captured from solution and placed against a MT. After successful binding, the optical trap was turned off and the excitation laser (λ = 488 nm, P = 0.8 mW; Blue Sky Research) was turned on. Fluorescence images were recorded at video rates (22–27 frames per second) by using a cooled EM-CCD camera (Cascade; Roper Scientific). Covalent cross-linking between kinesin motors and MTs was performed stepwise using a “zero-length” coupling reagent to minimize the introduction of undesired linkages (SI Materials and Methods). For tracking kinesin motility under limiting ATP conditions, overnight incubations of motor-bead complexes were performed at [ATP] = 200 nM, then adjusted to the desired ATP concentration (200–800 nM) before introduction to the flow chamber.
Data Analysis.
Video data were analyzed frame-by-frame using custom software written in LabView 7.0 (National Instruments). Standard centroid tracking was used to determine the (x, y) coordinates for each of the two fluorescent particles attached to the larger bead; the angle was computed from the line joining these positions. We estimated the angular resolution for a given frame to be ≈3°. When needed, the centroid of the 1.3-μm central bead was found by computing the pivot point about which the two fluorescent markers rotated over a short collection interval (typically, 5 s), assuming a rigid-body correlation of all three points throughout. The running angular variance as a function of lag time was computed for each video record by repeatedly calculating the variance within a growing time window of size τ, and averaging all values for that particular lag time (SI Materials and Methods). Last, the ensemble-averaged variance 〈θ2(τ)〉 was computed. Statistical and other analysis was performed using Igor Pro 5.0 (WaveMetrics).
Supplementary Material
Acknowledgments.
We thank N. Guydosh and M. Valentine for careful reading of the manuscript, and Block Laboratory (Stanford University) members, in particular N. Guydosh, for helpful discussions. This work was supported by a National Institutes of Health grant from the National Institute of General Medical Sciences (S.M.B.), and by a National Science Foundation fellowship (A.N.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907133106/DCSupplemental.
References
- 1.Asbury CL. Kinesin: World's tiniest biped. Curr Opin Cell Biol. 2005;17:89–97. doi: 10.1016/j.ceb.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Hua W, Young EC, Fleming ML, Gelles J. Coupling of kinesin steps to ATP hydrolysis. Nature. 1997;388:390–393. doi: 10.1038/41118. [DOI] [PubMed] [Google Scholar]
- 3.Schnitzer MJ, Block SM. Kinesin hydrolyses one ATP per 8-nm step. Nature. 1997;388:386–390. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- 4.Coy DL, Wagenbach M, Howard J. Kinesin takes one 8-nm step for each ATP that it hydrolyzes. J Biol Chem. 1999;274:3667–3671. doi: 10.1074/jbc.274.6.3667. [DOI] [PubMed] [Google Scholar]
- 5.Asbury CL, Fehr AN, Block SM. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaseda K, Higuchi H, Hirose K. Alternate fast and slow stepping of a heterodimeric kinesin molecule. Nat Cell Biol. 2003;5:1079–1082. doi: 10.1038/ncb1067. [DOI] [PubMed] [Google Scholar]
- 7.Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 8.Hua W, Chung J, Gelles J. Distinguishing inchworm and hand-over-hand processive kinesin movement by neck rotation measurements. Science. 2002;295:844–848. doi: 10.1126/science.1063089. [DOI] [PubMed] [Google Scholar]
- 9.Yildiz A, Selvin PR. Kinesin: Walking, crawling or sliding along? Trends Cell Biol. 2005;15:112–120. doi: 10.1016/j.tcb.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Jeney S, Stelzer EH, Grubmüller H, Florin EL. Mechanical properties of single motor molecules studied by three-dimensional thermal force probing in optical tweezers. Chem Phys Chem. 2004;5:1150–1158. doi: 10.1002/cphc.200301027. [DOI] [PubMed] [Google Scholar]
- 11.Hunt AJ, Howard J. Kinesin swivels to permit microtubule movement in any direction. Proc Natl Acad Sci USA. 1993;90:11653–11657. doi: 10.1073/pnas.90.24.11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howard J. The movement of kinesin along microtubules. Annu Rev Physiol. 1996;58:703–729. doi: 10.1146/annurev.ph.58.030196.003415. [DOI] [PubMed] [Google Scholar]
- 13.Vale RD, Fletterick RJ. The design plan of kinesin motors. Annu Rev Cell Dev Biol. 1997;13:745–777. doi: 10.1146/annurev.cellbio.13.1.745. [DOI] [PubMed] [Google Scholar]
- 14.Rice S, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 15.Crevenna AH. Secondary structure and compliance of a predicted flexible domain in kinesin-1 necessary for cooperation of motors. Biophys J. 2008;95:5216–5227. doi: 10.1529/biophysj.108.132449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue Y, et al. Movements of truncated kinesin fragments with a short or an artificial flexible neck. Proc Natl Acad Sci USA. 1997;94:7275–7280. doi: 10.1073/pnas.94.14.7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawaguchi K, Ishiwata S. Nucleotide-dependent single- to double-headed binding of kinesin. Science. 2001;291:667–669. doi: 10.1126/science.291.5504.667. [DOI] [PubMed] [Google Scholar]
- 18.Mori T, Vale RD, Tomishige M. How kinesin waits between steps. Nature. 2007;450:750–754. doi: 10.1038/nature06346. [DOI] [PubMed] [Google Scholar]
- 19.Romberg L, Pierce DW, Vale RD. Role of the kinesin neck region in processive microtubule-based motility. J Cell Biol. 1998;140:1407–1416. doi: 10.1083/jcb.140.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stryer L. Biochemistry. 4th Ed. New York: W. H. Freeman; 1995. [Google Scholar]
- 21.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 22.Hall IH. Deformation of Solids. London: Nelson; 1968. [Google Scholar]
- 23.Howard J. Mechanics of Motor Proteins and the Cytoskeleton. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- 24.Knowles TP. Role of intermolecular forces in defining material properties of protein nanofibrils. Science. 2007;318:1900–1903. doi: 10.1126/science.1150057. [DOI] [PubMed] [Google Scholar]
- 25.Carter NJ, Cross RA. Kinesin's moonwalk. Curr Opin Cell Biol. 2006;18:61–67. doi: 10.1016/j.ceb.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 26.Block SM. Kinesin motor mechanics: Binding, stepping, tracking, gating, and limping. Biophys J. 2007;92:2986–2995. doi: 10.1529/biophysj.106.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asenjo AB, Sosa H. A mobile kinesin-head intermediate during the ATP-waiting state. Proc Natl Acad Sci USA. 2009;106:5657–5662. doi: 10.1073/pnas.0808355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guydosh NR, Block SM. Direct observation of the binding state of the kinesin head to the microtubule. Nature. 2009;461:125–128. doi: 10.1038/nature08259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alonso MC, et al. An ATP gate controls tubulin binding by the tethered head of kinesin-1. Science. 2007;316:120–123. doi: 10.1126/science.1136985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yildiz A, Tomishige M, Gennerich A, Vale RD. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoenger A, et al. A new look at the microtubule binding patterns of dimeric kinesins. J Mol Biol. 2000;297:1087. doi: 10.1006/jmbi.2000.3627. [DOI] [PubMed] [Google Scholar]
- 32.Komori Y, Iwane AH, Yanagida T. Myosin-V makes two brownian 90° rotations per 36-nm step. Nat Struct Mol Biol. 2007;14:968–973. doi: 10.1038/nsmb1298. [DOI] [PubMed] [Google Scholar]
- 33.Block SM, Asbury CL, Shaevitz JW, Lang MJ. Probing the kinesin reaction cycle with a 2D optical force clamp. Proc Natl Acad Sci USA. 2003;100:2351–2356. doi: 10.1073/pnas.0436709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neuman KC, Block SM. Optical trapping. Rev Sci Instrum. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.