Abstract
A server (CheShift) has been developed to predict 13Cα chemical shifts of protein structures. It is based on the generation of 696,916 conformations as a function of the φ, ψ, ω, χ1 and χ2 torsional angles for all 20 naturally occurring amino acids. Their 13Cα chemical shifts were computed at the DFT level of theory with a small basis set and extrapolated, with an empirically-determined linear regression formula, to reproduce the values obtained with a larger basis set. Analysis of the accuracy and sensitivity of the CheShift predictions, in terms of both the correlation coefficient R and the conformational-averaged rmsd between the observed and predicted 13Cα chemical shifts, was carried out for 3 sets of conformations: (i) 36 x-ray-derived protein structures solved at 2.3 Å or better resolution, for which sets of 13Cα chemical shifts were available; (ii) 15 pairs of x-ray and NMR-derived sets of protein conformations; and (iii) a set of decoys for 3 proteins showing an rmsd with respect to the x-ray structure from which they were derived of up to 3 Å. Comparative analysis carried out with 4 popular servers, namely SHIFTS, SHIFTX, SPARTA, and PROSHIFT, for these 3 sets of conformations demonstrated that CheShift is the most sensitive server with which to detect subtle differences between protein models and, hence, to validate protein structures determined by either x-ray or NMR methods, if the observed 13Cα chemical shifts are available. CheShift is available as a web server.
Keywords: chemical shifts prediction, DFT calculations, validation server
Accurate and fast validation of protein structures constitutes a long-standing problem in NMR spectroscopy (1–3). Investigators have proposed a plethora of methods to determine the accuracy and reliability of protein structures in recent years (4–8). Despite this progress, there is a growing need for more sophisticated, physics-based and fast structure-validation methods (1, 2, 7). With these goals in mind, we recently proposed a new, physics-based solution of this important problem (9), viz., a methodology that makes use of observed and computed 13Cα chemical shifts (at the DFT level of theory) for an accurate validation of protein structures in solution (9) and in a crystal (10). Assessment of the ability of computed 13Cα chemical shifts to reproduce observed values for a single or an ensemble of structures in solution and in a crystal was accomplished by using the conformationally-averaged root-mean-square-deviation (ca-rmsd) as a scoring function (9). While computationally intensive, this methodology has several advantages: (i) it makes use of the 13Cα chemical shifts, not shielding, that are ubiquitous to proteins; (ii) it can be computed accurately from the ϕ, ψ, and χ torsional angles; (iii) there is no need for a priori knowledge of the oligomeric state of the protein; and (iv) no knowledge-based information or additional NMR data are required.
However, the primary and the most serious limitation of the method is the computational cost of such calculations, which prevents it from being adopted by spectroscopists and crystallographers as a standard validation routine (9). For this reason, we investigate here the dependence of the accuracy and speed of DFT calculations of the 13Cα chemical shifts in proteins on the size of the basis set used. The results of this analysis indicate that the 13Cα chemical shifts in proteins, computed at the DFT level of theory with a large basis set, can be reproduced accurately (within an average error of approximately 0.4 ppm) and approximately 9 times faster by using a small basis set. As a straightforward application of these findings, a server of the 13Cα chemical shifts (CheShift) for all 20 naturally occurring amino acid residues as a function of the φ, ψ, ω, χ1, and χ2 torsional angles was built. This server can be used to validate protein structures of any class or size at a high-quality level and, like the purely physics-based method (9) from which it was derived, it does not use any knowledge-based information. However, the CheShift server also provides accurate 13Cα chemical shift predictions for each amino acid residue in the sequence in a few seconds, on a single processor. These are the main advantages of this new quantum-mechanics-derived CheShift server over our previous approach (9).
There are several servers that provide fast and accurate predictions of 13Cα chemical shifts, namely SHIFTS (11, 12), SHIFTX (13), PROSHIFT (14), and SPARTA (15) A brief description of these servers, follows. SHIFTS (11, 12) is a DFT-computed server of chemical shifts for residues in α-helical or β-sheet conformations, plus a coil-database derived as a single average of the sheet and helix data (optimized by comparison with experimental data); SHIFTX13 is a hybrid predictive approach that employs precalculated empirically-derived chemical shift hypersurfaces in combination with classical or semiclassical equations (ring current, electric field, hydrogen bond, solvent effects, etc.). SHIFTX used 2 databases of 37 protein structures as input to generate the empirical constants, torsional angles and lookup tables; PROSHIFT14 is a neural-network-trained server, derived by using experimental 3D structures of proteins as input parameter; and SPARTA (15) is a server containing observed chemical shifts for 200 proteins for which a high resolution (≤2.4 Å) x-ray structure is available. The relative importance of the weighting factors for the φ,ψ, and χ1 torsional angles and sequence similarity was optimized empirically.
The existence of these servers raises the question as to whether a new server, such as CheShift, is necessary. What new information can we learn from its predictions? Even more important, if there are substantial differences among predictions from these servers and CheShift, what is the origin of such differences? Comparison of the 13Cα chemical shifts, computed for a large number of proteins using different servers, with the corresponding experimental data are very important because it can shed light on the strengths and weaknesses of each server. It will also enable us to determine which servers are sensitive enough to detect subtle differences between conformations and whether it is able to indicate to spectroscopists or crystallographers whether an ensemble, rather than a single conformation, is a better representation of the observed 13Cα chemical shifts in solution (9).
Several attempts have also been made recently to use 13Cα chemical shift data to facilitate protein structure determination and refinement (16, 17) and to derive initial protein models for molecular replacement in x-ray crystallography (18). The ability of a given server to guide protein structure refinement can be assessed by its discriminative power when applied to protein decoys generated from a given, native conformation.
To compare the performance of CheShift with that of other existing servers, the following 3 sets of proteins are analyzed in this work: 36 x-ray-derived protein structures solved at 2.3 Å resolution or better for which sets of 13Cα chemical shifts were also available (Section II, Table S3, SI Appendix); 15 pairs of x-ray and NMR-derived protein conformations (Section II, Table S4, SI Appendix); and decoys from the ROSETTA@HOME set (19), namely 1AIL (20), 1RNB (21), and 1UBI (22) from the Protein Data Bank (PDB) (23), showing an rmsd of up to 3 Å from the corresponding x-ray structure.
The most important results related to the performance of CheShift, and 4 other servers, are discussed here. Additional material related to the dependence of the accuracy and speed of DFT calculations of the 13Cα chemical shifts of proteins on the size of the basis set used; analysis of x-ray and x-ray-NMR pairs of structures; and approximations used to interpolate computed 13Cα chemical shift values are provided in Sections I, II, and III, respectively of the SI Appendix.
Results and Discussion
In the absence of a “gold standard” against which to compare the predictions obtained from any servers, we adopted the 13Cα chemical-shift values computed at the DFT level of theory by using a large basis set as an ‘internal standard reference’ (see Materials and Methods).
Determination of the Sensitivity of All Servers for Members of a Set of 36 X-Ray-Derived Protein Structures.
Results of the analysis of the 13Cα chemical-shift predictions for each of 36 x-ray-derived protein models, based on the correlation coefficient R (24), obtained by using SHIFTS, SHIFTX, PROSHIFT, SPARTA, and CheShift are shown in Table S3 (Section II, SI Appendix). The differences in the R ranges are small among all of the servers, around 0.04 depending on the protein, albeit these small differences could be important for an accurate prediction. Besides, these results indicate that, for all of the proteins, the R value obtained from any server is greater than the one obtained from CheShift. This raises the following question: do these servers provide a more sensitive validation method than CheShift? To answer this question, 2 of the 36 validation results are analyzed here in detail.
Protein 1RGE (Ribonuclease Sa).
The structure of this protein was solved (25) at 1.15 Å resolution with an R-factor of 10.9%. The corresponding crystal structure contains 2 chemically identical but crystallographically independent molecules in the asymmetric unit, named here as A and B (25). The main-chain torsional angles (φ and ψ) of the independent molecules are very similar (25) with the Cα rmsd between them of 0.4 Å. On the other hand, the all-heavy-atom rmsd is 1.1 Å due to differences in side chains, especially those on the protein surface, occupying different rotameric states.
Comparison of the predicted 13Cα chemical shifts, computed as the conformational-average (ca) (9) between the 2 chains, with the observed 13Cα chemical shifts yields R values of 0.95, 0.98, 0.99, 0.97, and 0.97 for CheShift, SHIFTX, SPARTA, SHIFTS, and PROSHIFT, respectively.
At first glance, all servers appear to be more accurate than CheShift. However, it is necessary to determine whether all servers are sensitive enough to detect differences between the independent molecules A and B. To answer this question, we carried out an additional test that does not require a comparison with the observed 13Cα chemical shifts. Thus, we computed the correlation coefficient R between the 13Cα chemical-shift predictions obtained for molecules A and B, respectively, by using each of the 5 servers. The results of this test give the following R values: 0.96, 1.00, 1.00, 0.98, and 1.00 for CheShift, SHIFTX, SPARTA, SHIFTS, and PROSHIFT, respectively (see Table S3, SI Appendix). Except for CheShift (0.963) and SHIFTS (0.981), none of the servers is able to discriminate, beyond doubt, between molecules A and B. From a statistical point of view, the R values obtained from SHIFTX (0.997), SPARTA (0.997), and PROSHIFT (0.996) servers indicate that molecules A and B are practically indistinguishable protein models with which to compute the 13Cα chemical shifts. In other words, these 3 servers cannot detect the conformational difference between molecules A and B.
This test enables us to conclude that a lower R value between predicted and observed 13Cα chemical shifts does not necessarily mean poorer accuracy; on the contrary, it could mean higher sensitivity to detect subtle structural differences.
If this were a valid conclusion, a similar analysis carried out with a larger basis set, namely using the results from the more-accurate “internal standard reference,” should lead to a lower correlation, R, between 13Cα chemical shifts predicted for molecules A and B. Indeed, this is the case. The R value (0.93) computed with the larger basis set is significantly lower than the R value obtained with CheShift (0.96) or any other server, namely, 1.00, 1.00, 0.98, and 1.00 for SHIFTX, SPARTA, SHIFTS, and PROSHIFT, respectively.
The previous analysis demonstrates that CheShift is a more sensitive server to detect subtle structural differences than any other servers, although this analysis does not reveal the origin of such sensitivity. To detect this origin, we first carried out a graphic analysis of the correlation between corresponding torsional angles in molecules A and B, and, second, from these graphs we determined the distribution of differences between predicted 13Cα chemical shifts for each of these 2 molecules by using the CheShift server. Fig. S2 (Section II, SI Appendix) shows the well-known (25) strong correlation between the corresponding backbone torsional angles derived from molecules A and B of protein 1RGE. Consistently, with the low value of the Cα rmsd (0.4 Å) between these 2 molecules, the correlation coefficient, R, computed between backbone torsional angles of the molecules A and B of 1RGE, is greater than 0.99.
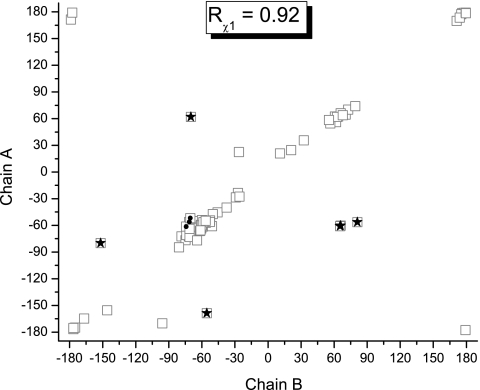
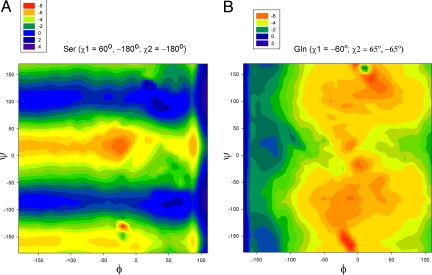
On the other hand, Fig. 1 shows the correlation between corresponding side-chain torsional angles χ1 for molecule A and B. The R value (0.92, obtained after removing the, approximately, identical χ1 = ≈180° and ∼−180°, see Fig. 1) is lower than the one obtained for the backbone torsional angles (>0.99), indicating the significantly higher side-chain all-heavy-atom rmsd between molecules A and B of 1RGE (1.1 Å). To determine whether the observed differences in the side-chain χ1 torsional angles, shown in Fig. 1, are the origin of the R values yielded by CheShift for molecules A and B, we highlighted those residues showing differences between predicted 13Cα chemical shifts for molecules A and B of 1RGE, greater than 2.0 ppm. Among all 8 residues, 5 (highlighted as black-filled stars in Fig. 1) show a significant departure from the linear regression. These 5 residues possess significantly different side-chain χ1 torsional angles in molecules A and B and, hence, significantly different 13Cα chemical shift predictions. Two out of these 5 highlighted residues in Fig. 1, namely Asp-25 and Arg-40, were reported (25) to have higher temperature factors or partial disorder of the side-chains. Another 2 (Ser-48 and Thr-76) of these 5 residues are in different crystal environment, i.e., these residues of molecule A are part of a well-ordered hydrogen-bond network, while they are oriented toward the solvent in molecule B.25 In particular, the influence of 2 different torsional angles χ1, for a fixed χ2, on the predicted CheShift value of any Ser residue is illustrated in Fig. 2A. Finally, the remaining residue, Arg-63, of these 5 is located in the loop region (Gly-61-Thr-64) whose conformation is different between the 2 molecules (25).
Fig. 1.
Plot of the χ1 torsional angles in degrees (as open-squares) from chain A versus chain B of the x-ray-determined structure of PDB ID 1RGE. We highlighted those residues showing differences greater than 2.0 ppm between predicted 13Cα chemical shifts from molecule A and B of 1RGE by CheShift with filled stars and filled circles. For details about the distribution of the black-filled symbols, see Protein 1RGE (Ribonuclease Sa).
Fig. 2.
Map of the differences in the computed 13Cα chemical shifts (in ppm according to the color scale) between an arbitrarily selected pair of side-chain torsional angles. The color indicates the difference in 13Cα chemical shifts (in ppm) for any pair of φ and ψ. (A) for Ser with χ1 = 160° and −180°, and a fixed χ2 = −180°; and panel (B) for Gln with χ2 = 65° and −65°, and a fixed χ1 = −60°.
There are 3 other residues in Fig. 1, namely Gln-32, Ile-71 and Gln-77, close to the regression line and highlighted as filled-black circles. Their χ1 torsional angles are very similar, although all 3 residues show significant differences in the side-chain χ2 torsional angles, namely (≈g+, ≈g−) for Gln-32 and Ile-71, and (≈g−, ≈t) for Gln-77. To illustrate the influence of 2 different torsional angles χ2 for a fixed χ1 on the predicted CheShift value, we selected Gln (in Fig. 2B).
Finally, it is important to note that, for a given molecule, A or B of 1RGE, some residues are reported to show 2 discrete side-chain conformations (25). Differences up to 4.0 ppm between CheShift-predicted chemical shifts are obtained by using these alternative side-chain conformations, as for Thr5A, Val6A, and Ser42A from molecule A or Ser3B and Thr5B from molecule B. For some of these residues, the alternative side-chain conformational dilemma can be resolved easily by inspection of the occupancy, e.g., Thr5A shows 1 of the 2 conformations with much higher occupancy (approximately 80%) (25). However, in other residues, such as Val6A and Ser42A of molecule A or Ser3B and Thr5B of molecule B, the alternative conformations show very similar occupancies (approximately 50%) (25) although significantly different chemical shifts; if the occupancy does not offer conclusive evidence, and if it is necessary to select one conformation, then the CheShift predictions could be a useful criterion with which to decide which 1 of the 2 conformations should be selected.
The results derived from the analysis of 2 chains of protein 1RGE enable us: (i) to illustrate that a higher correlation coefficient, R, obtained for the 13Cα chemical shift prediction between molecules A and B could mean less sensitivity to detect subtle structural differences, rather than more accurate predictions; and (ii) to determine the origin of the difference between 13Cα chemical shift predictions for the 2 molecules; i.e., although the main contribution determining the predicted 13Cα chemical shifts comes from backbone torsional angles, a proper consideration of the side-chain torsional angles (χ1 and χ2) is very important for an accurate 13Cα chemical shift validation (see Fig. 2 A–B). The latter conclusion is in agreement with evidence (11, 26–28) indicating the role of side-chain conformations in the computation of accurate 13Cα chemical-shift values.
Protein Interleukin 1β (Human).
The computed 13Cα chemical shifts for 2 different x-ray structures of this protein solved at 2.0 Å resolution and refined to a crystallographic R-factor of 19.0% (4I1B) (29) and 17.2% (2I1B) (30), are compared with the observed 13Cα chemical shifts in solution [Biological Magnetic Resonance data Bank (BMRB) accession no. 1061(31)]. The all-heavy-atom rmsd between these 2 x-ray structures is 1.1 Å with a difference, mainly, in the loop regions (29), e.g., the all-heavy-atom rmsd between loop residues His-30-Val-41, Val-47-Asp-54, Val-85-Glu-96, and Gly-136-Asp-145 are 1.5 Å, 1.6 Å, 1.3 Å, and 0.9 Å, respectively. The results for R obtained with CheShift, SHIFTX, and SPARTA point to 2I1B, rather than 4I1B, as a better representation of the observed 13Cα chemical shifts in solution (see Section II, Table S3, SI Appendix). However, only CheShift indicated that 2I1B (R = 0.91) is a significantly better model than 4I1B (R = 0.87) to reproduce the observed 13Cα chemical shifts. In fact, the CheShift R value indicated that approximately 83% of the observed 13Cα chemical shifts in solution are reproduced by the 2I1B protein model, compared to only approximately 76% of protein 4I1B. A similar analysis, carried out with SHIFTX and SPARTA, indicates that approximately 92% and approximately 96% of the observed 13Cα chemical shifts in solution are reproduced by the 2I1B protein model and, a slightly smaller, approximately 90% and approximately 94% by 4I1B, respectively. The SHIFTS server points to protein 4I1B (approximately 90%), rather than 2I1B (approximately 88%), as a better representation of the observed 13Cα chemical shifts. On the other hand, according to PROSHIFT predictions, both proteins are equivalent models with which to reproduce (approximately 92% of) the observed 13Cα chemical shifts (despite the differences between these 2 structures, mainly, in the loop regions). Clearly, for this protein too, CheShift provides a more sensitive discrimination between different models.
As an additional test, the 13Cα chemical shifts computed by using the internal standard reference indicated that protein 2I1B is, in fact, a significantly better representation of the observed chemical shifts in solution (R = 0.87), than protein 4I1B (R = 0.81), in agreement with the CheShift predictions.
Regarding the disagreement between CheShift and SHIFTS, the latter server contains, besides a database of DFT-computed shifts for residues populating helical and sheet conformations, a “coil” database (with coil designating residues belonging to neither helical nor β-sheet region) computed as an average of helix and sheet data. Conceivably, this could be the reason for this disagreement, since the 4I1B and 2I1B proteins differ, mainly, in the loop (i.e., coil) regions. In other words, although quantum-mechanical calculations are a common feature of both CheShift and SHIFTS, these calculations were limited to only some regions of the Ramachandran map for SHIFTS but not for the CheShift server.
Are the Servers Sensitive Enough to Determine Differences Between X-Ray and NMR Models? Test on 15 Pairs of X-Ray and NMR-Derived Sets of Protein Conformations.
The results obtained from the validation analysis involving 15 pairs of x-ray and sets of NMR-derived protein models are shown in Table S4 (Section II, SI Appendix). For the NMR-derived conformations, the R values were computed between the observed 13Cα chemical shifts and the predicted conformational-averaged ones (9), i.e., among all structures of the NMR-derived ensemble (see Computation of the Conformationally-Averaged rmsd in Section I of SI Appendix). As already noted for an x-ray set of structures, all R values computed by CheShift are systematically lower than those of the other servers. However, for several pairs of x-ray and NMR-derived conformations most of the servers, but not CheShift, do not show differences between x-ray and NMR-derived structures, or the differences are very small, in terms of the correlation coefficient, R. To understand whether these results reflect real similarity between the x-ray and NMR models or arise from the low sensitivity of the servers, 2 cases have been selected for further analysis, namely protein PDB ID 3LZT (32) solved by x-ray diffraction at 0.92 Å resolution, and 50 conformations of PDB ID 1E8L (33) (solved by NMR spectroscopy), and protein PDB ID 1UBQ (34) (x-ray derived structure at 1.8 Å resolution) and 128 conformations of PDB ID 1XQQ (35) (NMR-derived ensemble).
A Comparative Validation Analysis of Proteins 1E8L and 3LZT.
The NMR solution structure of hen Lysozyme (PDB ID 1E8L) (33), determined with the aid of residual dipolar coupling data, shows “…conformational disorder within the NMR ensemble in some regions of the structure, most notably in the long loop and involving residues in the turns between helices A and B and between the first two strands in the β-sheet (33).” Model 1 of the 50 models of the NMR-derived ensemble (PDB ID 1E8L) (33) has an all-heavy-atom rmsd of 2.20 Å from the corresponding x-ray structure (PDB ID 3LZT), solved at 0.92 Å resolution (32), indicating major conformational differences between these 2 structures. The results shown in Table S4 (Section II, SI Appendix) indicate that only CheShift, SHIFTX, and SPARTA point to the x-ray structure, but not the NMR-derived ensemble, as a better representation of the observed 13Cα chemical shifts in solution. However, only CheShift shows a significant difference between these 2 protein models, namely R = 0.89 and R = 0.94 for proteins 1E8L and 3LZT, respectively (i.e., in agreement with the existence of significant differences between these 2 models in some regions of the structure, such as the long loop, and also involving residues in the turns between helices), while the differences obtained using SHIFTX or SPARTA are minimal, i.e., R = 0.95 and 0.96, and R = 0.96 and 0.97 for proteins 1E8L and 3LZT, respectively. On the other hand, SHIFTS does not discriminate between these 2 proteins (R = 0.95 for both 1E8L and 3LZT, respectively).
If the 8 cysteines are excluded (to make a fair comparison with the results obtained from the SHIFTS server which does not consider cysteines), the following results are obtained: (1) for CheShift, the difference is slightly smaller than the one obtained with the cysteines included, i.e., R = 0.91 and 0.95 for proteins 1E8L and3LZT, respectively, although the x-ray structure remains as a much better representation of the observed 13Cα chemical shifts in solution; (2) SHIFTX does not discriminate between these protein models (R = 0.97); and (3) SPARTA provides improved agreement for both proteins, keeping the difference to a minimum, i.e., R = 0.97 and 0.98 for proteins 1E8L and 3LZT, respectively.
Overall, the CheShift and SPARTA servers point to the same conclusion, with or without cysteines, but only CheShift shows higher discriminative power, as was obtained in the analyses carried out for the 2 proteins whose structures were determined by x-ray diffraction. In other words, CheShift indicates that the x-ray-derived structure, 3LZT, is a significantly better representation of the observed 13Cα chemical shifts in solution than the NMR-derived ensemble of 1E8L.
A Comparative Validation Analysis of Proteins 1UBQ and 1XQQ.
The structure of ubiquitin solved by x-ray diffraction at 1.8 Å resolution, PDB ID 1UBQ (34), and 128 conformations obtained using NMR-derived information, PDB ID 1XQQ (35), were compared according to their ability to reproduce the observed 13Cα chemical shifts in solution. Among all servers (see Table S4, Section II, SI Appendix), CheShift and SHIFTS predictions indicate that the NMR-derived ensemble (1XQQ) is a better representation of the observed 13Cα chemical shifts in solution than the x-ray structure (1UBQ), but only CheShift shows a significant difference between them, i.e., R = 0.95 (NMR) and 0.91 (x-ray). Even more important, the results obtained with CheShift are consistent with previous calculations (36) carried out by using the internal standard reference, indicating that the 1XQQ ensemble is a significantly better representation of the observed 13Cα chemical shifts in solution than the 1UBQ single protein model.
On the other hand, SHIFTX does not discriminate between these x-ray and NMR models, but SPARTA shows slightly better agreement between observed and predicted 13Cα chemical shifts for the x-ray-derived structure (1UBQ), rather than the NMR-derived ensemble (1XQQ). This result should not be surprising since the x-ray structure of ubiquitin (1UBQ) is included in the SPARTA database.
Are the Servers Sensitive Enough to Discriminate Decoys from Native Conformations?
To answer this question, sets of decoys for proteins 1AIL (20), 1RNB (21), and 1UBI (22) for which the 13Cα chemical shifts of the proteins are available, were taken from the ROSETTA@HOME decoys set (19), and are considered here. All decoys analyzed here are close to the x-ray determined conformation, i.e., within an arbitrary rmsd cutoff of 3 Å. The x-ray-determined conformations from which the decoys were generated are termed “native” conformations here, although an x-ray structure may, or may not, be the best model with which to represent the observed 13Cα chemical shifts in solution (9).
If an attempt to discriminate decoys from the native structure, based only on 13Cα chemical shift information, would include conformations with an rmsd beyond the 3 Å cutoff value, then addition of other selection criteria, such as NOE-derived distance constraints, would be necessary because the 13Cα chemical shift is only a local property of the residue, i.e., a given 13Cα chemical shift can correspond to more than one set of backbone and side-chain torsional angles.
The ability to discriminate decoys from the native structures, using CheShift, SHIFTS, SHIFTX and SPARTA for the proteins PDB ID 1AIL (20) and 1RNB (21) is illustrated in Figs. S3A-D and S4A-D, respectively (SI Appendix). The results show that only CheShift and SPARTA are able to discriminate the decoys from the native conformation for both proteins. On the other hand, SHIFTS fails for both proteins while SHIFTX was able to discriminate the native structure of only 1AIL. Analysis of the 1UBI decoys is discussed in detail in the next subsection.
Further analysis of the results shown in Figs. S3 and S4 (SI Appendix) indicates that none of the servers are able to discriminate among all of the decoys, i.e., as to which one is closest to the “native” structure, indicating that, for this purpose, another scoring function is necessary.
Is the X-Ray “Native” Conformation the Best Model with Which to Represent the Observed 13Cα Chemical Shifts in Solution? Test on Decoys Derived from Ubiquitin (1UBI).
The ability of different servers to discriminate decoys of ubiquitin from the native structure [1UBI, solved at 1.8 Å resolution (22)] is illustrated in Fig. S5A–D (SI Appendix). Only SHIFTS and SPARTA discriminate all of the protein decoys from the native conformation, PDB ID 1UBI. In other words, CheShift and SHIFTX do not recognize the native conformation (i.e., the x-ray model) as the best representation of the observed 13Cα chemical shifts in solution. This failure poses the question whether the x-ray native conformation is indeed the best structure with which to represent the observed 13Cα chemical shifts in solution. To answer this important question, we computed the agreement between the observed and predicted 13Cα chemical shifts for 10 NMR-derived, high-resolution structures of ubiquitin, namely protein PDB ID 1D3Z (37); see results in Fig. S5 (SI Appendix). The prediction of CheShift indicates that any model from the 1D3Z ensemble is a better representation of the observed 13Cα chemical shifts in solution than the native (PDB ID 1UBI) or any protein decoy. This is not a surprising result since previous calculations carried out with the internal standard reference indicated that the 1D3Z conformations are a better representation of the observed 13Cα chemical shifts than a single x-ray structure (9). A similar conclusion is obtained from the analysis with the SHIFTS and SHIFTX servers, but not with SPARTA which favors the 1UBI model over any of the 1D3Z conformations. The latter is not an unexpected result because the SPARTA database contains an x-ray model of ubiquitin (1UBQ).
Conclusions
We have shown that the quantum-mechanical basis of the CheShift server enables us to predict the 13Cα chemical shifts with reasonable accuracy in seconds and, hence, provides a standard with which to evaluate the quality of any reported protein structure solved by either x-ray crystallography or NMR-spectroscopy, provided that the experimentally observed 13Cα chemical shifts are available. These conclusions are supported here by an extensive analysis of a large number of x-ray-determined structures, pairs of x-ray and NMR-determined conformations, and the power to discriminated protein decoys from “native” conformations. Moreover, a detailed comparison with the results obtained for these sets of conformations using other available servers illustrates one of the main advantages of CheShift predictions: these predictions are significantly more sensitive than those of any of the tested servers to conformational differences between protein models. This was verified, in most cases, by comparing CheShift predictions with those obtained using the internal standard reference.
Even though the CheShift server has somewhat lower sensitivity to detect subtle conformational differences between protein models than the highly-accurate internal standard reference predictions, it is a thousand times faster and, hence, it overcomes the main limitation of this purely physics-based, 13Cα-based method (9). Even more important, the CheShift-server predictions can now be adopted as a validation routine by spectroscopists and crystallographers. In fact, members of the scientific community are invited to use it by uploading their protein models on a new web server.
Materials and Methods
Experimental Set of Structures.
All of the information concerning the x-ray and NMR-derived set of conformations used, as well as the BMRB accession number from which the observed 13Cα chemical shifts were obtained, are listed on Tables S3 and S4 (Section II, SI Appendix). It is worth noting that no 13Cα chemical shift reference correction (15) was applied to the experimentally observed values.
Reproducing Observed 13Cα Chemical Shifts of Proteins: Dependence on the Basis Set Size.
Five basis sets using the locally-dense basis-set approximation (38) (see Table S1, SI Appendix), viz., 6–31G/3–21G, 6–31G(d)/3–21G, 6–311G(d,p)/3–21G, 6–311+G(d,p)/3–21G, and 6–311+G(2d,p)/3–21G, and the uniform 3–21G/3–21G basis set were initially applied to 10 NMR-derived conformations of the 76-residue α/β protein ubiquitin [PDB ID 1D3Z (37)]. The results of this analysis for 3 proteins (see Table S2, SI Appendix) indicate that, first, the 13Cα chemical shifts in proteins, computed at the DFT level of theory with the large [6–311+G(2d,p)/3–21G] basis set, can be reproduced accurately (within an average error of approximately 0.4 ppm) and approximately 9 times faster by using the small (6–31G/3–21G) basis set with an effective TMS value of 195.4 ppm and extrapolating it with: 13Cα = − 1.597 + 1.040 × 13Cμα, where 13Cμα represents the 13Cα chemical shifts computed for a given residue μ with the small basis set, and, second, the results provide evidence that the conclusions reached apply to proteins of any size or class. Moreover, in Section I of the SI Appendix, an analysis of the magnitude of the errors is provided.
Internal Standard Reference.
As an internal standard reference, the values computed at the DFT level of theory by using a large basis set [6–311+G(2d,p)/3–21G], as a “basis set limit result,” were adopted. This arbitrary reference was chosen because this physics-based method is extremely sensitive to small conformational changes (10) and, hence, it represent a very accurate (9, 36) method with which to computed the 13Cα chemical shifts for a given protein structure model.
Building the CheShift Database.
For the generation of the 696,916 conformations, as function of the φ, ψ, χ1, and χ2 torsional angles, for all 20 naturally occurring amino acids, the following sampling procedure was used: (i) the backbone torsional angles φ and ψ were sampled every 10°; (ii) all ω torsional angles were assumed to be 180°, except for Pro residues for which the cis conformation (0°) was also considered; (iii) all cysteines were considered nonbonded; (iv) all χ1 side-chain torsional angles were sampled every 30°; (v) all χ2 side-chain torsional angles were sampled according to the most ‘frequently-seen’ torsional values (39); and (vi) any conformation with a total ECEPP05 (40) internal energy >30 kcal/mol was rejected. This cutoff value in the total internal energy was chosen because it is large enough to cover a broad range of conformations populating the Ramachandran map, i.e., to account for the existence of conformations generated with several different force-fields used to determine x-ray and NMR-derived structures. For each of these 696,916 conformations, the 13Cα chemical shifts were computed using a small basis set and linearly extrapolated to the large basis set by using the above mentioned linear regression.
Approximations Used to Interpolate Computed 13Cα Chemical Shift Values.
Since the database is a 13Cα chemical shift coarse-grained representation of the continuum variable space in φ, ψ, χ1, and χ2 torsional angles, an accurate interpolation method must be used to compute 13Cα chemical shift for any arbitrary combination of these 4 torsional angles. Among all possible options that do not require adjustable parameters, we tested 2: a Gaussian (41) and a linear interpolation, respectively (see Section III, SI Appendix). To decide whether the Gaussian or the linear interpolation provides a more accurate representation of the 13Cα chemical shift hypersurface, the following test was carried out. An arbitrary selected fraction of the accessible torsional angle space (φ = [−150°, −160°]; ψ = [160°, 170°]; and χ1 = [−180°, 180°]) for the tripeptide Ac-GXG-NMe, with X = Ser, was sampled by using a fine grid, namely 2° for φ and ψ backbone torsional angles, and 5° for the χ1 side-chain torsional angle. For this fine grid, the 13Cα chemical shift map was computed using a small basis set and linearly extrapolated to a large basis set. The ability of the Gaussian and linear interpolations to reproduce these fine-grid values by using the corresponding coarse grain data, namely at 10° steps for φ and ψ backbone torsional angles, and 30° for χ1, was analyzed graphically (see Fig. S6, SI Appendix) and by the frequency of the error distribution (see Fig. S7, SI Appendix). Both, the graphic analysis and the standard deviation of the frequency of the error distribution indicate that the linear interpolation is a significantly better approximation (σ = 0.09 ppm for the fine grid mesh results) than the Gaussian interpolation (σ = 0.27 ppm) and, hence, the linear interpolation was adopted. The accuracy of the linear interpolation to reproduce the values obtained for the fine-grid mesh is due to the small-torsional-angle variations chosen to build the coarse-grained CheShift database which captures the most important dependence of the 13Cα chemical shift on the backbone and side-chain torsional angles.
Supplementary Material
Acknowledgments.
We thank Dr. H.A. Baldoni of the Universidad Nacional de San Luis, Instituto de Matemática Aplicada San Luis, Consejo Nacional de Investigaciones Científicas y Técnicas de Argentina for suggestions and discussion regarding the material presented in Section I of the SI Appendix. We also thank all spectroscopists, crystallographers, and those in charge of both the PDB and the BRMB databases because, without their effort, this work would not be possible. This research was supported by grants from the National Institutes of Health (GM-14312 and GM-24893), and the National Science Foundation (MCB05–41633). Support was also received from the CONICET, FONCyT-ANPCyT (PAV 22642/22672), and from the Universidad Nacional de San Luis (P-328501), Argentina. The research was conducted using the resources of Pople, a facility of the National Science Foundation Terascale Computing System at the Pittsburgh Supercomputer Center.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908833106/DCSupplemental.
References
- 1.Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 2.Billeter M, Wagner G, Wüthrich K. Solution NMR structure determination of proteins revisited. J Biomol NMR. 2008;42:155–158. doi: 10.1007/s10858-008-9277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williamson MP, Craven CJ. Automated protein structure calculation from NMR data. J Biomol NMR. 2009;43:131–143. doi: 10.1007/s10858-008-9295-6. [DOI] [PubMed] [Google Scholar]
- 4.Vriend G. WHAT IF: A molecular modeling and drug design program. J Mol Graphics. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 5.Lüthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 6.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 7.Huang YJ, Powers R, Montelione GT. Protein NMR Recall, Precision, and F-measure scores (RPF scores): Structure quality assessment measures based on information retrieval statistics. J Am Chem Soc. 2005;127:1665–1674. doi: 10.1021/ja047109h. [DOI] [PubMed] [Google Scholar]
- 8.Davis IW, et al. MolProbity: All atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vila JA, Scheraga HA. Assessing the accuracy of protein structures by quantum mechanical computations of 13Cα chemical shifts. Acc Chem Res. 2009 doi: 10.1021/ar900068s. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnautova YA, Vila JA, Martin OA, Scheraga HA. What can we learn by computing 13Cα chemical shifts for X-ray protein models? Acta Crystallogr D. 2009;65:697–703. doi: 10.1107/S0907444909012086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu X-P, Case DA. Automated prediction of 15N, 13Ca, 13Cb and 13C′ chemical shifts in proteins using a density functional database. J Biomol NMR. 2001;21:321–333. doi: 10.1023/a:1013324104681. [DOI] [PubMed] [Google Scholar]
- 12.Xu X-P, Case DA. Probing multiple effects on 15N, 13Ca, 13Cb and 13C′ chemical shifts in peptides using density functional theory. Biopolymers. 2002;65:408–423. doi: 10.1002/bip.10276. [DOI] [PubMed] [Google Scholar]
- 13.Neal S, Nip AM, Zhang H, Wishart DS. Rapid and accurate calculation of protein 1H, 13C, and 15N chemical shifts. J Biomol NMR. 2003;26:215–240. doi: 10.1023/a:1023812930288. [DOI] [PubMed] [Google Scholar]
- 14.Meiler J. PROSHIFT: Protein chemical shift prediction using artificial neural networks. J Biomol NMR. 2003;26:25–37. doi: 10.1023/a:1023060720156. [DOI] [PubMed] [Google Scholar]
- 15.Shen Y, Bax Ad. Protein backbone chemical shifts predicted from searching a database for torsional angle and sequence homology. J Biomol NMR. 2007;38:289–302. doi: 10.1007/s10858-007-9166-6. [DOI] [PubMed] [Google Scholar]
- 16.Shen Y, et al. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci USA. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vila JA, et al. Quantum chemical 13Cα chemical shift calculations for protein NMR structure determination, refinement, and validation. Proc Natl Acad Sci USA. 2008;105:14389–14394. doi: 10.1073/pnas.0807105105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramelot TA, et al. Improving NMR protein structure quality by Rosetta refinement: A molecular replacement study. Proteins. 2008;75:147–167. doi: 10.1002/prot.22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.All Atom Decoy Sets from Rosetta@home. 2007. [Accessed on May 2009]. Available at http://depts.washington.edu/bakerpg/
- 20.Liu J, et al. Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat Struct Biol. 1997;4:896–899. doi: 10.1038/nsb1197-896. [DOI] [PubMed] [Google Scholar]
- 21.Baudet S, Janin J. Crystal structure of a barnase-d(GpC) complex at 1.9 Å resolution. J Mol Biol. 1991;219:123–132. doi: 10.1016/0022-2836(91)90862-z. [DOI] [PubMed] [Google Scholar]
- 22.Ramage R, et al. Synthetic, structural and biological studies of the ubiquitin system: The total chemical synthesis of ubiquitin. Biochem J. 1994;299:151–158. doi: 10.1042/bj2990151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berman HM, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Press HW, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipes in FORTRAN 77. The Art of Scientific Computing. 2nd Ed. Cambridge, UK: Cambridge Univ Press; 1992. pp. 630–633. [Google Scholar]
- 25.Sevcik J, Dauter Z, Lamzin VS, Wilson KS. Ribonuclease from streptomyces aureofaciens at atomic resolution. Acta Crystallogr D. 1996;52:327–344. doi: 10.1107/S0907444995007669. [DOI] [PubMed] [Google Scholar]
- 26.Havlin RH, Le H, Laws DD, deDios AC, Oldfield E. An ab initio quantum chemical investigation of carbon-13 NMR shielding tensors in glycine, alanine, valine, isoleucine, serine, and threonine: Comparisons between helical and sheet tensors, and effects of χ1 on shielding. J Am Chem Soc. 1997;119:11951–11958. [Google Scholar]
- 27.Iwadate M, Asakura T, Williamson MP. Cα and Cβ carbon-13 chemical shifts in protein from an empirical database. J Biomol NMR. 1999;13:199–211. doi: 10.1023/a:1008376710086. [DOI] [PubMed] [Google Scholar]
- 28.Villegas ME, Vila JA, Scheraga HA. Effects of side-chain orientation on the 13C chemical shifts of antiparallel β-sheet model peptides. J Biomol NMR. 2007;37:137–146. doi: 10.1007/s10858-006-9118-6. [DOI] [PubMed] [Google Scholar]
- 29.Veerapandian B, et al. Functional implications of interleukin-1β based on three-dimensional structure. Proteins. 1992;12:10–23. doi: 10.1002/prot.340120103. [DOI] [PubMed] [Google Scholar]
- 30.Priestle JP, Chär H-P, Grütter MG. Crystallographic refinement of interleukin 1β at 2.0 Å resolution. Proc Natl Acad Sci USA. 1989;86:9667–9671. doi: 10.1073/pnas.86.24.9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ulrich EL, et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walsh MA, et al. Refinement of triclinic hen egg-white Lysozyme at atomic resolution. Acta Crystallogr D. 1998;54:522–546. doi: 10.1107/s0907444997013656. [DOI] [PubMed] [Google Scholar]
- 33.Schwalbe H, et al. NMR solution structure of hen Lysozyme. Protein Sci. 2001;10:677–688. doi: 10.1110/ps.43301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 Å resolution. J Mol Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 35.Lindorff-Larsen K, Best RB, Depristo MA, Dobson CM, Vendruscolo M. Simultaneous determination of protein structure and dynamics. Nature. 2005;433:128–132. doi: 10.1038/nature03199. [DOI] [PubMed] [Google Scholar]
- 36.Vila JA, Villegas ME, Baldoni HA, Scheraga HA. Predicting 13Cα chemical shifts for validation of protein structures. J Biomol NMR. 2007;38:221–235. doi: 10.1007/s10858-007-9162-x. [DOI] [PubMed] [Google Scholar]
- 37.Cornilescu G, Marquardt JL, Ottiger M, Bax A. Validation of protein structure from anisotropic carbonyl chemical shifts in a dilute liquid crystalline phase. J Am Chem Soc. 1998;120:6836–6837. [Google Scholar]
- 38.Chesnut DB, Moore KD. Locally dense basis-sets for chemical-shift calculations. J Comp Chem. 1989;10:648–659. [Google Scholar]
- 39.Lovell SC, Word JM, Richardson JS, Richardson DC. The penultimate rotamer library. Proteins. 2000;40:389–408. [PubMed] [Google Scholar]
- 40.Arnautova YA, Jagielska A, Scheraga HA. A new force field (ECEPP05) for peptides proteins and organic molecules. J Phys Chem B. 2006;110:5025–5044. doi: 10.1021/jp054994x. [DOI] [PubMed] [Google Scholar]
- 41.Kuszewski J, Qin J, Gronenborn AM, Clore MG. The impact of direct refinement against 13Cα and 13Cβ chemical shifts on protein structure determination by NMR. J Magn Reson B. 1995;106:92–96. doi: 10.1006/jmrb.1995.1017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.