Abstract
Arthropod phenoloxidase (PO) generates quinones and other toxic compounds to sequester and kill pathogens during innate immune responses. It is also involved in wound healing and other physiological processes. Insect PO is activated from its inactive precursor, prophenoloxidase (PPO), by specific proteolysis via a serine protease cascade. Here, we report the crystal structure of PPO from a lepidopteran insect at a resolution of 1.97 Å, which is the initial structure for a PPO from the type 3 copper protein family. Manduca sexta PPO is a heterodimer consisting of 2 homologous polypeptide chains, PPO1 and PPO2. The active site of each subunit contains a canonical type 3 di-nuclear copper center, with each copper ion coordinated with 3 structurally conserved histidines. The acidic residue Glu-395 located at the active site of PPO2 may serve as a general base for deprotonation of monophenolic substrates, which is key to the ortho-hydroxylase activity of PO. The structure provides unique insights into the mechanism by which type 3 copper proteins differ in their enzymatic activities, albeit sharing a common active center. A drastic change in electrostatic surface induced on cleavage at Arg-51 allows us to propose a model for localized PPO activation in insects.
Keywords: innate immune, tyrosinase, melanization, zymogen activation, hemocyanin
Phenoloxidase (PO), a critical component of the innate immune system in insects and crustaceans, is present as a zymogen [prophenoloxidase (PPO)] in hemolymph and becomes activated on wounding or infection (1, 2). Possessing o-hydroxylase (EC 1.14.18.1) and o-di-PO (EC 1.10.3.1) activities, PO converts a variety of monophenolic and o-diphenolic substrates to o-quinones (3). Quinones can act as cross-linkers for wound healing, and they also polymerize to form melanin capsules around parasites and parasitoids (4–6). Quinones and other reactive intermediates (e.g., 5,6-dihydroxyindole) directly kill microbial pathogens (7). Although PO-generated compounds are powerful weapons against pathogens, they could also cause damage to host tissues and cells. Consequently, the activation of PPO is mediated by a cascade of highly specific serine proteases and regulated as a local transient reaction against invading organisms (8).
The proteolytic activation of PPO requires a trypsin-like serine protease, known as PPO activating protease (PAP) or PPO activating enzyme, which cuts the protein substrate next to an Arg residue near its amino-terminus (9–13). PAP contains regulatory clip domain(s) followed by a catalytic domain that hydrolyzes synthetic substrates at various ionic strengths and cleaves PPO in low-salt buffers preferably. In some insects, PAP generates active PO in the presence of an auxiliary factor consisting of clip-domain serine protease homolog (SPH), which lacks catalytic activity because of the substitution of the active site Ser by Gly (14, 15). Unlike its precursor, active PO self-associates into oligomers, binds to other proteins, and sticks to column matrices; consequently, it has never been purified from any arthropod species to near homogeneity for structural analysis (16). PPO is synthesized by hemocytes; released to plasma presumably because of cell lysis; and, in part, transported to cuticle (17). When wounding or infection occurs, recognition proteins associate with aberrant tissues or pathogens to trigger an extracellular serine protease pathway. At the end of this cascade, active PAP and, in some insects, high Mr SPHs are generated to activate PPO (18–20). Interestingly, certain chemical compounds (e.g., cetylpyridinium chloride) that do not cleave peptide bond can activate highly purified PPO, perhaps by inducing a conformational change at the active site (21).
PO belongs to a class of metalloproteins with type 3 copper centers (22), which also includes hemocyanin, catechol oxidase, and tyrosinase. Tyrosinase and PO catalyze 2 reactions: the o-hydroxylation of monophenols to catechols and oxidization of catechols to o-quinones (22, 23). Catechol oxidase catalyzes exclusively in the latter reaction (24). The differences in enzymatic activities could not be predicted from their sequences (25, 26). Unlike these enzymes, hemocyanins are oxygen carrier proteins in molluscs and some arthropods. Arthropod hemocyanins, greatly different from those of molluscs in sequence, tertiary structure, and quaternary organization (reviewed in ref. 22), are built up in vivo as oligomers of hexamers, with each hexamer composed of heterogeneous subunits with a Mr of ≈72 kDa (27). Arthropod hemocyanins are homologous to PPOs and storage hexamerins in insect hemolymph (26). In comparison, mollusc hemocyanins are decameric or di-decameric hollow cylindrical structures with large Mrs up to 9 MDa. Each subunit has a Mr of ≈400 kDa and appears as a string of 7 to 8 monomers of ≈50 kDa arranged in a “beads-on-a-string” structure (28, 29). Despite their differences in overall structure and function, all type 3 copper proteins share a similar di-copper active center (reviewed in ref. 23). Under certain conditions, such as limited proteolysis or binding to small molecules (e.g., SDS), hemocyanins can be converted to enzymes displaying PO activities (30, 31). Although o-di-PO activity of hemocyanins was observed most of the time (32), mono-oxygenase activity was observed from hemocyanins of a scorpion (33), a tarantula (34), and an octopus (35).
The structure and function of hemocyanins and catechol oxidases have been well studied in the past 2 decades (reviewed in ref. 22). The crystal structure of a bacterial tyrosinase was determined recently (36). However, the structural basis for the functional differences between these type 3 copper proteins remains elusive. Here, we report the crystal structure of PPO from Manduca sexta at a resolution of 1.97 Å, which represents the initial structure determined from any POs to date. The structural data provide unique insights into the mechanism by which this class of proteins differs in enzymatic activities. In addition, we propose a model for insect PPO activation during innate immune responses.
Results
Overall Structure of M. sexta PPO.
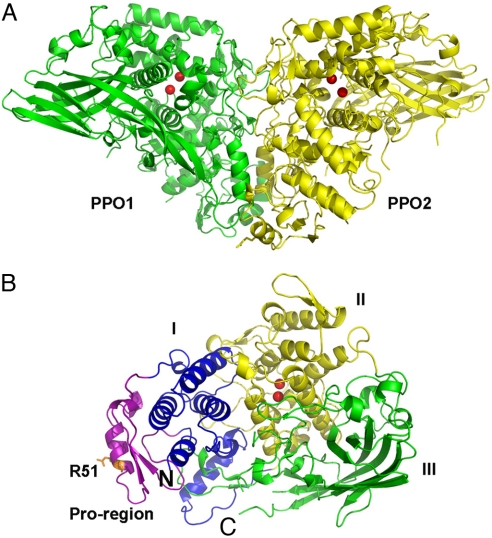
The structure of M. sexta PPO was determined by molecular replacement by using the structure of an arthropod hemocyanin as a search model. The heterodimeric PPO consists of 2 polypeptides: PPO1 of 685 residues and PPO2 of 695 residues (37) (Fig. 1). The final structure was refined to a resolution of 1.97 Å with excellent statistics (Table S1). Most of the residues are well defined in the final model, with only 25 disordered residues in the flexible regions (PPO1: 559–577, 618–619, and 4 residues at the carboxyl-terminus; PPO2: 567–584, 624–628, and 2 residues at the carboxyl-terminus). The structures of PPO1 and PPO2 closely resemble each other, displaying a 0.96-Å rmsd over 636 aligned residues with ≈48% sequence identity. Both structures adopt a fold similar to that observed in arthropod hemocyanins (rmsd between PPO1 and 1OXY is 1.27 Å over 526 aligned residues, with 41.4% sequence identity). There are 2 disulfide bonds found in each monomer (PPO1: Cys-580/Cys-622 and Cys-582/Cys-629; PPO2: Cys-586/Cys-630 and Cys-588/Cys-637).
Fig. 1.
Overall structure of M. sexta PPO. (A) The heterodimeric PPO is formed in a back-to-back mode. PPO1 and PPO2 are shown in green and yellow, respectively. (B) Domains of PPO2 are colored as follows: pro-region, purple; domain I, blue; domain II, yellow; domain III, green. The di-copper atoms are located in domain II and are shown as red spheres. The proteolytic site R51 residue is shown in the stick. The amino-terminus and carboxyl-terminus of PPO2 are indicated as N and C, respectively.
The PPO subunits adopt a compact structure that can be divided into 4 domains (Fig. 1B): the pro-region (PPO1: 17–67, PPO2: 17–70), the noncontiguous domain I (PPO1: 1–16 and 68–182, PPO2: 1–16 and 71–188), domain II (PPO1: 183–421, PPO2: 189–427), and domain III (PPO1: 422–680, PPO2: 428–693). The pro-region is mainly composed of an α-helix containing the proteolytic cleavage site Arg-51 flanked by a short, 2-stranded, parallel β-sheet. Domains I and II are predominantly α-helical. Domain III is mainly composed of a twisted, 7-stranded, antiparallel β-sheet. PPO1 and PPO2 are related by a pseudo–2-fold symmetry and interact with each other in a back-to-back fashion, displaying a butterfly shape with dimensions of ≈140 Å × 70 Å × 80 Å (Fig. 1A).
Active Site: the Di-Nuclear Copper Center.
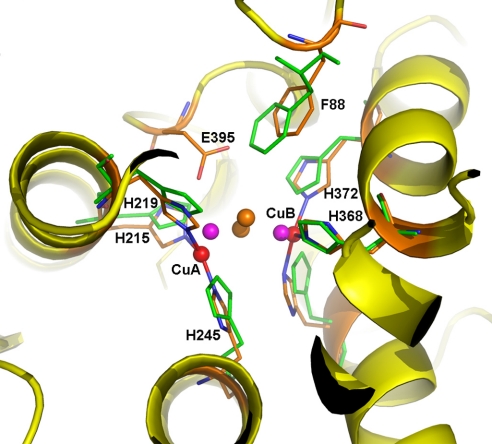
The active site is composed of 2 copper atoms (CuA and CuB) forming the di-copper center, with each copper atom coordinated with the Nε2 atoms of 3 conserved His residues in a distorted, trigonal, planar geometry (Fig. 2). The di-nuclear copper center is buried in the center of domain II and is inaccessible to the exterior solvent. In PPO1, CuA is coordinated with His-209, His-213, and His-239, whereas CuB is coordinated with His-366, His-370, and His-406. In PPO2, CuA is coordinated with His-215, His-219, and His 245, whereas CuB is coordinated with His-368, His-372, and His-408. There is an additional solvent molecule coordinated to both of the copper atoms in PPO2 only, at an average distance of 2.57 Å. The 2 copper atoms in both PPO1 and PPO2 are separated by a large distance (CuA-CuB distances in PPO1 and PPO2 are 4.53 Å and 4.87 Å, respectively), which resembles the deoxy form of other type 3 copper proteins, such as arthropod hemocyanins (38). Therefore, the current PPO structure represents the inactive deoxy state of the enzyme.
Fig. 2.
Di-copper center in M. sexta PPO2. The active site of PPO2 can be superimposed well with that of oxygenated Limulus polyphemus hemocyanin (Lp-HC, PDB ID code 1OXY). The secondary structures of PPO2 are shown in the ribbon and colored in yellow. The 6 copper-coordinating His ligands are shown as sticks, with those from Lp-HC colored green. The di-copper atoms are shown as spheres: PPO2, red; Lp-HC, purple. The peroxide ion in Lp-HC is shown as brown spheres. Notice the unique E395 in PPO2, which is located near the substrate place holder F88. E395 could be a base for phenol deprotonation, which is key to the ortho-phenol hydroxylation activity of PPO.
In addition to the di-nuclear copper center, a conserved Phe residue in each PPO subunit (Phe-85 in PPO1 and Phe-88 in PPO2) located on a loop in domain I protrudes into the active site and stacks onto the imidazole ring of one of the CuB-coordinating His ligands (His-370 in PPO1 and His-372 in PPO2, Fig. 2). The Phe residue is considered as the “place holder” for phenolic substrates (see below). Surprisingly, there is a unique Glu residue in PPO2 (Glu-395) but not in PPO1, which is located near CuA site and in close vicinity to Phe-88, the place holder. Glu-395 is flexible, and its carboxylate head group adopts dual conformations, one of which occupies a position at a distance of ≈3 Å from Cζ of Phe-88. We postulate that this unique residue, Glu-395, is the basic residue responsible for deprotonation of the hydroxyl group in monophenolic substrates, which is essential for the o-hydroxylase (i.e., mono-oxygenase) activity of the enzyme (see below).
Dimer Interface.
PPO was found to exist mainly as a heterodimer at physiological ionic strength (37). We reanalyzed the purified protein sample used for crystallization on a high-resolution size exclusion column under physiological conditions and found PPO displaying a single sharp peak on the chromatograph, corresponding to a dimer with an Mr of ≈160 kDa. In the crystal structure, we found 2 types of dimer association: a very tight heterodimer in the asymmetric unit and a loose heterodimer formed by a PPO1 and a symmetry-related PPO2 molecule through crystal lattice contacts. The tight dimer is formed between PPO1 and PPO2 in a back-to-back fashion along a pseudo–2-fold axis (Fig. 1). The association mode closely resembles that of the tight dimer found in an arthropod hemocyanin (39). The dimer interface can be separated into 2 regions (Fig. S1). Interactions in region 1 mainly involve the pro-region, domain I, and a loop region between 2 β-strands extended from domain III (Fig. S1). Interactions in region 2 mainly involve domain II from both PPO1 and PPO2. The tight PPO heterodimer is “glued” together through extensive hydrophobic and charge-charge interactions, burying a solvent-accessible surface area of ≈4,660 Å2, which is approximately twice the size of that in the arthropod hemocyanin (PDB ID code 1hcy). In comparison, the loose dimer formed through crystallography symmetry has a much smaller interface, with only ≈915 Å2 buried. Therefore, we consider the tight dimer as the biological dimer, which is consistent with the suggestion from the PISA web server (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html).
Discussion
Mechanism of Tyrosine Hydroxylation Activity: Catalytic Residues for Phenol Deprotonation.
Crystal structures of many type 3 copper proteins are available, including arthropod and mollusc hemocyanins (29, 38–40), a sweet potato catechol oxidase (41), and a Streptomyces tyrosinase (36). In all these structures, the entrances to the di-copper center are blocked by hydrophobic residues that need to be dislocated for activation (reviewed in refs. 22 and 42). The blocker is a Leu residue in the mollusc hemocyanin and a highly conserved Phe residue in all known arthropod hemocyanins. In the tyrosinase structure, a Tyr residue is provided by an associated caddie protein (ORF378) (36). This Tyr residue was kept away from the active center by the caddie protein at a sufficient distance to avoid a reaction. In the catechol oxidase structure, an inhibitor (1-phenyl-2-thiourea) was located in the equivalent position (41). Because the aromatic rings of these blocking residues could be well superimposed on each other, they were suggested as the place holders for incoming substrates and are stabilized by stacking interactions with a His residue at the CuB site (reviewed in ref. 23). In the PPO structure, Phe-85 and Phe-88 act as the place holders and stack on His-370 and His-372 in PPO1 and PPO2, respectively. Therefore, the activation of PPO probably involves pulling out of the 2 bulky residues from their current positions.
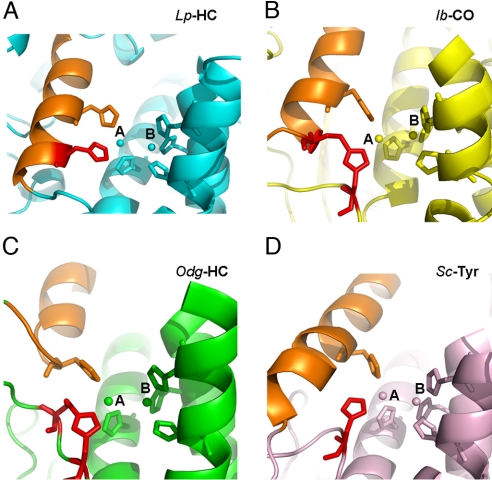
Although the di-copper active sites of all these type 3 copper proteins could be well superimposed, interesting differences occur mainly at the CuA site. In the mollusc hemocyanin, catechol oxidase, and tyrosinase structures, one of the copper-coordinating histidine ligands at the CuA site is provided from a loop. This is distinctively different from that in arthropod hemocyanins: the corresponding His residue is located on the same α-helix where a second CuA-coordinating histidine resides (Fig. 3). Interestingly, an unusual thioether bond was observed between the Cε atom of a unique His residue on the loop and a neighboring Cys residue in mollusc hemocyanin (29), catechol oxidase (41), and Neurospora crassa tyrosinase (43). A closer analysis of these unique covalent linkages reveals a fundamental difference. In the catechol oxidase structure, the thioether bond tethers the His on the loop to an α-helix, where a second CuA-coordinating histidine is located (Fig. 3B), to stabilize the residue at the CuA site (41), yielding essentially the same CuA coordination as in arthropod hemocyanins (Fig. 3 A and B). In comparison, the thioether bonds in both mollusc hemocyanin and Neurospora tyrosinase link the unique histidine to a flanking Cys residue within the same flexible loop (29, 43) (Fig. 3C). Therefore, it seems that these histidines in the latter proteins are flexible, similar to the equivalent histidine (His-54) found in the Streptomyces tyrosinase structure, which lacks the thioether bond (36) (Fig. 3D). We propose that these unique flexible His residues are involved in deprotonation of the hydroxyl group of monophenols before o-hydroxylation of the substrates (i.e., mono-oxygenase activity) (see below).
Fig. 3.
Different histidine coordination at CuA site. The active sites of 4 representative type 3 copper proteins are shown in the ribbons. Limulus polyphemus hemocyanin (Lp-HC, PDB ID code 1oxy), cyan (A); sweet potato Ipomoea batatas catechol oxidase (Ib-CO, PDB ID code 1bug), yellow (B); Octopus dofleini hemocyanin (Odg-HC, PDB ID code 1js8), green (C); and Streptomyces castaneoglobisporus tyrosinase (Sc-Tyr, PDB ID code 1wx5), salmon (D). The di-copper atoms and their 6-histidine ligands are shown in spheres and sticks, respectively. The unique histidine ligands at CuA sites are colored in red for comparison. The structurally conserved α-helix, where a second histidine locates, is colored in orange for reference in each structure. In Lp-HC, the histidine in comparison is located on the same α-helix where another histidine ligand resides. Notice that the thioether bond in Ib-CO is formed between 2 secondary structures, tethering the histidine residue at the CuA site, which resembles the CuA coordination in Lp-HC. In comparison, the equivalent histidine residues in both Odg-HC and Sc-Tyr are flexible. The thioether bond in Odg-HC is formed within the flexible loop, whereas that in Sc-Tyr does not form any thioether bonds.
Previous studies collectively suggest the CuA site to be critical for the mono-oxygenase activity of type 3 copper proteins (22, 23, 36, 44). In the crystal structure of catechol oxidase, a bulky Phe residue covers the CuA site and blocks its access to the substrate, which was suggested as the basis for the lack of mono-oxygenase activity. This could now be additionally supplemented with the unique thioether bond between 2 secondary structures (Fig. 3B). By using model and enzymatic systems, the mechanism of the mono-oxygenase activity was demonstrated to be electrophilic aromatic substitution (44). Deprotonation of the phenol is an essential step for coordination of the phenolic oxygen to one of the copper atoms and subsequent o-hydroxylation of phenols to catechols (44, 45). The flexible His-54 in the Streptomyces tyrosinase may act as a base for deprotonation of monophenolic substrates (36). We propose that the flexible CuA-coordinating histidines in mollusc hemocyanin (Fig. 3C) and N. crassa tyrosinase could function in the same way, and thus be responsible for their mono-oxygenase activities (35, 43). Such a flexible CuA-coordinating His ligand cannot be found in PPO structures, which adopt near-identical di-nuclear copper centers as in most arthropod hemocyanins (Figs. 2 and 3A). However, we found the unique acidic residue Glu-395 in PPO2 structures at the CuA site close to the substrate holder Phe-88 (Fig. 2). Similar acidic residues were not observed in any other type 3 copper proteins. We propose that Glu-395 may be the base to abstract a proton from the phenolic substrate, allowing the phenolate to coordinate with CuA, which is necessary for the subsequent ortho-hydroxylation. The equivalent position in PPO2 is occupied by a Ser residue. Therefore, it is likely that only PPO2 is responsible for the tyrosine hydroxylation activity of M. sexta PPO. Our proposed mechanism could thus explain why only di-PO activities were found from most arthropod hemocyanins, which do not have a residue acting as a general base at the active site. To explain why mono-oxygenase activities are found in hemocyanins from the scorpion (33) and tarantula (34) after activation, we suggest that the residues responsible for phenol deprotonation might be contributed by neighboring subunits in the oligomers after conformational changes induced by limited proteolysis or detergent treatment. This is consistent with the data showing that only oligomeric forms of these hemocyanins possess mono-oxygenase activities (33, 34).
Mechanism of PPO Activation in Insects.
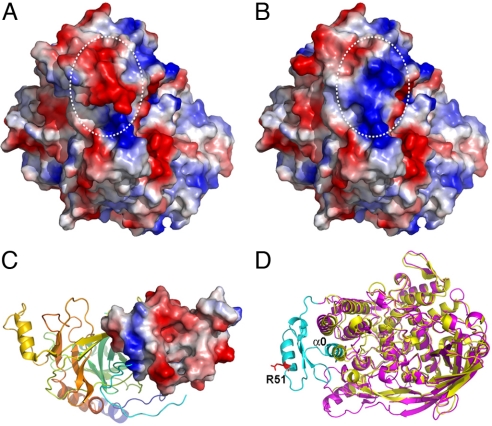
PPO is synthesized by hemocytes and released to plasma, and some of the precursors are transported to cuticles (17). In the silkworm, Bombyx mori, oxidation of certain methionine residues deprives the potential of the precursor to be transported (46). The previously proposed surface methionines of B. mori PPO (46) can now be confirmed from the current crystal structure. The activation of M. sexta PPO requires the presence of PPO, PAP, and SPHs simultaneously (20). It has been shown that without SPHs, PPO can be cleaved by PAP at the correct position but does not display PO activities. The clip domains in PAP and SPHs seem to be essential for PPO activation (18, 47). The solution structure of PAP-2 dual-clip domains suggests a PPO-binding site, a bacteria-interacting region, and a negatively charged surface for activator/adaptor docking in each domain. The purified complexes of M. sexta SPHs have an average Mr of 790 kDa (19). Based on these observations, we hypothesize that PPO activation is carried out by a PAP on the surface of a large complex of SPHs (19). Although a direct interaction between SPHs and PO has not yet been confirmed, SPHs probably play a role in conformational change of PO, which is a part of its activation process to dislodge the substrate place holders Phe-85 and Phe-85 in PPO1 and PPO2, respectively. In the current PPO structure, Arg-51 at the proteolytic site is located in an α-helix (45–54) in the pro-region, which might be disrupted on cleavage. We compared the surface properties of PPO before and after the presumed disruption of the α-helix containing the cleavage site by computer modeling. Interestingly, there is a drastic electrostatic switch on cleavage, resulting in a change of surface potential from largely negative to mainly positive in this region (Fig. 4 A and B). We postulate that this electrostatic switching might be important for interactions of PPO with the PAP and SPHs, perhaps through their clip domains. Indeed, the crystal structure of an SPH from Holotrichia diomphalia (18) displayed a predominantly negatively charged surface on its clip domain central cleft region, which was suggested to be the functional unit for binding PO (Fig. 4C). The PAP-2 dual-clip domains (47) contain 3 positively charged areas, one of which may be involved in initial association with the PPO. On proteolytic cleavage, the remarkable change in electrostatic potential may lead to an exchange of partner, with the PPO-interacting clip domain of PAP passing the cleaved PPO (i.e., PO) to a clip domain of SPHs. The association of SPHs with PO could trigger the conformational changes in PO domain I that lead to the subsequent dislocation of the substrate place holder Phe-85/PPO1 or Phe-88/PPO2, resulting in PO activation. The high Mr complex of M. sexta SPH-1 and SPH-2 was found to associate loosely with PAP-1/PAP-3 and PPO to form a ternary activation complex (19, 20). The SPH oligomers in H. diomphalia displayed a 2-tiered hexameric ring structure from cryogenic electron microscopy study, which might serve as the scaffold for PPO activation (18). Whether the clip domain SPHs in M. sexta also adopt a similar quaternary organization is not known. Nevertheless, the crystal structure described here provides a solid foundation for further investigation of PPO activation.
Fig. 4.
Electrostatic surface potential switching of M. sexta PPO2 on cleavage. The negatively and positively charged surfaces are colored in red and blue, respectively. (A) The intact PPO2 structure. (B) Computer-modeled PPO2 structure after simulated cleavage at R51 and removal of the α-helix where it locates. Notice the oppositely charged surface after cleavage as indicated in white circles. (C) Crystal structure of an SPH from H. diomphalia (PDB ID code 2B9L). The clip domain is shown as an electropotential surface, whereas the rest of the protein is shown in the ribbon. Notice the predominantly negative-charged surface of the clip domain. (D) Superimposition of the structures of PPO2 (purple) and Lp-HC (PDB ID code 1OXY, yellow). The N-terminus of PPO2 flanking the proteolytic site R51 (shown in the red stick) is colored in cyan. Notice that α0 (1–16) of PPO2 superimposes well with one of the conserved α-helices in domain I of Lp-HC. Therefore, α0 is considered as part of domain I in PPO2, which may still be associated with the rest of the PO structure even after the proteolytic cleavage.
It was shown that PPO could be alternatively activated by in vitro treatment of cationic detergent without proteolytic enzymes (21). The current PPO structure displays a predominantly negative charge at the proteolytic site (Fig. 4A). The cationic detergents might interact with this region through electrostatic interactions, which could cause conformational changes of PPO. As a result, the pro-region and domain I could move as a single piece because of their strong association (Fig. 4D), resulting in disposition of the substrate place holders and subsequent PPO activation without peptide bond cleavage. However, the exact detergent binding sites on PPO are not known; therefore, the detailed mechanism by which detergent activates PPO needs further investigation.
Materials and Methods
Structure Determination and Analysis.
PPO from M. sexta was purified as described in ref. 37 and was concentrated to 10 mg/mL. Crystals were obtained under conditions containing 20% PEG10K and 100 mM Hepes (pH 7.5) at room temperature for a week. Twenty percent glycerol was added to the crystallization condition for cryoprotection. A set of data was collected to a resolution of 1.97 Å at 100 K at the Advanced Photon Source, beam-line 19-ID, Argonne National Laboratory (Argonne, IL). The crystal belongs to space group P21212 with 2 polypeptides in the asymmetrical unit. The data were processed at the beam-line with program HKL3000 (48). The initial phasing was obtained by molecular replacement by using the Phaser program of the CCP4 suit (49). Subsequent manual modeling was carried out with the COOT program (50). The structure was refined with REFMAC5 (51), and the MolProbity server (52) was used for the final model analysis. The current model has excellent geometry and refinement statistics (Table S1). All molecular graphic figures were generated with PyMol (53).
Supplementary Material
Acknowledgments.
We thank the staff of beam-line 19ID at the Advanced Photon Source for their support, Drs. Michael Kanost and Karl Kramer for their helpful comments on the manuscript. This work was supported by the Oklahoma Agricultural Experiment Station at Oklahoma State University under Project OKL02618 (to J.D.) and National Institutes of Health Grant GM58634 (to H.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 3HHS).
This article contains supporting information online at www.pnas.org/cgi/content/full/0906095106/DCSupplemental.
References
- 1.Ashida M, Brey PT. Recent advances on the research of the insect prophenoloxidase cascade. In: Brey PT, Hultmark D, editors. Molecular Mechanisms of Immune Response in Insects. London: Chapman & Hall; 1998. pp. 135–172. [Google Scholar]
- 2.Kanost MR, Gorman MJ. Phenoloxidases in insect immunity. In: Beckage NE, editor. Insect Immunity. New York: Academic; 2008. pp. 69–96. [Google Scholar]
- 3.Nappi AJ, Christensen BM. Melanogenesis and associated cytotoxic reactions: Applications to insect innate immunity. Insect Biochem Mol Biol. 2005;35(5):443–459. doi: 10.1016/j.ibmb.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 4.Gillespie JP, Kanost MR, Trenczek T. Biological mediators of insect immunity. Annu Rev Entomol. 1997;42:611–643. doi: 10.1146/annurev.ento.42.1.611. [DOI] [PubMed] [Google Scholar]
- 5.Lavine MD, Strand MR. Insect hemocytes and their role in immunity. Insect Biochem Mol Biol. 2002;32:1295–1309. doi: 10.1016/s0965-1748(02)00092-9. [DOI] [PubMed] [Google Scholar]
- 6.Cerenius L, Lee BL, Söderhäll K. The proPO-system: Pros and cons for its role in invertebrate immunity. Trends Immunol. 2008;29:263–271. doi: 10.1016/j.it.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Zhao P, Li J, Wang Y, Jiang H. Broad-spectrum antimicrobial activity of the reactive compounds generated in vitro by Manduca sexta phenoloxidase. Insect Biochem Mol Biol. 2007;37:952–959. doi: 10.1016/j.ibmb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang H. The biochemical basis of antimicrobial responses in Manduca sexta. Insect Science. 2008;15:53–66. [Google Scholar]
- 9.Jiang H, Wang Y, Kanost MR. Pro-phenol oxidase activating proteinase from an insect, Manduca sexta: A bacteria-inducible protein similar to Drosophila easter. Proc Natl Acad Sci USA. 1998;95:12220–12225. doi: 10.1073/pnas.95.21.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang H, Wang Y, Yu X-Q, Kanost MR. Prophenoloxidase-activating proteinase-2 from hemolymph of Manduca sexta. J Biol Chem. 2003;278:3552–3561. doi: 10.1074/jbc.M205743200. [DOI] [PubMed] [Google Scholar]
- 11.Jiang H, Wang Y, Yu X-Q, Zhu Y, Kanost MR. Prophenoloxidase-activating proteinase-3 (PAP-3) from Manduca sexta hemolymph: A clip-domain serine proteinase regulated by serpin-1J and serine proteinase homologs. Insect Biochem Mol Biol. 2003;33:1049–1060. doi: 10.1016/s0965-1748(03)00123-1. [DOI] [PubMed] [Google Scholar]
- 12.Lee SY, et al. Molecular cloning of cDNA for prophenoloxidase-activating factor I, a serine protease, is induced by lipopolysacharide or 1,3-beta-glucan in coleopteran insect, Holotrichia diomphalia larvae. Eur J Biochem. 1998;257:615–621. doi: 10.1046/j.1432-1327.1998.2570615.x. [DOI] [PubMed] [Google Scholar]
- 13.Satoh D, Horri A, Ochiai M, Ashida M. Prophenoloxidase-activating enzyme of the silkworm, Bombyx mori: Purification, characterization and cDNA cloning. J Biol Chem. 1999;274:7441–7453. doi: 10.1074/jbc.274.11.7441. [DOI] [PubMed] [Google Scholar]
- 14.Lee SY, et al. In vitro activation of pro-phenol-oxidase by two kinds of pro-phenol-oxidase-activating factors isolated from hemolymph of coleopteran, Holotrichia diomphalia larvae. Eur J Biochem. 1998;254:50–57. doi: 10.1046/j.1432-1327.1998.2540050.x. [DOI] [PubMed] [Google Scholar]
- 15.Yu XQ, Jiang H, Wang Y, Kanost MR. Nonproteolytic serine proteinase homologs are involved in prophenoloxidase activation in the tobacco hornworm, Manduca sexta. Insect Biochem Mol Biol. 2003;33:197–208. doi: 10.1016/s0965-1748(02)00191-1. [DOI] [PubMed] [Google Scholar]
- 16.Sugumaran M. Comparative biochemistry of eumelanogenesis and the protective roles of phenoloxidase and melanin in insects. Pigm Cell Res. 2002;15:2–9. doi: 10.1034/j.1600-0749.2002.00056.x. [DOI] [PubMed] [Google Scholar]
- 17.Asano T, Ashida M. Cuticular pro-phenoloxidase of the silkworm, Bombyx mori. Purification and demonstration of its transport from hemolymph. J Biol Chem. 2001;276:11100–11112. doi: 10.1074/jbc.M008426200. [DOI] [PubMed] [Google Scholar]
- 18.Piao SP, et al. Crystal structure of a clip-domain serine protease and functional roles of the clip domains. EMBO J. 2005;24:4404–4414. doi: 10.1038/sj.emboj.7600891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Jiang H. Prophenoloxidase (proPO) activation in Manduca sexta: An analysis of molecular interactions among proPO, proPO-activating proteinase-3, and a cofactor. Insect Biochem Mol Biol. 2004;34:731–742. doi: 10.1016/j.ibmb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Gupta S, Wang Y, Jiang H. Manduca sexta prophenoloxidase (proPO) activation requires proPO-activating proteinase (PAP) and serine proteinase homologs (SPHs) simultaneously. Insect Biochem Mol Biol. 2005;35:241–248. doi: 10.1016/j.ibmb.2004.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall M, Scott T, Sugumaran M, Söderhäll K, Law JH. Proenzyme of Manduca sexta phenol oxidase: Purification, activation, substrate specificity of the active enzyme, and molecular cloning. Proc Natl Acad Sci USA. 1995;92:7764–7768. doi: 10.1073/pnas.92.17.7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Decker H, et al. Similar enzyme activation and catalysis in hemocyanins and tyrosinases. Gene. 2007;398:183–191. doi: 10.1016/j.gene.2007.02.051. [DOI] [PubMed] [Google Scholar]
- 23.Decker H, Schweikardt T, Tuczek F. The first crystal structure of tyrosinase: All questions answered? Angew Chem Int Ed. 2006;45:4546–4550. doi: 10.1002/anie.200601255. [DOI] [PubMed] [Google Scholar]
- 24.Gerdemann C, Eicken C, Krebs B. The crystal structure of catechol oxidase: New insight into the function of type-3 copper proteins. Acc Chem Res. 2002;35:183–191. doi: 10.1021/ar990019a. [DOI] [PubMed] [Google Scholar]
- 25.Sánchez-Ferrer A, Rodríguez-López JN, García-Cánovas F, García-Carmona F. Tyrosinase: A comprehensive review of its mechanism. Biochim Biophys Acta. 1995;1247:1–11. doi: 10.1016/0167-4838(94)00204-t. [DOI] [PubMed] [Google Scholar]
- 26.Burmester T. Molecular evolution of the arthropod hemocyanin superfamily. Mol Biol Evol. 2001;18:184–195. doi: 10.1093/oxfordjournals.molbev.a003792. [DOI] [PubMed] [Google Scholar]
- 27.Ballweber P, Markl J, Burmester T. Complete hemocyanin subunit sequences of the hunting spider Cupiennius salei: Recent hemocyanin remodeling in entelegyne spiders. J Biol Chem. 2002;277:14451–14457. doi: 10.1074/jbc.M111368200. [DOI] [PubMed] [Google Scholar]
- 28.Miller KI, Schabtach E, van Holde KE. Arrangement of subunits and domains within the Octopus dofleini hemocyanin molecule. Proc Natl Acad Sci USA. 1990;87:1496–1500. doi: 10.1073/pnas.87.4.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuff ME, Miller KI, van Holde KE, Hendrickson WA. Crystal structure of a functional unit from Octopus hemocyanin. J Mol Biol. 1998;278:855–870. doi: 10.1006/jmbi.1998.1647. [DOI] [PubMed] [Google Scholar]
- 30.Jaenicke E, Decker H. Conversion of crustacean hemocyanin to catecholoxidase. Micron. 2004;35:89–90. doi: 10.1016/j.micron.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 31.Decker H, Terwilliger N. Cops and robbers: Putative evolution of copper oxygen-binding proteins. J Exp Biol. 2000;203:1777–1782. doi: 10.1242/jeb.203.12.1777. [DOI] [PubMed] [Google Scholar]
- 32.Decker H, Jaenicke E. Recent findings on phenoloxidase activity and antimicrobial activity of hemocyanins. Dev Comp Immunol. 2004;28:673–687. doi: 10.1016/j.dci.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Nillius D, Jaenicke E, Decker H. Switch between tyrosinase and catecholoxidase activity of scorpion hemocyanin by allosteric effectors. FEBS Lett. 2008;582:749–754. doi: 10.1016/j.febslet.2008.01.056. [DOI] [PubMed] [Google Scholar]
- 34.Decker H, Rimke T. Tarantula hemocyanin shows phenoloxidase activity. J Biol Chem. 1998;273:25889–25892. doi: 10.1074/jbc.273.40.25889. [DOI] [PubMed] [Google Scholar]
- 35.Morioka C, Tachi Y, Suzuki S, Itoh S. Significant enhancement of monooxygenase activity of oxygen carrier protein hemocyanin by urea. J Am Chem Soc. 2006;128:6788–6789. doi: 10.1021/ja061631h. [DOI] [PubMed] [Google Scholar]
- 36.Matoba Y, Kumagai T, Yamamoto A, Yoshitsu H, Sugiyama M. Crystallographic evidence that the dinuclear copper center of tyrosinase is flexible during catalysis. J Biol Chem. 2006;281:8981–8990. doi: 10.1074/jbc.M509785200. [DOI] [PubMed] [Google Scholar]
- 37.Jiang H, Wang Y, Ma C, Kanost MR. Subunit composition of pro-phenol oxidase from Manduca sexta: Molecular cloning of subunit proPO-P1. Insect Biochem Mol Biol. 1997;27:835–850. doi: 10.1016/s0965-1748(97)00066-0. [DOI] [PubMed] [Google Scholar]
- 38.Hazes B, et al. Crystal structure of deoxygenated Limulus polyphemus subunit II hemocyanin at 2.18 A resolution: Clues for a mechanism for allosteric regulation. Protein Sci. 1993;2:597–619. doi: 10.1002/pro.5560020411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volbeda A, Hol WG. Crystal structure of hexameric haemocyanin from Panulirus interruptus refined at 3.2 A resolution. J Mol Biol. 1989;209:249–279. doi: 10.1016/0022-2836(89)90276-3. [DOI] [PubMed] [Google Scholar]
- 40.Magnus KA, et al. Crystallographic analysis of oxygenated and deoxygenated states of arthropod hemocyanin shows unusual differences. Proteins. 1994;19:302–309. doi: 10.1002/prot.340190405. [DOI] [PubMed] [Google Scholar]
- 41.Klabunde T, Eicken C, Sacchettini JC, Krebs B. Crystal structure of a plant catechol oxidase containing a dicopper center. Nat Struct Biol. 1998;5:1084–1090. doi: 10.1038/4193. [DOI] [PubMed] [Google Scholar]
- 42.Decker H, et al. Recent progress in hemocyanin research. Integr Comp Biol. 2007;47:631–644. doi: 10.1093/icb/icm063. [DOI] [PubMed] [Google Scholar]
- 43.Lerch K. Primary structure of tyrosinase from Neurospora crassa. II. Complete amino acid sequence and chemical structure of a tripeptide containing an unusual thioether. J Biol Chem. 1982;257:6414–6419. [PubMed] [Google Scholar]
- 44.Itoh S, Fukuzumi S. Monooxygenase activity of type 3 copper proteins. Acc Chem Res. 2007;40:592–600. doi: 10.1021/ar6000395. [DOI] [PubMed] [Google Scholar]
- 45.Itoh S, et al. Oxygenation of phenols to catechols by a (mu-eta 2:eta 2-peroxo)dicopper(II) complex: Mechanistic insight into the phenolase activity of tyrosinase. J Am Chem Soc. 2001;123:6708–6709. doi: 10.1021/ja015702i. [DOI] [PubMed] [Google Scholar]
- 46.Asano T, Ashida M. Transepithelially transported pro-phenoloxidase in the cuticle of the silkworm, Bombyx mori. Identification of its methionyl residues oxidized to methionine sulfoxides. J Biol Chem. 2001;276:11113–11125. doi: 10.1074/jbc.M008425200. [DOI] [PubMed] [Google Scholar]
- 47.Huang R, et al. The solution structure of clip domains from Manduca sexta prophenoloxidase activating proteinase-2. Biochemistry. 2007;46:11431–11439. doi: 10.1021/bi7010724. [DOI] [PubMed] [Google Scholar]
- 48.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: The integration of data reduction and structure solution—From diffraction images to an initial model in minutes. Acta Crystallogr D. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 49.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Murshudov G, Vagin A, Dodson E. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 52.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeLano W. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.