Abstract
Plasma cells can synthesize and secrete thousands of Ig molecules per second, which are folded and assembled in the endoplasmic reticulum (ER) and are likely to place unusually high demands on the resident chaperones and folding enzymes. We have discovered a new resident ER protein (pERp1) that is a component of the BiP chaperone complex. PERp1 is substantially up-regulated during B to plasma cell differentiation and can be induced in B cell lines by some UPR activators, arguing that it represents a potentially new class of conditional UPR targets. In LPS-stimulated murine splenocytes, pERp1 interacted covalently via a disulfide bond with IgM monomers and noncovalently with other Ig assembly intermediates. Knockdown and overexpression experiments revealed that pERp1 promoted correct oxidative folding of Ig heavy chains and prevented off-pathway assembly intermediates. Although pERp1 has no homology with known chaperones or folding enzymes, it possesses a thioredoxin-like active site motif (CXXC), which is the signature of oxidoreductases. Mutation of this sequence did not affect its in vivo activity, suggesting that pERp1 is either a unique type of oxidoreductase or a previously unidentified class of molecular chaperone that is dedicated to enhancing the oxidative folding of Ig precursors.
Keywords: chaperone, endoplasmic reticulum
The environment of the endoplasmic reticulum (ER) lumen is specialized for the folding and assembly of proteins destined for the extracellular space. The ER is populated by a large number of molecular chaperones and folding enzymes that serve to prevent the aggregation of nascent polypeptide chains that entered the ER cotranslationally and guide their proper maturation. Although the ER exists in all eukaryotic cells, it is easy to imagine that the demands on this organelle would be much greater in secretory tissues, which synthesize and secrete large quantities of proteins. For instance, a single plasma cell can synthesize and secrete thousands of IgM pentamers (H2L2)5 per second, which requires the folding of 70 Ig domains, the assembly of 10 heavy and light chains each with a J chain, the addition of 26 N-linked glycans, and the formation of ≈100 disulfide bonds for each pentamer (1)! To meet these enormous demands, the quiescent B cell undergoes dramatic and carefully orchestrated morphological changes upon activation by antigen to become an antibody-producing factory. This is accomplished by a large expansion of the ER membranes, which is supported by up-regulation of most resident ER chaperones, folding enzymes, and components of the ER quality control and degradation systems (2–4). XBP-1, which is activated by the unfolded protein response (UPR) pathway, plays an essential role in this process (4–6). To aid in the oxidative folding and assembly of massive quantities of Ig, protein disulfide isomerase (PDI) and ERp57, two oxidoreductases that catalyze disulfide bond formation, reduction, and isomerization, are also highly up-regulated during plasma cell differentiation (3). Recent expansion of the PDI family to nearly 20 members (7) argues that oxidative folding may be more complex than previously appreciated and may require cell type or substrate specific factors that can catalyze and/or assist oxidative folding to ultimately enhance the efficiency of folding and assembly of secretory pathway proteins.
We have identified a plasma cell–induced, resident ER protein, pERp1. This highly conserved protein is expressed at very low levels in the more mature B cell subpopulations and is significantly up-regulated during plasma cell differentiation. Our in vivo analyses demonstrate that pERp1 is an unusual type of resident ER protein that specifically assists Ig biosynthesis.
Results
pERp1 Is a Lymphocyte-Specific ER Protein that Interacts with the BiP Complex.
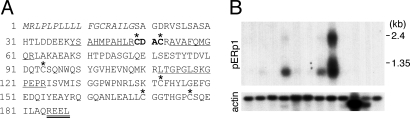
We identified a multichaperone complex that is associated with unassembled Ig heavy chains (HC) in a mouse plasmacytoma cell line Ag8(8) (γ+, κ−) (8). Further mass spectophometric analysis of this complex revealed an additional protein comigrating at ≈23kDa with cyclophilin B and SPF-2, which we named pERp1 (Fig. 1A). A splice variant of pERp1, PACAP, which is predicted to encode a very different protein, was identified in B cells induced to undergo apoptosis (9). Alignment of pERp1 amino acid sequences from various species identified six conserved cysteine residues including a single CXXC motif (red letters in Fig. 1A) (10), which is commonly the active sites of oxidoreductases that have a specific structure called the thioredoxin fold (11). pERp1 has an N-terminal signal sequence and a C-terminal ER retention signal (Fig. 1A), suggesting that it is a soluble resident ER protein. Immunofluorescence staining data revealed that pERp1 colocalized with ERp57, another resident ER protein (Fig. S1). We found that pERp1 transcripts were most abundant in lymphoid tissues, such as thymus and spleen (Fig. 1B), and the vast majority of ESTs recorded for it are from lymphoid cells (38). A significant signal for pERp1 mRNA was also detected in the small intestine, which probably reflects the presence of gut associated lymphoid tissues that contain large numbers of IgA secreting plasma cells (Fig. 1B), and may also explain its detection in the microarray study of gastric tumors (12).
Fig. 1.
Characterization of pERp1. (A) Amino acid sequence of mouse pERp1. The peptide sequences obtained from mass spec analysis are underlined. The 6 conserved cysteine residues are marked with asterisks, and the CXXC motif is in bold. The signal sequence is italicized, and the ER retention signal is double underlined. (B) Tissue distribution of pERp1 mRNA. A human multitissue Northern blot was probed with pERp1 or β-actin cDNA. The transcripts were detected by autoradiography. β-actin was used as a loading control. 1, peripheral blood leukocyte; 2, lung; 3, placenta; 4, small intestine; 5, liver; 6, kidney; 7, spleen; 8, thymus; 9, colon (no mucosa); 10, skeletal muscle; 11, heart; 12, brain.
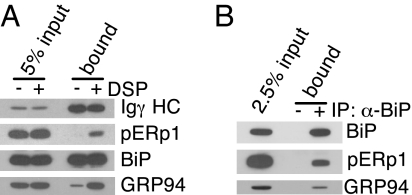
We confirmed that pERp1 was part of the BiP complex bound to Ig HC by isolating Ig HC and immunoblotting with a polyclonal anti-pERp1 antiserum. Unlike BiP binding, which is stable to nonionic detergent lysing conditions, we found that the association of pERp1 with Ig HC was only readily detected when cells were pretreated with dithiobis(succinimidyl propionate) (DSP) (Fig. 2A), a membrane permeable, thiol-cleavable cross-linking agent. When BiP was immunoprecipitated from DSP-treated Ag8.653 cells, which are Ig negative, pERp1 was coprecipitated with BiP (Fig. 2B), demonstrating that it is a component of the ER chaperone complex even in the absence of Ig HC.
Fig. 2.
pERp1 is a component of the BiP chaperone complex. (A) Ag8(8) cells were treated with or without DSP and γ HC were precipitated with Protein A-Sepharose. Isolated proteins were analyzed by immunoblotting as indicated. Total cellular lysate (5% input) was used to assess the amount of proteins coprecipitating with HC. (B) Ag8.653 cells were treated with DSP, lysed, and the BiP complex was immunoprecipitated with anti-BiP. The same procedure was performed with Protein A Sepharose alone as a control.
pERp1 Expression Is Greatly Induced During Plasma Cell Differentiation.
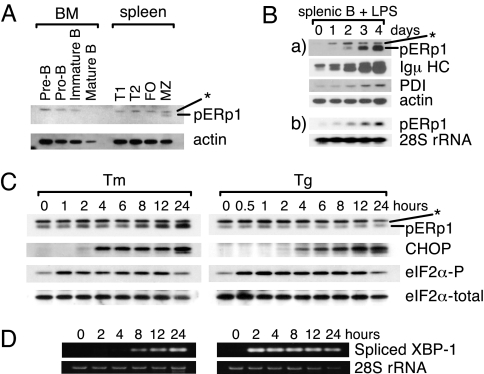
We next examined the expression of pERp1 in mouse primary B cell subpopulations. Cells were collected from bone marrow and spleen, and the indicated B lineage subpopulations were isolated (13, 14). Marginal zone B cells were the only B cell population in which we were able to detect any pERp1 protein, and the level of expression in these cells was well below that observed in any of the plasmacytoma cell lines (Fig. 3A and data not shown). We next examined its expression during plasma cell differentiation by treating mouse splenic B cells with lipopolysaccharide (LPS), a B cell mitogen. We found that both pERp1 protein and mRNA levels increased significantly during differentiation, with kinetics similar to those observed for the up-regulation of Igμ HC (Fig. 3B). As expected, protein disulfide isomerase (PDI), a well-characterized ubiquitously expressed ER oxidoreductase was also up-regulated during LPS-induced plasma cell differentiation (Fig. 3B) (3). To determine whether the induction of pERp1 was generally linked to plasma cell differentiation or whether it was specific to LPS stimulation, we induced plasma cell differentiation through the Ig receptor (15). Again, pERp1 was induced with kinetics similar to those of μ HC and LC (Fig. S2A).
Fig. 3.
Induction of pERp1 expression in response to terminal differention of mouse primary B cells to plasma cells. (A) Mouse B cells were collected from bone marrow (BM) or spleen, followed by fractionation into subpopulations using the cell surface markers (see SI Materials and Methods). Cells were lysed, and proteins were analyzed by immunoblotting with anti-pERp1. Actin was used as a loading control. BM, bone marrow; FO, folicular B cells; MZ, marginal zone B cells. (B) Mouse splenic B cells were stimulated with LPS for the indicated times. Cells were collected, lysed, and proteins (a) were analyzed by immunoblotting and total RNA (b) was subjected to Northern blot analysis for pERp1 transcripts. 28S ribosomal RNA levels serve as a loading control. (C) I.29μ+ cells were treated with tunicamycin (Tm) or thapsigargin (Tg) for the indicated times. Cells were collected, lysed, and proteins were analyzed by immunoblotting as indicated. Total eIF2α serves as a loading control. Asterisks indicate a nonspecific band detected by the anti-pERp1 antiserum. (D) I.29μ+ cells were treated with tunicamycin (Tm) or thapsigargin (Tg) for the indicated times. Spliced XBP1 mRNA was amplified from total RNA by RT-PCR. An aliquot of input RNA used in the RT-PCRs was run on agarose gels and the 28S rRNA was detected by ethidium bromide staining for the loading control
Plasma cell differentiation depends on activation of a modified unfolded protein response (UPR), which is necessary to produce the transcriptionally active form of XBP-1 and to up-regulate molecular chaperones and folding enzymes (4, 6). To determine whether pERp1 was a target of the UPR, we used the I.29μ+ lymphoid cell line, which induces IgM and pERp1 expression in response to LPS treatment (Fig. S2B). We found that tunicamycin treatment led to a modest induction of pERp1, but thapsigarin had no effect on pERp1 expression (Fig. 3C). However, eIF-2α phosphorylation, CHOP expression, and XBP-1 splicing were readily induced by both treatments (Fig. 3 C and D), arguing that the failure to observe pERp1 induction in response to thapsigargin was not because the cells did not respond to this UPR inducer.
pERp1 Interacts Covalently and Noncovalently with Assembling IgM in Plasma Cells.
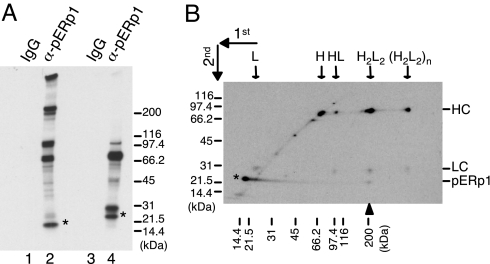
Based on its expression pattern and association with unassembled γ heavy chains, the data thus far were consistent with pERp1 playing a role in Ig biosynthesis. IgM monomers, consisting of two HC and two LC, assemble into pentamers with the aid of J chain for secretion. To determine whether pERp1 interacted with any of the IgM assembly intermediates, purified mouse splenic B cells were stimulated with LPS. pERp1 was immunoprecipitated from metabolically labeled cell lysates, and the precipitated proteins were analyzed by SDS/PAGE. Because Ig assembly intermediates are disulfide linked and must be identified under nonreducing conditions, we were unable to use DSP to stabilize potential interactions with pERp1, which was likely to underestimate any association. Under nonreducing conditions, free pERp1, and several slower migrating proteins, were detected (Fig. 4A, lane 2). Analysis of a similar sample under reducing conditions resolved all of the slower migrating bands into two predominant bands migrating at ≈75 and 25 kDa, which are the size of μ HC and LC, respectively (Fig. 4A, lane 4), which is consistent with data obtained from I.29μ+ cells observed in the accompanying article (38). pERp1 migrated slower under reducing conditions (indicated with asterisks), suggesting that it possesses at least one long-range intrachain disulfide bond that linked two distantly located cysteine residues in the primary structure.
Fig. 4.
pERp1 associates with assembling IgM intermediates. (A) Mouse splenic B cells were stimulated with LPS for 3 days to induce plasma cell differentiation and metabollically labeled for 3 h. TCA precipitated proteins were redisolved in pH6 lysis buffer, immunoprecipitated with either anti-pERp1 or rabbit IgG, and separated by SDS/PAGE under nonreducing (lanes 1 and 2) or reducing conditions (lanes 3 and 4). The bands were visualized by autoradiography. Asterisks indicate pERp1. (B) Two-dimensional SDS/PAGE was performed using a portion of the same sample that was immunoprecipitated with anti-pERp1 in A. Free pERp1 is indicated with an asterisk, and an additional pERp1 spot that migrated slower in the first dimension is shown with an upward arrowhead. Assembly intermediates of IgM are indicated with downward arrows. Molecular weight markers (kDa) for the first dimension are indicated across the bottom and for the second dimension to the right.
To determine the composition of the bands observed under nonreducing conditions, a portion of the anti-pERp1 immunoprecipitated material was subjected to two-dimensional SDS/PAGE (Fig. 4B). The sample was electrophoresed under nonreducing conditions in the first dimension, and then under reducing conditions to separate Ig assembly intermediates into HC and LC. We detected a spot corresponding to free pERp1 just to the left of the diagonal (indicated with an asterisk), which is indicative of a protein with internal disulfide bonds. In addition, there were several spots migrating at ≈75 and 25 kDa, which were identified as μ HC and LC by immunoblotting (data not shown). The major spot on the diagonal at ≈75 kDa included BiP and free μ HC, which is in keeping with the fact that a number of other lighter spots on the diagonal corresponded to members of the ER chaperone complex (Fig. S3). Based on the migration of the various proteins on nonreducing gels, we deduced that pERp1 interacts with all of the IgM assembly intermediates, including free HC and LC. pERp1 was also detected just below the IgM monomers (shown with an upward arrowhead), which demonstrated that unlike its association with the other intermediates, pERp1 appeared to interact covalently with IgM monomers via a disulfide bond. An interchain disulfide bond between pERp1 and IgM monomers was also detected when we examined LPS-treated I.29μ+ cells (data not shown). This type of analysis has been used to detect trapped mixed disulfide intermediates between other oxidoreductases (i.e., PDI-family members) and their substrates (16).
pERp1 Promotes Oxidation and Stability of Ig HC In Vivo.
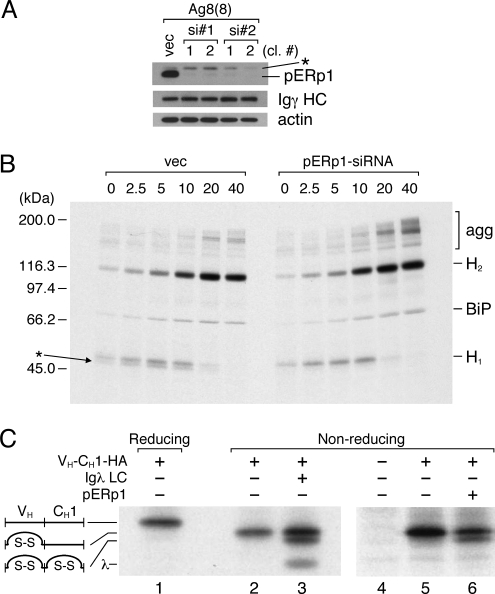
To better understand the role of pERp1 in antibody biosynthesis, we established stable subclones of the Ag8(8) cell line with decreased pERp1 expression. Two different RNA sequences were targeted with siRNA, and two independent single-cell clones were isolated for each sequence that had significantly reduced level of pERp (Fig. 5A). We examined the folding and assembly of γ HC in pulse–chase experiments in control and pERp1 knockdown clones (Fig. 5B). In the vector control line, two closely migrating bands were detected initially in the region where free HC were expected to migrate that chased into a single band migrating at ≈110 kDa, which represented HC dimers (H2). When pERp1 expression was significantly reduced, the assembly of HC monomers into dimers appeared to be unaffected. However, in all four clones we observed two consistent differences. First, there was only a single band present where HC monomers should migrate, and second we observed high molecular weight complexes near the top of the gel at later time points. Analysis of the same immunoprecipitated material under reducing conditions revealed the presence of BiP and only a single band for γ HC in both vector control and knockdown clones (Fig. S4). Together these data suggested that the doublet band observed at early time points in the vector control line was likely to represent two different oxidation intermediates of the HC and that the very slow migrating bands present in greater quantities in the knockdown clones were disulfide bonded HC oligomers, which represent off-pathway products.
Fig. 5.
Decreasing pERp1 expression affected oxidization and stability of Ig molecules. (A) Clones with reduced expression of pERp1 were established in the Ag8(8) cell line using two different siRNA sequences or with an empty vector to make the control line (vec). The levels of pERp1 and Igγ HC were analyzed by immunobloting. Actin was also detected as a loading control. (B) The cell lines were metabolically labeled with [35S] methionine and cysteine for 5 min and chased for the indicated times. Cells were lysed in the presence of NEM, and Igγ HC were isolated with Protein A-Sepharose beads. Precipitated proteins were separated by SDS/PAGE under nonreducing conditions, and the bands were visualized by autoradiography. An astersisk indicates a further oxidized form of the monomeric Igγ HC, and the disulfide-linked, larger oligomeric forms of the Igγ HC are indicated. (C) The indicated vectors were introduced into 293T cells. Cells were metabolically labeled with [35S] methionine and cysteine for two hours, and the mini-HC was immunoprecipitated with anti-HA antibody and analyzed by SDS/PAGE under reducing (lane 1) or nonreducing (lanes 2–6) conditions. Bands were visualized by autoradiography.
To more directly examine the effects of pERp1 expression on the oxidation of Ig domains, we used a simplified mini-HC that is composed of only a VH and CH1 domain, which makes changes in the oxidation of individual Ig domains much easier to detect (17). As reported in ref. 17, only the VH domain was oxidized when this mini-HC was expressed alone, and LC coexpression led to oxidation of the CH1 domain in those HC that assembled with LC (Fig. 5C, lanes 1–3). Overexpression of pERp1 induced oxidation of the CH1 domain in a portion of the HC even in the absence of LC (Fig. 5C, lane 6). To determine whether pERp1 affected HC stability, we examined γ HC turnover in Ag8(8) control and knockdown cells (Fig. S5 A and B) and in 293T cells transfected with γ HC alone or together with pERp1 (Fig. S5 C and D). In both cases, we found that expression of pERp1 consistently increased the stability of HC compared with cells that were not expressing it, although the effects were fairly modest.
Oxidative Folding of Antibody Domains Does Not Depend on the CXXC Motif.
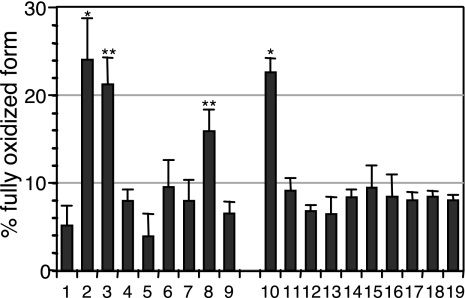
The data described thus are consistent with pERp1 acting either as a new class of chaperone that induced release of BiP from Ig domains to allow them to fold and form their intradomain disulfide bond or as an unusual type of oxidoreductase, which directly oxidized the HC domains. We first examined the possibility that pERp1 is an oxidoreductase, because it contains a CXXC sequence. We mutated each of the cysteines of pERp1 individually, as well as various disulfide pairs to serine according to the intrachain disulfide bond pattern of pERp1 (Fig. S6, see also ref. 38) and determined the effect on HC oxidation. Like wild-type pERp1, which oxidized ≈24% of the CH1 domains, we found that the C50S and C178S single mutants and the C50S-C178S double mutant all maintained nearly full activity (≈21%, 16%, and 23% respectively) (Fig. 6). However, all of the other pERp1 cysteine mutants had no significant activity (Fig. 6). This was not because they were poorly expressed (Fig. S7), but unlike wild-type pERp1 many of the mutants formed dimers or higher order disulfide linked oligomers, suggesting that the mutations were likely to be affecting the overall structure of pERp1. Therefore, the lack of activity in these mutants is hard to interpret, because it is impossible to discriminate between disruption of the active site and alteration of the structure. Of note, the C53S and C53S-C171S mutants were well behaved in terms of their migration on nonreducing SDS gels and had no activity. The significance of this disulfide bond in pERp1's function is currently under investigation.
Fig. 6.
Overexpression of pERp1 induces oxidative folding and stability of Ig HC in a CXXC motif independent manner. The mini-HC was expressed alone or with the indicated pERp1 constructs. Oxidation of the HC was analyzed as in Fig. 5C. The intensity of the bands was measured using the phosphorimager, and the percentage of the fully oxidized form was calculated and plotted. The mean values and standard errors are calculated from at least three independent experiments. *, P < 0.001; **, P < 0.005. 1, vector control; 2, WT; 3, C50S; 4, C53S; 5, C95S; 6, C143S; 7, C171S; 8, C178S; 9, no cys; 10, C50-C178S; 11, C53-C171S; 12, C95-C143S; 13, C50-C178 only; 14, C53-C171 only; 15, C95-C143 only; 16, C50S-C53S; 17, C171S-C178S; 18, C50-C53 only; 19, C171-C178 only. A diagram of the cysteines and the disulfide bond patterns present in WT and the various mutant is shown in Fig. S6.
To determine whether pERp1 might act indirectly to oxidize Ig domains, we examined the possibility that it served as a nucleotide exchange factor for BiP, which would trigger its release and allow Ig domains to fold (17, 18). However, there was no indication that recombinant pERp1 significantly stimulated BiP's ATPase activity either alone or in conjunction with ERdj3 (Fig. S8). The very modest effects it had on BiP's ATPase activity were also observed under reducing conditions, which would abrogate pERp1 structure, so this is most likely due to a minor contaminant in our preparation (Fig. S8). We also measured its ability to synergize with PDI in an in vitro folding assay, but again found no indication of an interaction (data not shown). Thus, we conclude that its activity on oxidative folding is likely to be direct.
Discussion
Lymphoid cells, and particularly B lineage cells, have been unusually rich sources for identifying new chaperones. BiP, the first ER chaperone to be discovered in any organism, was identified in association with the free HC synthesized in an Abelson virus transformed pre-B cell line (19). Calnexin (20) and ERdj3 (21) were also discovered due to their interaction with Ig HC, and Ig HC were among the first substrates to be found for PDI (22), GRP170 (23), PPI (24), and GRP94 (25). This is likely due in part to the unique biology of B cell development. Unlike most other multimeric proteins, there is a developmentally ordered production of Ig subunits with HC being expressed earlier than LC (26). In addition, the mechanisms for generating antibodies are particularly unusual. For most genes, great care is taken to ensure that mutations are rapidly detected and repaired to maintain primary amino acid sequences that fold efficiently into biologically active proteins. Instead, the DNA rearrangements that give rise to Ig variable regions are characterized by imprecise joining of V, D, and J gene segments, addition of nontemplated bases at the site of joining of these segments, and finally, during later stages of differentiation, directed hypermutation of the variable region exons of HC genes (26). Although these mechanisms are essential to generate antibody diversity and allow affinity maturation of the immune response, they clearly increase the likelihood of producing a protein that is incapable of folding and assembling properly. Thus, B lineage cells are likely to be particularly dependent on the ER quality control system, and it is possible that specific factors are required for this.
In the absence of association with LC, the CH1 domain of HC remains unoxidized and bound to BiP (17, 27). Using a very short pulse labeling, two oxidation forms of free γ HC were detected in Ag8(8) cells, but only the less oxidized form was present in the pERp1 knockdown clones. It is likely that the less oxidized form of HC represents one in which both the CH1 domain and a second domain are in the reduced state and that pERp1 plays a role in the oxidation of this domain. Because we see no evidence of two molecular forms for the dimeric state of HC, this would suggest that either these two forms are not readily separated on these gels or that oxidation of this second domain is enhanced by self-assembly in the absence of pERp1. Dr. Ingrid Haas and coworkers demonstrated that folding and oxidation of LC domains was assembly dependent and argued that this might be true for some other Ig domains (28). Thus, we would speculate that the second unoxidized domain is likely to be either CH2 or CH3, because the VH domain remains unpaired until it associates with the VL domain of a LC. It is somewhat surprising that overexpression of pERp1 in nonlymphoid cells induced the oxidation of the CH1 domain in ≈24% of the mini HC, because this domain remains unoxidized in Ag8(8) cells where pERp1 is expressed (29). This could reflect its ability to promiscuously assist in the oxidation of other domains when it is overexpressed. Alternatively, it is possible that pERp1 normally acts in concert with LC to fold this domain, but can do so less efficiently without LC when its expression is sufficiently high.
Although we have demonstrated that pERp1 assists in the oxidative folding of Ig domains, it is not clear whether it does so as an oxidoreductase or as a chaperone. The strongest data in support of it playing a direct role in disulfide bond formation is our detection of mixed disulfide bonds between pERp1 and IgM monomers. Disulfide bonds are catalyzed via the transfer of a bond from PDI-like molecules to the substrate, which involves an intermediate step where the bond exists between the oxidase and the substrate (30). A second argument for it being an oxidoreductase is the presence of a CXXC motif in the sequence. However, the fact that these cysteines are involved in disulfide bonds with cysteines at the C terminus and not with each other (38), and our finding that one of the cysteines can be mutated without affecting activity argue against this possibility. In fact, mutation of either cysteine in the active site of other oxidoreductases is sufficient to abrogate their activity. In the case of PDI, which possess two CXXC active sites, mutation of both sites simultaneously to either SXXC or CXXS, considerably reduced oxidation of a substrate to <5% of wild-type activity (31, 32). In addition, the reductase activity of ERp57 was diminished significantly when both of its sites were similarly mutated (33, 34). Thus, if pERp1 is an oxidoreductase it is not a conventional one. The other possibility is that pERp1 represents a new class of chaperone, which might be most compatible with the finding that it binds noncovalently with most IgM assembly intermediates. The fact that pERp1 did not stimulate BiP's ATPase activity (Fig. S8) or PDI's oxidoreductase activity (data not shown) suggests that the oxidation of the Ig domain we observed (Fig. 5) was likely to be a direct effect of pERp1, because BiP and PDI are the only ER proteins known to participate in the oxidative folding of Ig in vitro (35) or in vivo (19, 22). However, we cannot rule out the possibility that pERp1 acts through some unknown ER resident protein. Finally, it is conceivable that like PDI, pERp1 possess both oxidase and chaperone activities (36). In vitro studies are underway to distinguish between these possibilities.
Although the folding of individual Ig domains and assembly of HC and LC subunits occurs throughout B cell development, we did not detect expression of pERp1 at significant levels until B cells were stimulated to undergo terminal plasma cell differentiation. Other ER chaperones and folding enzymes are also significantly up-regulated during this process and are targets of the modified UPR (4). We found that tunicamycin induced pERp1 expression in a number of B cell lines, and a previous microarray study detected an increase of pERp1/PACAP transcripts upon enforced expression of XBP-1(S) in a Burkitt's lymphoma line (4). Indeed, inspection of the pERp1 promoter reveals the presence of potential XBP-1 binding sites in both mouse and human promoters. However, we failed to observe up-regulation of pERp1 in response to thapsigargin, even though a full UPR was activated. Thus, it is possible that pERp1 is a conditional UPR target that requires an additional lineage-specific component that is either suppressed by thapsigargin or somehow supplied by tunicamycin. This concept might also apply to the expression of other lineage specific chaperones and folding enzymes.
We identified a new type of resident ER protein in lymphoid cells that is significantly induced during plasma cell differentiation. It binds to all Ig assembly intermediates, assists in the oxidative folding of Ig domains, and prevents the formation of off-pathway, disulfide-linked HC oligomers. However, pERp1 shows no homology to known chaperones or oxidoreductases and does not appear to act indirectly through either BiP or PDI, arguing that it represents a unique member of the folding/quality control apparatus of lymphoid cells. Further in vitro studies are required to determine its precise mechanism of action.
Materials and Methods
Detailed materials and methods can be found in SI Materials and Methods.
Cell Lines and Antibodies.
The mouse plasmacytoma cell lines Ag8(8) (γ+,κ−) and Ag8.653 (γ−,κ−), the human Burkitt's lymphoma cell line Ramos, the I.29μ+ murine B cell line, and 293T cell lines were cultured as described in SI Materials and Methods. The UPR inducers tunicamycin (2.5 μg/mL) and thapsigargin (1 μM) (Sigma) were used for indicated times. A polyclonal anti-pERp1 antiserum was raised against recombinant pERp1 and affinity purified. The production of this antibody and the source of all other antibodies used in this study are described in SI Materials and Methods.
Identification and Cloning of pERp1.
Ag8(8) cells were treated with DSP and γ HC were isolated. Bound proteins were identified using mass-spectrometry and peptide sequencing of trypsin-digested fragments as described in ref. 8. Human and mouse pERp1 were cloned from Ramos and Ag8(8) cell lines, respectively, using primer pairs described in SI Materials and Methods.
Transfection of Cells and Detection of Proteins and RNA.
Cells were prepared for immunoblotting or immunoprecipitation as described in SI Materials and Methods. Information on recombinant plasmids, methods for transfection of cells with recombinant plasmids, and immunostaining can also be found there. Cross-linking with DSP, metabolic labeling, pulse–chase, and immunoprecipitation were performed as described in refs. 8 and 29. Isolation of total RNA and Northern blot analysis were performed by standard methods (37). A condition to amplify spliced XBP-1 from total RNA by reverse transcriptase-PCR (RT-PCR) is described in SI Materials and Methods.
Isolation and Fractionation of Primary B Cells and Induction of Plasma Cell Differentiation.
Mouse B cells were isolated from the bone marrow or spleen and individual subpopulations were isolated as described in refs. 13 and 14. Plasma cell differentiation of splenic B cells was induced with LPS or with anti-IgM and cytokines as described in SI Materials and Methods.
Detection of Mixed Disulfides.
Cells were washed with ice-cold PBS containing 10 mM NEM for 5 min and treated with 10% TCA to prevent postlysis disulfide bond formation/reduction. The resulting pellet was washed twice with 70% acetone, and proteins were dissolved in pH6 lysis buffer, followed by immunoprecipitation and two-dimentional SDS/PAGE as described in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank the members of the L.M.H. laboratory; Drs. Johannes Buchner (Technical University of Munchen, Munich, Germany), Akira Inoue, and Walid Awad for helpful discussions and for technical support; Dr. Peter Burrows (University of Alabama Birmingham, Birmingham, AL) for critical comments on the manuscript; Dr. Janet Stavnezer (University of Massachusetts Medical School, Worchester, MA) for kindly providing the I.29μ+ cell line; and Drs. Richard Ashmun, Ann Marie Hamilton-Easton, Michael Nash, and Jim Houston for cell sorting. This work is supported by National Institutes of Health Grant GM54068, Cancer Center CORE Grant CA21765, and the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811591106/DCSupplemental.
References
- 1.Hendershot LM, Sitia R. Immunoglobulin assembly and secretion. In: Honjo T, Alt FW, Neuberger M, editors. Molecular Biology of B cells. Elsevier Science; 2004. pp. 261–273. [Google Scholar]
- 2.Wiest DL, et al. Membrane biogenesis during B cell differentiation: Most endoplasmic reticulum proteins are expressed coordinately. J Cell Biol. 1990;110:1501–1511. doi: 10.1083/jcb.110.5.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Anken E, et al. Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity. 2003;18:243–253. doi: 10.1016/s1074-7613(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 4.Shaffer AL, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 5.Reimold AM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 6.Iwakoshi NN, et al. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- 7.Appenzeller-Herzog C, Ellgaard L. The human PDI family: Versatility packed into a single fold. Biochim Biophys Acta. 2008;1783:535–548. doi: 10.1016/j.bbamcr.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonfoco E, Li E, Kolbinger F, Cooper NR. Characterization of a novel proapoptotic caspase-2- and caspase-9-binding protein. J Biol Chem. 2001;276:29242–29250. doi: 10.1074/jbc.M100684200. [DOI] [PubMed] [Google Scholar]
- 10.Katoh M, Katoh M. MGC29506 gene, frequently down-regulated in intestinal-type gastric cancer, encodes secreted-type protein with conserved cysteine residues. Int J Oncol. 2003;23:235–241. [PubMed] [Google Scholar]
- 11.Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: Substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasegawa S, et al. Genome-wide analysis of gene expression in intestinal-type gastric cancers using a complementary DNA microarray representing 23,040 genes. Cancer Res. 2002;62:7012–7017. [PubMed] [Google Scholar]
- 13.Hardy RR, Shinton SA. Characterization of B lymphopoiesis in mouse bone marrow and spleen. Methods Mol Biol. 2004;271:1–24. doi: 10.1385/1-59259-796-3:001. [DOI] [PubMed] [Google Scholar]
- 14.Rolink AG, Andersson J, Melchers F. Molecular mechanisms guiding late stages of B-cell development. Immunol Rev. 2004;197:41–50. doi: 10.1111/j.0105-2896.2004.0101.x. [DOI] [PubMed] [Google Scholar]
- 15.Phillips C, Klaus GG. Soluble anti-mu monoclonal antibodies prime resting B cells to secrete immunoglobulins in response to interleukins-4 and -5. Eur J Immunol. 1992;22:1541–1545. doi: 10.1002/eji.1830220629. [DOI] [PubMed] [Google Scholar]
- 16.Jessop CE, et al. ERp57 is essential for efficient folding of glycoproteins sharing common structural domains. EMBO J. 2007;26:28–40. doi: 10.1038/sj.emboj.7601505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YK, Brewer JW, Hellman R, Hendershot LM. BiP and immunoglobulin light chain cooperate to control the folding of heavy chain and ensure the fidelity of immunoglobulin assembly. Mol Biol Cell. 1999;10:2209–2219. doi: 10.1091/mbc.10.7.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung KT, Shen Y, Hendershot LM. BAP, a mammalian BiP-associated protein, is a nucleotide exchange factor that regulates the ATPase activity of BiP. J Biol Chem. 2002;277:47557–47563. doi: 10.1074/jbc.M208377200. [DOI] [PubMed] [Google Scholar]
- 19.Haas IG, Wabl M. Immunoglobulin heavy chain binding protein. Nature. 1983;306:387–389. doi: 10.1038/306387a0. [DOI] [PubMed] [Google Scholar]
- 20.Hochstenbach F, David V, Watkins S, Brenner MB. Endoplasmic reticulum resident protein of 90 kilodaltons associates with the T- and B-cell antigen receptors and major histocompatibility complex antigens during their assembly. Proc Natl Acad Sci USA. 1992;89:4734–4738. doi: 10.1073/pnas.89.10.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen Y, Hendershot LM. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP's interactions with unfolded substrates. Mol Biol Cell. 2005;16:40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roth RA, Koshland ME. Role of disulfide interchange enzyme in immunoglobulin synthesis. Biochemistry. 1981;20:6594–6599. doi: 10.1021/bi00526a012. [DOI] [PubMed] [Google Scholar]
- 23.Lin HY, et al. The 170-kDa glucose-regulated stress protein is an endoplasmic reticulum protein that binds immunoglobulin. Mol Biol Cell. 1993;4:1109–1119. doi: 10.1091/mbc.4.11.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lang K, Schmid FX, Fischer G. Catalysis of protein folding by prolyl isomerase. Nature. 1987;329:268–270. doi: 10.1038/329268a0. [DOI] [PubMed] [Google Scholar]
- 25.Melnick J, Aviel S, Argon Y. The endoplasmic reticulum stress protein GRP94, in addition to BiP, associates with unassembled immunoglobulin chains. J Biol Chem. 1992;267:21303–21306. [PubMed] [Google Scholar]
- 26.Meffre E, Casellas R, Nussenzweig MC. Antibody regulation of B cell development. Nat Immunol. 2000;1:379–385. doi: 10.1038/80816. [DOI] [PubMed] [Google Scholar]
- 27.Hendershot L, Bole D, Kohler G, Kearney JF. Assembly and secretion of heavy chains that do not associate posttranslationally with immunoglobulin heavy chain-binding protein. J Cell Biol. 1987;104:761–767. doi: 10.1083/jcb.104.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leitzgen K, Knittler MR, Haas IG. Assembly of immunoglobulin light chains as a prerequisite for secretion. A model for oligomerization-dependent subunit folding. J Biol Chem. 1997;272:3117–3123. doi: 10.1074/jbc.272.5.3117. [DOI] [PubMed] [Google Scholar]
- 29.Vanhove M, Usherwood YK, Hendershot LM. Unassembled Ig heavy chains do not cycle from BiP in vivo but require light chains to trigger their release. Immunity. 2001;15:105–114. doi: 10.1016/s1074-7613(01)00163-7. [DOI] [PubMed] [Google Scholar]
- 30.Tu BP, Weissman JS. Oxidative protein folding in eukaryotes: Mechanisms and consequences. J Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LaMantia ML, Lennarz WJ. The essential function of yeast protein disulfide isomerase does not reside in its isomerase activity. Cell. 1993;74:899–908. doi: 10.1016/0092-8674(93)90469-7. [DOI] [PubMed] [Google Scholar]
- 32.Walker KW, Lyles MM, Gilbert HF. Catalysis of oxidative protein folding by mutants of protein disulfide isomerase with a single active-site cysteine. Biochemistry. 1996;35:1972–1980. doi: 10.1021/bi952157n. [DOI] [PubMed] [Google Scholar]
- 33.Antoniou AN, et al. The oxidoreductase ERp57 efficiently reduces partially folded in preference to fully folded MHC class I molecules. EMBO J. 2002;21:2655–2663. doi: 10.1093/emboj/21.11.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beynon-Jones SM, Antoniou AN, Powis SJ. Mutational analysis of the oxidoreductase ERp57 reveals the importance of the two central residues in the redox motif. FEBS Lett. 2006;580:1897–1902. doi: 10.1016/j.febslet.2006.02.055. [DOI] [PubMed] [Google Scholar]
- 35.Mayer M, Kies U, Kammermeier R, Buchner J. BiP and PDI cooperate in the oxidative folding of antibodies in vitro. J Biol Chem. 2000;275:29421–29425. doi: 10.1074/jbc.M002655200. [DOI] [PubMed] [Google Scholar]
- 36.Puig A, Gilbert HF. Protein disulfide isomerase exhibits chaperone and anti-chaperone activity in the oxidative refolding of lysozyme. J Biol Chem. 1994;269:7764–7771. [PubMed] [Google Scholar]
- 37.Sambrook JF, Russell DW. Molecular Cloning. 3rd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- 38.van Anken E, et al. Efficient IgM assembly and secretion requires the plasma cell induced ER protein pERp1. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0903036106. 10.1073/PNAS.0903036106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.