Abstract
The delivery of nucleic acids with polycations offers tremendous potential for developing highly specific treatments for various therapeutic targets. Although materials have been developed and studied for polynucleotide transfer, the biological mechanisms and fate of the synthetic vehicle has remained elusive due to the limitations with current labeling technologies. Here, we have developed polymer beacons that allow the delivery of nucleic acids to be visualized at different biological scales. The polycations have been designed to contain repeated oligoethyleneamines, for binding and compacting nucleic acids into nanoparticles, and lanthanide (Ln) chelates [either luminescent europium (Eu3+) or paramagnetic gadolinium (Gd3+)]. The chelated Lns allow the visualization of the delivery vehicle both on the nm/μm scale via microscopy and on the sub-mm scale via MRI. We demonstrate that these delivery beacons effectively bind and compact plasmid (p)DNA into nanoparticles and protect nucleic acids from nuclease damage. These delivery beacons efficiently deliver pDNA into cultured cells and do not exhibit toxicity. Micrographs of cultured cells exposed to the nanoparticle complexes formed with fluorescein-labeled pDNA and the europium-chelated polymers reveal effective intracellular imaging of the delivery process. MRI of bulk cells exposed to the complexes formulated with pDNA and the gadolinium-chelated structures show bright image contrast, allowing visualization of effective intracellular delivery on the tissue-scale. Because of their versatility, these delivery beacons posses remarkable potential for tracking and understanding nucleic acid transfer in vitro, and have promise as in vivo theranostic agents.
Keywords: intracellular imaging, nucleic acid delivery, theranostic, lanthanide
The delivery of therapeutic nucleic acids such as siRNA, antisense agents, transcription factor decoys, and plasmid (p)DNA offers an unprecedented opportunity for developing highly specific treatments for many devastating diseases (1, 2). The use of synthetic materials, such as polymers, for polynucleotide delivery has rapidly grown, and presents a wealth of promising alternatives to conventional viral vectors (3–5), which have caused serious problems in the clinic. Although much is known about the infection pathways, advantages, and troubles of viral vectors (6), researchers in the field of nonviral delivery are just beginning to understand the transfection mechanisms, benefits, and potential issues with the multitude of materials being developed as macromolecular drug carriers (7, 8). Considering that the delivery vehicle has a central role in the mechanisms, kinetics, efficacy, and toxicity of nucleic acid medicines, little is known about how the vehicle structure affects drug fate both in vitro and in vivo. For this reason, smart biomaterials termed “theranostic” agents are being developed that provide diagnostic imaging, therapeutic delivery, and the ability to monitor treatment efficacy (9). Indeed, the parallel development of novel nucleic acid drugs and theranostic vehicles that offer disease diagnosis, treatment, and the ability to understand the delivery mechanisms/kinetics on a range of biological scales will advance this field toward the discovery of personalized treatment strategies.
Tracking the delivery of nucleic acids within cells and tissues has traditionally been accomplished by a number of methods such as labeling nucleotides with fluorescent dyes (8), radiotracers (10), quantum dots (11, 12), and/or with various reporter gene assays (4). Although these methods have yielded a means of monitoring the presence and location of nucleic acids, many issues have surfaced with labeling polymeric delivery vehicles with these techniques. For example, the polymer labeling efficiency is often poor, nonuniform, irreproducible, and difficult to characterize. Also, dyes can alter the delivery mechanisms and/or increase side-effects (11). For this reason, the development of new material-based delivery systems that allow monitoring of both the nucleic acid and the delivery vehicle, on the cellular and tissue scales, is essential to improve the delivery efficiency, optimize the vehicle structure, and monitor treatment efficacy in living systems.
Lanthanide (Ln) metals are endowed with several unique properties that can be exploited for the development and study of nonviral delivery vehicles. Complexes housing luminescent Lns, such as europium (Eu3+), offer many unique advantages over the aforementioned labeling methods due to their small hydrophilic structures, long luminescence lifetimes, large Stokes shifts, and their stability from quenching and photobleaching (13, 14). The chelates of Lns are relatively nontoxic; structures containing gadolinium (Gd3+) are FDA-approved as MRI contrast agents due to their paramagnetic nature and slow electronic relaxation time (15). Polymers and macromolecules containing gadolinium chelates are being studied as new contrast agents, because their large structures increase the rotational correlation time, which improves resolution and sensitivity (16). Also, MRI is advantageous for following drug delivery in vivo with very high resolution, because it is a noninvasive and safe imaging method (17).
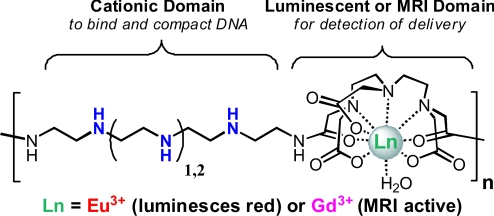
The polymer delivery vehicles developed here offer a creative and powerful method for tracking the delivery of nucleic acids. As shown in Fig. 1, we have designed these previously undescribed delivery beacons to contain systematically repeated Ln chelates within an oligoethyleneamine backbone. We have found that these materials can bind and compact pDNA into nanoparticles (termed polyplexes) that are taken up into cultured human cervix adenocarcinoma (HeLa) cells in an effective and nontoxic manner. We reveal that these delivery beacons offer a dual imaging modality for tracking transport in vitro on the nm/μm scale via microscopy (using the luminescent Eu3+-chelated structures) and for detection via MRI within bulk cultured tissues on the sub-mm scale (using the paramagnetic Gd3+-chelated materials). These scaffolds offer a unique motif that can be readily tailored and optimized as both luminescent and MRI theranostic delivery agents.
Fig. 1.
Schematic representations of the two analogous polymeric imaging beacons that differ in the ethyleneamine length (3a or 3b contain 3 or 4 ethyleneamines). The structures can be chelated with either Eu3+ or Gd3+ for microscopy and MRI imaging, respectively.
Results and Discussion
Polymer Synthesis and Polyplex Formation.
The design of these materials (Fig. 1) was inspired by the characteristics of successful cationic nucleic acid delivery vehicles and the many elegant macromolecular imaging agents being examined for disease diagnosis. The amine-containing comonomer precursors were synthesized using a series of protection/deprotection reactions previously published by our lab to obtain 1a and 1b (Scheme S1) (5). Next, DTPA-BA was reacted with 1a or 1b in dimethyl sulfoxide at room temperature for 24 h to form two polymer structures 2a and 2b with protected oligoethyleneamine units and pendant carboxylates from the anhydride ring opening during polymerization (Scheme S1). The Boc protecting groups were removed yielding 3a and 3b, which were confirmed by NMR analysis (SI Appendix). The deprotected precursors were then chelated with the chloride salts of Eu3+ and Gd3+ (confirmed via FT-IR and ICP-MS) (Tables S1 and S2). The final Ln-chelated polymers were analyzed via GPC using a triple detection system (refractive index, static light scattering, and viscometry). A similar molecular mass, degree of polymerization, and polydispersity were achieved for all polymer structures 3a, 3b, Eu3a, Eu3b Gd3a, and Gd3b (Table S3), which was expected because all of the Ln-containing polymers were synthesized from the same parent batches of 3a and 3b. Because of similarities in polymer characteristics and the similar chemical properties of Lns in the 3+ oxidation state, sound comparison of polymer biological activity was possible between the different analogs.
The polymers, 3a, 3b, Eu3a, Eu3b, Gd3a, and Gd3b, were examined for their ability to complex pDNA using an electrophoretic gel shift assay (Fig. S1). Polyplexes were formulated at N/P ratios (n = polymer secondary amine no.; P = DNA phosphate no.) between 0 and 40 before being electrophoresed in an agarose gel. Polymers Eu3a, Eu3b, Gd3a, and Gd3b began to charge-neutralize pDNA at an N/P ratio of 5, and pDNA migration was mostly suppressed at N/P of 20. With these polymers, the N/P ratio needed to fully inhibit gel migration of pDNA was higher than other polyamidoamine vehicles created in our laboratory (5). The presence of the carboxylate groups likely lowers the cationic charge on the polymer backbone. It is interesting to note that the nonchelated polymers (3a and 3b) do not bind with pDNA even at an N/P ratio of 100. This result is likely due to the fact that the nonchelated polymers have negatively-charged carboxylates, which likely neutralize the positive charge from the amine groups on the polymer backbone (Fig. S1).
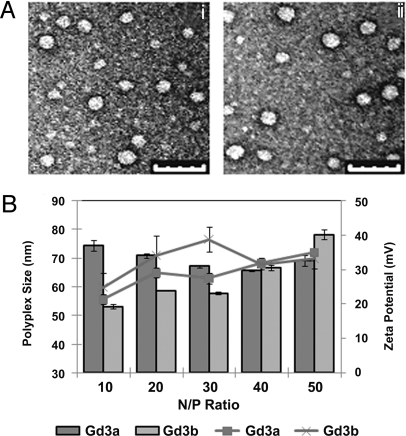
The size and morphology of the polyplexes were then examined via transmission electron microscopy (TEM) and dynamic light scattering (DLS). TEM micrographs indicate that the polyplexes formulated with Eu3a and Eu3b and pDNA at N/P of 40 exhibit spherical morphology with particle diameters between 35 and 60 nm (Fig. 2A). DLS and zeta potential data (Fig. 2B) generally showed slightly larger particle sizes resulting from measuring the hydrodynamic radii (sphere of hydration). In the DLS studies, the size of Gd3a polyplexes slightly decreased with the N/P ratio (from 74 to 66 nm); however, Gd3b polyplexes generally increased in size with the N/P ratio (from 53 to 78 nm). Zeta potential measurements reveal that polyplexes exhibited a positive surface charge (between 20 and 40 mV) that increased with the N/P ratio. We were unable to analyze polyplexes formed with Eu3a and Eu3b via DLS and zeta potential measurements due to laser/detector interference with the absorption/luminescence emission bands of Eu3+. The stability of these polyplexes from nuclease degradation was examined by exposure to high concentrations of FBS (33%), and pDNA integrity was observed via gel electrophoresis. These data show there is no evidence of pDNA degradation in the polyplexes at any time point assayed (shown as the lack of formation of band 4, Fig. S2, Gels Eu3a, Eu3b, Gd3a, and Gd3b). Also, the brightness of the pDNA band in the polyplex gels (band 2, Fig. S2) remains constant as a function of exposure time. In the control gel of naked pDNA (gel pDNA, lanes 9–12), band 2 was absent, and band 4 (degraded DNA) was clearly observed, indicating full degradation after only 1 h of incubation in 25% FBS.
Fig. 2.
Polyplex size and charge as observed by transmission electron microscopy, dynamic light scattering, and zeta potential measurements. (A) TEM micrographs of (Ai) Eu3a and (Aii) Eu3b polyplexes (N/P = 40). Polyplexes were negatively stained with uranyl acetate. (Scale bar, 100 nm.) (B) DLS (bars) and zeta potentials (lines) of polyplexes at various N/P ratios (the average and SD of three measurements are shown).
Cellular Delivery and Toxicity Studies.
Toxicity assays were conducted to assess the cellular uptake, toxicity, and transfection efficiency of the polyplexes formulated with the polymer beacons. HeLa cells were transfected with polyplexes formulated with Cy5-pDNA and either Eu3a, Eu3b, Gd3a, or Gd3b. Four hours after transfection, cellular uptake efficiency was determined by monitoring the fluorescence intensity of Cy5-labeled pDNA via flow cytometry. Cell viability experiments were conducted by assaying the cells for protein content 48 h after transfection. Toxicity was assessed by normalizing the results of the protein assay to a control of untreated cells; this assay was then used to calculate the fraction of viable cells in each well. The transfection efficiency was also monitored 48 h after transfection using a luciferase reporter gene expression assay (Fig. S4). These results are reported as relative light units (RLUs) per milligram of protein. It should be mentioned that these conventional assays only yield pDNA uptake and transcription efficiency, but do no yield information about the fate of the polymer (thus, the development and study of our intracellular delivery beacons; see below).
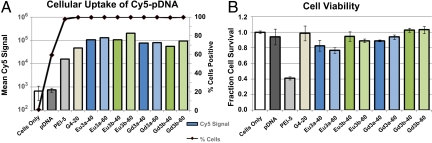
All polyplexes formed with these polymers promoted high cellular uptake while maintaining high viability, as depicted in Fig. 3A. In this study, negative controls consisted of cells only and pDNA only, whereas positive controls consisted of polyplexes formed with Jet-PEI at an N/P = 5 and polyplexes formed with G4 at N/P of 20. Polymer G4 is a polyamidoamine delivery vehicle previously developed by our group that consists of alternating meso-galactaramide units and four ethyleneamine groups (4). As shown, the delivery beacons yielded high pDNA uptake (≈100% of cells), and the intensity of Cy5 fluorescence was higher than our positive controls, Jet-PEI and G4, indicating that these vehicles are promising for further studies. Fig. 3B reveals that the toxicity of these structures is very low; even at very high polymer concentrations (high N/P ratios), cell viability remained very high (between 80 and 100% cell viability) when compared with our positive control Jet-PEI (cell viability only 40% at low N/P = 5). Similar high cell viability (>80%) is verified when studied by an MTT assay with cells cultured under the same conditions (Fig. S3). When the luciferase expression was compared (Fig. S4), these systems displayed slightly lower gene expression (107-108 RLU/mg) compared with our positive controls, G4 (109 RLU/mg) and Jet-PEI (1010 RLU/mg). However, as previously discussed, these polymer vehicles yielded higher cellular uptake than both positive controls and much lower toxicity than Jet-PEI (18, 19). Although we currently do not fully understand the small discrepancy between the uptake and gene expression data, the imaging experiments (see below) indicate that a much higher fraction of the internalized polyplexes are located in the cytoplasm, whereas a lower fraction is found in the nucleus. Thus, nuclear entry may be the main barrier for gene expression. Indeed, for many forms of nucleic acid therapeutics, such as siRNA, high cytoplasmic delivery is the ultimate goal. These delivery vehicles could be extremely useful in these therapeutic methodologies.
Fig. 3.
The effect of lanthanide chelate and N/P ratio on polyplex uptake into Hela cells and cell viability. (A) Cellular uptake of polyplexes formulated using Cy5-pDNA. The percentage of cells (line) containing Cy5 pDNA and mean fluorescence intensity (bar) of Cy5 in a population of at least 30,000 cells. (B) Cell viability after exposure to polyplexes using unlabeled pDNA. The N/P ratio of the polyplex used is indicated after the polymer name on the x axis.
Cellular Imaging of the Polymer Beacons.
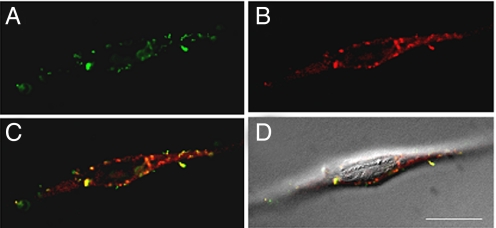
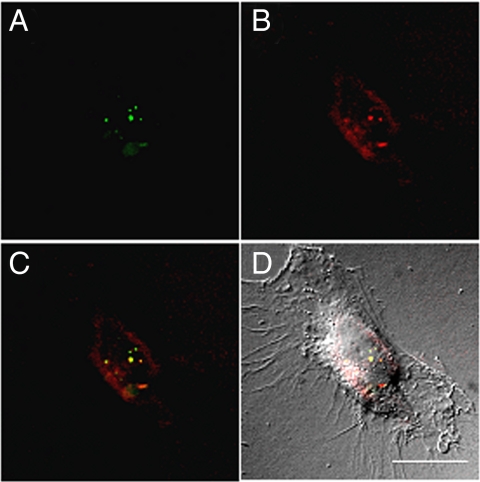
Further investigation of the polymer beacons chelated with luminescent Eu3+ was examined via fluorescence microscopy. HeLa cells were transfected with polyplexes formulated with FITC-labeled pDNA and either Eu3a or Eu3b at an N/P ratio of 40. Figs. 4 and 5 each represent one vertical optical slice obtained by deconvolution of a Z-stack series. The intracellular location of both FITC-pDNA (Figs. 4A and 5A) and the polymers (Figs. 4B and 5B) can be clearly observed. By overlaying the pDNA and polymer images (Figs. 4C and 5C), colocalization can be qualitatively observed by yellow punctate staining. Most of the staining resulting from FITC-pDNA or Eu3+-polymer luminescence is localized to the cytoplasm and a lower fraction appears in the nucleus. As previously stated, this observation could be a contributing factor to the lower luciferase gene expression, but could prove useful in the development of cytoplasmic-targeted nucleic acid therapies. Also, it is clear that a small amount of uncomplexed pDNA is present within the cells whereas a larger fraction of free polymer is observed in the cellular cytoplasm. We have observed a similar result by formulating polyplexes at a lower N/P ratio of 20, where a high amount of free polymer was also observed in the cells (Fig. S5). These phenomena could be attributed to the excess used to formulate polyplexes at high N/P ratios, but may indicate premature pDNA release during cellular uptake and early intracellular trafficking (both are concerns in this area, but not well understood). Also, for Eu3a, the intracellular staining pattern is more punctate, whereas diffuse staining is more apparent for Eu3b. These data could indicate that Eu3a is more localized to endocytic vesicles, whereas Eu3b (with more secondary amines per repeat unit) could promote higher endocytic release. These data reveal that our “proof of concept” polymer beacons (and future derivatives) will be useful in understanding intracellular polymer fate, as well as elucidating the mechanisms of delivery.
Fig. 4.
Deconvoluted micrographs of a HeLa cell transfected with FITC-pDNA/Eu3a polyplexes. (A) FITC-pDNA fluorescence (green). (B) Eu3+ luminescence within the Eu3a beacons (red). (C) An overlay of the FITC-pDNA and Eu3a images. Yellow pixels can be qualitatively used to visualize regions of colocalization. (D) An overlay of image in C and a DIC image to show contrast of the cell. (Scale bar, 20 μm.)
Fig. 5.
Deconvoluted micrographs of a HeLa cell transfected with FITC-pDNA/Eu3b polyplexes. (A) FITC-pDNA fluorescence (green). (B) Eu3+ luminescence within the Eu3b beacons (red). (C) An overlay of the FITC-pDNA and Eu3b images. Yellow pixels can be qualitatively used to visualize regions of colocalization. (D) An overlay of image in C and A DIC image to show contrast of the cell. (Scale bar, 20 μm.)
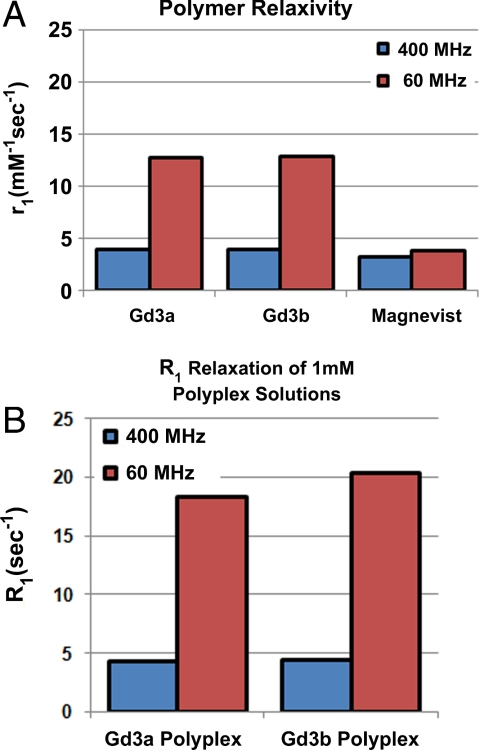
Whereas the Eu3+ polymer analogs can be visualized on the nm/μm scale within cells, the Gd3+ analogs provide the ability to understand nucleic acid delivery within tissues on the sub-mm scale. First, their relaxivity (r1) and relaxation rate constants (R1) were determined. This characteristic of the polymers (r1, determined by plotting 1/T1 versus concentration) or polyplexes (R1, equivalent to 1/T1) is used to understand the contrast enhancement of the delivery beacons. We performed the relaxivity experiments on both the free Gd3a and Gd3b polymers (Fig. 6A) in solution and commercial contrast agent, Gd3+-DTPA (Magnevist) at two different magnetic field strengths (60 MHz/1.41 T and 400 MHz/9.4 T at 37 °C). Also, we tested the ability of Gd3a and Gd3b polyplexes to enhance longitudinal water proton relaxation (Fig. 6B). The relaxivities of both polymers in solution were higher than the clinically-used chelate Gd3+-DTPA, and a dramatic enhancement was observed at 1.41 T, likely due to the slower rotational correlation time of the delivery beacons. When comparing the longitudinal relaxation rates (R1) of free polymer to polyplexes at 1.41 T (Gd3+ concentration is identical) a dramatic relaxivity increase was observed. This difference in relaxivity between free polymer and polyplex could provide a unique method to track nucleic acid release within tissues.
Fig. 6.
The relaxivities and relaxation rate constants of water solutions containing dissolved polymer and polyplexes, respectively. (A) Relaxivity (r1) of aqueous solutions containing free polymer (Gd3a and Gd3b only) and Magnevist at 400 MHz (9.4 T) and 60 MHz (1.41 T). Relaxivity was calculated from four concentrations (5, 2.5, 1.25, and 0 mM) of each agent in water. (B) The relaxation rate constants (R1) of polyplexes formed with Gd3a and Gd3b at a concentration of 1 mM at 400 MHz (9.4 T) and 60 MHz (1.41 T).
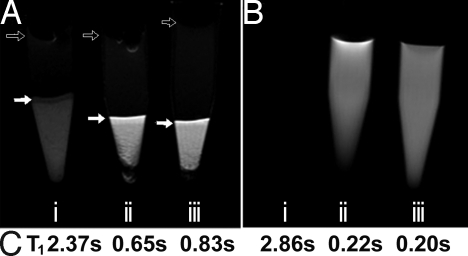
To further investigate the MRI contrast capabilities of the Gd3+-chelate materials, HeLa cells were transfected with the polyplexes formulated with Gd3a and Gd3b. As shown in Fig. 7A, tubes containing live transfected or untransfected cells were subjected to a T1-weighted pulse sequence, imaged for tissue contrast, and then analyzed for water 1H T1 values. Untransfected cells that do not contain Gd3a or Gd3b demonstrate minimal image brightness/contrast (Fig. 7Ai), and this control cell pellet has a higher T1 value (2.4 s; Fig. 7C) than cells transfected with Gd3a and Gd3b polyplexes. Cells transfected with Gd3a and Gd3b (Fig. 7 Aii and Aiii) show a clear enhancement in the cellular image brightness and substantially lower T1. Fig. 7B shows the T1-weighted images, using the same pulse conditions as in Fig. 7A, with 1 mM (based on the Gd3+ concentration) solutions (Fig. 7 Bii and Biii) versus a PBS control. Because the beacon solutions are so bright relative to the saline control, the control becomes unobservable. This is confirmed by their T1 analysis at 9.4 T, which agrees with the MR imaging experiment. These promising data clearly show the potential of our imaging beacons to trace the spatial and temporal delivery of nucleic acids within bulk cells and/or tissues.
Fig. 7.
MRI data for cells transfected with Gd3a/pDNA and Gd3b/pDNA polyplexes. The cells were thoroughly washed and gravity pelleted into Eppendorf tubes. (A) Transfected cell pellets and controls: (Ai) Pellet of untreated HeLa cells, (Aii) cell pellets transfected with Gd3a polyplexes (N/P = 40), and (Aiii) cells transfected with Gd3b polyplexes (N/P = 40). Solid arrows indicate the buffer-cell interface in each sample. The perturbations at the top of the buffer (open arrows) are due to bubbles at the buffer-air interface. The darker spots in the cell pellet are due to cell density gradients. (B) MR images of (Bi) PBS buffer, (Bii) Gd3a ([Gd3+] = 1 mM), and (Biii) Gd3b ([Gd3+] = 1 mM) using the same pulse parameters as above. PBS is completely dark because of the high T1 contrast relative to Gd3a/b solutions. (C) T1 quantification of cell and solution samples imaged. Cell samples were allowed to settle into NMR tubes for 24 h and then analyzed with an inversion recovery pulse sequence.
This proof-of-concept study has revealed that our nucleic acid delivery beacons show great promise in the development of innovative techniques to understand the delivery, trafficking mechanisms, and fate of nonviral delivery vehicles on differing biological scales. The Ln metals offer many unique properties, which can be exploited to enhance these imaging techniques. The Eu3+-chelated polymers can be visualized by their luminescence within cultured cells on the nm/μm scale for tracking the intracellular delivery of polyplexes. For larger-scale MR imaging, the paramagnetic nature of the Gd3+-chelated polymers offers a safe and noninvasive probe for following nucleic acid delivery within tissues on the sub-mm scale. These scaffolds have potential for monitoring in vivo delivery in a spatial and temporal manner and can be used intact (without removal of the imaging probe) due to their nontoxic and effective delivery profile. Indeed, these creative and powerful materials can be broadly applied and exploited by researchers for the discovery of unique nucleic acid drug/vehicle conjugates and to understand and treat many devastating diseases.
Materials and Methods
Materials.
Diethylenetriaminepentacetic acid (DTPA) and ethyl trifluoroacetate were obtained from Alfa Aesar Chemical. All other reagents and solvents used in the synthesis were obtained from Aldrich and were used without further purification. Monomers 1a and 1b were synthesized following a previously published procedure developed in our lab (5). DTPA-bisanhydride (BA) was prepared using a standard procedure. Data for the full characterization of all previously uncharacterized compounds can be found in SI Appendix.
Cell Culture Materials.
Media and supplements were purchased from Gibco/Invitrogen. HeLa cells were purchased from ATCC and cultured according to specified conditions. Plasmid DNA, pCMVb and gWiz-Luc, were purchased from PlasmidFactory and Aldevron, respectively. Nuclease free water, Opti-MEM, and DMEM (supplemented with 10% FBS/100 units/mL penicillin/100 μg/mL streptomycin/0.25 μg/mL amphotericin) and PBS were all purchased from Invitrogen. Luciferase substrate reagent and cell culture lysis buffer used in the transfection efficiency assays were purchased from Promega. A DC protein assay kit used in the viability assay was purchased from Bio-Rad. FITC-labeled pDNA and the Cy5 nucleic acid labeling kit was purchased from Mirus.
Synthesis of Poly([N2,N3,N4-tris(tert-butoxycarbonyl)tetraethylenetriamine] amido diethylenetriaminetriaacetic acid), 2a.
Polymer 2a was prepared by dissolving monomer 1a (3.63 g, 7.41 mmol) in 25 mL of DMSO at room temperature. DTPA-BA (2.64 g, 7.41 mmol) was dissolved in 5 mL of DMSO and added directly to the solution of 1a under constant stirring. The polymerization was carried out for 18 h, after which the solution was pipetted into a 6,000- to 8,000-Da MWCO dialysis bag (Spectrum laboratories) and dialyzed extensively against methanol for 18 h, after which the solution was removed from the bag and evaporated in vacuo to yield 2a as an orange viscous oil (yield = 3.88 g, 47%).
Synthesis of Poly([N2,N3,N4,N5-tetrakis(tert-butoxycarbonyl)pentaethylenetetra mine] amidodiethylenetriaminetriaacetic acid), 2b.
Polymer 2b was prepared according to an identical procedure used to prepare 2a except monomer 1b (4.77 g, 7.54 mmol) was polymerized with DTPA-BA (2.69 g, 7.54 mmol) yielding a deep amber oil (yield = 4.17 g, 56%).
General Synthesis of Poly[(tetraethylenetriamine)amido diethylene tri amine triaacetic acid], 3a and Poly[(pentaethylenetetramine) amido diethylene tri amine triaacetic acid], 3b.
An aliquot of 5 mmol of each protected polymer (2a or 2b) was dissolved in 20 mL of CH2Cl2 and cooled to 0 °C. Next, 20 mL of trifluoroacetic acid was added to each solution, and the reaction was warmed to room temperature. The reactions were allowed to proceed for 3 h, after which the solvents were removed in vacuo and each mixture was redissolved in water. Both water solutions were brought to a pH = 6 using a 1 M sodium bicarbonate. The solutions were each deposited in a separate 6,000- to 8,000-Da MWCO dialysis bag (Spectrum laboratories) and dialyzed against ultrapure water for 24 h. The dialyzed solutions were lyophilized to yield the fluffy, unchelated polymers 3a and 3b (yields = 1.88 g, 76%; 1.97 g, 80%, respectively).
General Synthesis of Poly [(tetra ethyl ene triamine) amido (Ln3+) diethyl ene triaminetriacetate], Eu3a or Gd3a, and Poly [(penta ethyl ene tetramine) amido (Ln3+)diethylenetriaminetriacetate], Eu3b or Gd3b.
A 2-mmol aliquot of each unchelated polymer, 3a and 3b, was dissolved in 20 mL of ultrapure water at room temp. Next, 2 mmol of LnCl3 (either EuCl3 or GdCl3) was dissolved in 5 mL of water and dripped into the respective polymer solution in three separate aliquots. The pH was adjusted to pH = 6 after each aliquot addition. The solution was allowed to stir for 2 h and then was dialyzed in a 6,000- to 8,000-Da MWCO bag against ultrapure water for 24 h. Dialyzed solutions were lyophilized to yield fluffy off-white polymers Gd3a, Gd3b, Eu3a, and Eu3b (yields = Gd3a = 1.20 g, 86%; Gd3b = 1.19 g, 80%; Eu3a = 1.13 g, 81%; Eu3b = 1.17 g, 79%).
Polymer and Polyplex Characterization.
The molecular mass and polydispersity for polymers 3a, 3b, Eu3a, Eu3b, Gd3a, and Gd3b were measured with a Viscotek GPCmax instrument equipped with a GMPWXL (aqueous phase) column coupled to a triple detection system (static light scattering, viscometry, and refractive index) as shown in Table S3. A solution of 0.5 M sodium acetate containing 20% acetonitrile was used as the mobile phase. Agarose gel electrophoreses shift assays and TEM studies (Philips EM 420 Scanning TEM) were performed according to previously published methods (4, 5).
Nuclease Degradation Assay.
The ability of the polymers to protect pDNA from nuclease degradation was analyzed according to modified form of a previously published procedure (20). Each polymer (5 μL) was combined with pCMVβ (1 μg in 5 μL of water) to form polyplexes at N/P = 40. After 30 min, FBS (5 μL) was added to each polyplex solution, and the samples were incubated for 0, 1, 2, 4, and 6 h, at 37 °C. The samples were then treated with 10% SDS (2 μL), and then stored at 4 °C until all samples were finished incubating. All trials were then incubated at 60 °C for 18 h to release the polymer from the pDNA. Loading buffer (2 μL, Blue Juice; Invitrogen) was added to an aliquot of each polyplex solution and loaded onto an agarose gel (0.6%, 3 μL ethidium bromide) in 20 μL aliquots, and electrophoresed to analyze for plasmid degradation. Plasmid DNA only and FBS were used as negative controls.
DLS and Zeta Potential Measurements.
Polyplex sizes and zeta potential were measured using a Zetasizer (Nano ZS) DLS instrument with a 633 nm laser (Malvern Instruments). Polyplexes were formed at N/P ratios at 10, 20, 30, 40, and 50 by combining equal volumes of a polymer solution in ultrapure water with a 0.02 mg mL−1 pDNA solution and allowing the polyplexes to form for 1 h.
Cellular Delivery of pDNA and Cell Viability Analysis.
Twenty-four hours before transfection, HeLa cells were seeded in six well plates at a density of 250,000 cells per well and incubated in supplemented DMEM (10% FBS) at 37 °C and 5% CO2. Polyplexes were formulated by adding 250 μL solutions of each polymeror control dissolved in water (concentration calculated based on N/P ratios of 40 and 60) to 5 μg of Cy5-labeled pDNA (250 μL solution). Just before transfection, 1 mL Opti-MEM was added to each polyplex solution. Cells were aspirated of old media, washed with 1 mL PBS, and the appropriate polyplex solution was added to each well. Cells were then incubated at 37 °C under a 5% CO2 atmosphere for 4 h. The cell suspensions were then prepared for analysis as previously described (5). HeLa cell suspensions were analyzed on a FACS Canto II flow cytometer equipped with a 633 nm helium-neon laser. Mean Cy5 fluorescence intensity was measured using the appropriate forward and side scatter gates. A control of untransfected cells was used to create a gate such that <1% of cell-associated autofluorescence is detected in the Cy5 channel. This gating strategy was used for subsequent samples of transfected cells to determine the percentage of cells transfected. For gene expression analysis, polyplexes were formulated in an identical manner as above except with GWiz-luc pDNA and allowed to transfect cells for 48 h before assaying for luciferase activity according to previously published methods. Cell viability was characterized by measuring the amount of protein in cell lysates using a Bio-Rad DC protein assay kit in triplicate. Viability is reported as the fraction of protein in each sample normalized to a control of untransfected cells.
Fluorescence Microscopy.
Twenty-four hours before transfection, cells were seeded in six well plates containing 25 mm no. 1 glass coverslips at a density of 50,000 cells per well. Just before transfection, polyplexes were formed as noted above for the cellular uptake assays except 200 μL of polymer solution was added to 200 μL of FITC-labeled pDNA ([pDNA] = 0.02 mg/mL; N/P = 40). Cells were removed from the incubator, aspirated of old media, washed with 2 mL PBS, and 2 mL Opti-MEM was added to each well. To transfect cells, 1 mL Opti-MEM from each well was added to the appropriate polyplex solution, pipetted up and down to mix, and returned to the well to deliver a total of 4 μg DNA per well. Four hours after transfection, 2.4 mL of supplemented DMEM was added to each well. Twenty-four hours after transfection, cells were aspirated of old media, washed three times with 1 mL PBS, and fixed for 2 h using 2% paraformaldehyde in PBS (pH 7.4) at 4 °C. Afterward, coverslips were removed from each well and carefully washed four times with 0.5 mL PBS. Coverslips were then mounted in Prolong antifade mounting media (Molecular Probes) and allowed to dry at room temperature overnight. Cells were observed using a Zeiss Axioplan Imaging 2 infinity-corrected, upright scope, a 63× oil objective (N.A. 1.4), and standard filter sets for FITC and Rhodamine. To visualize Europium luminescence, a custom filter set (excitation max 405 nm ± 40 nm, dichroic 440 nm LP, emission max 610 ± 75 nm) was built from filters purchased from Chroma. Images of each cell were collected as z-stacks with a z-spacing of 0.27 μm (Eu3a and Eu3b) using an Orca-ER CCD camera (Hamamatsu). The resulting images were processed using AutoQuant Autodeblur (Media Cybernetics). Data correction for each z-stack was applied for bias and flatfield frame and optical density. Then, stacks were processed using 3D blind deconvolution over 50 iterations. The final fluorescence images were minimally processed for background subtraction, brightness, and contrast. To increase fine detail in some DIC images, filters for pseudoflatfield and kalman stack were applied. After deconvolution, all image processing was completed using ImageJ open source software (National Institutes of Health) (21).
Inversion Recovery Experiments of Polyplexes in Solution.
T1 measurements were carried out on Bruker AMX-400MHz (9.4 T) and Anasazi FT-NMR 60MHz (1.41 T) NMR spectrometers for all samples. Polyplexes were formed at an N/P = 40 such that the concentration of Gd3+ was 1 mM. After an incubation time of 30 min the longitudinal relaxation rate constant (T1) for each solution was measured using an inversion recovery pulse sequence (180°–dt–90°–acquire) at 298 K. Arrayed data [n(dt) = 10] were processed using Acorn NUTS software and fit to a three-parameter model. The inverse of the T1 data were used to determine the longitudinal relaxation rate constants (R1) imparted by the polyplexes. Solution concentration data for beacons Gd3a and Gd3b, as well as the Magnevist control were used to calculate relaxivity (r1) by generating a curve fit to (1/T1) = [Gd3+](r1) + b, where b = (1/T1) for the working solvent (water).
MRI of Transfected HeLa Cells.
Twenty-four hours before transfection, HeLa cells were seeded in 10 flasks (with a surface area of 75 cm2) at a density of 4.0 × 106 cells per flask. Polyplexes were prepared 1 h before transfection using βCMV pDNA and the respective polymer at an N/P ratio of 40. Cells were transfected in 15 mL Opti-MEM using 80 μg pDNA/flask. Nontransfected cells were subjected to the same media changes as the transfected cells. Four hours after transfection, 15 mL of DMEM containing 10% FBS was added to each well. Twenty-four hours after transfection, cells were aspirated of media, washed extensively with PBS and collected via trypsination. Cells were pelleted and resuspended in PBS twice, then pelleted again in 0.5 mL Eppendorf tubes for analysis. All MR images were acquired with a Bruker Avance III NMR spectrometer equipped with a Micro2.5 imaging probe featuring a 3 cm rf coil. The images were acquired using a RARE-inversion recovery T1-weighted pulse sequence. Acquisition parameters are as follows: TR (repetition time) = 3,500 msec; TI (inversion time) = 1,200 msec; TE (echo time) = 8.5 msec; FOV = 3 × 3 cm; and resolution = 256 × 256. T1 measurements of these same cell pellets and controls were carried out using the same parameters on a Bruker AMX-400 MHz spectrometer. For the T1 experiments, the HeLa cell pellets used for the MRI experiment were suspended in PBS buffer to make turbid solutions that were deposited into NMR tubes and allowed to rest for 24 h. On settling, each tube contained ≈4 cm of sedimented HeLa cell pellets. NMR tubes were placed so the entirety of the HeLa pellet was within the NMR coil. The longitudinal relaxation rate constant (T1) for each pellet and the control solutions were measured using an inversion recovery pulse sequence (180° − dt − 90° − acquire) at 298 K. Arrayed data [n(dt) = 10] was processed using Acorn NUTS software and fit to a three-parameter model.
Supplementary Material
Acknowledgments.
We thank Prof. Chris Gulgas (Longwood University, Farmville, VA) for his help in performing the relaxivity experiments at 60 MHz. This work was supported by the Camille Dreyfus Teacher-Scholar Award program. T.M.R. is a fellow of the Alfred P. Sloan Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904860106/DCSupplemental.
References
- 1.Leong PL, et al. Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. Proc Natl Acad Sci USA. 2003;100:4138–4143. doi: 10.1073/pnas.0534764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heidel JD, et al. Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA. Proc Natl Acad Sci USA. 2007;104:5715–5721. doi: 10.1073/pnas.0701458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schaffert D, Wagner E. Gene therapy progress and prospects: Synthetic polymer-based systems. Gene Ther. 2008;15:1131–1138. doi: 10.1038/gt.2008.105. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y, Reineke TM. Hydroxyl stereochemistry and amine number within poly(glycoamidoamine)s affect intracellular DNA delivery. J Am Chem Soc. 2005;127:3004–3015. doi: 10.1021/ja0436446. [DOI] [PubMed] [Google Scholar]
- 5.Srinivasachari S, Liu Y, Zhang G, Prevette L, Reineke TM. Trehalose click polymers inhibit nanoparticle aggregation and promote pDNA delivery in serum. J Am Chem Soc. 2006;128:8176–8184. doi: 10.1021/ja0585580. [DOI] [PubMed] [Google Scholar]
- 6.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 7.Karmali PP, Chaudhuri A. Cationic liposomes as non-viral carriers of gene medicines: Resolved issues, open questions, and future promises. Med Res Rev. 2007;27:696–722. doi: 10.1002/med.20090. [DOI] [PubMed] [Google Scholar]
- 8.Sonawane ND, Szoka FC, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J Biol Chem. 2003;278:44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 9.Pan D, et al. Ligand-directed nanobialys as theranostic agents for drug delivery and manganese-based magnetic resonance imaging of vascular targets. J Am Chem Soc. 2008;130:9186–9187. doi: 10.1021/ja801482d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malik N, et al. Dendrimers: Relationship between the structure and biocompatability in vitro, and preliminary studies on the biodistribution of 125I-labelled polyamidoamine dendrimers in vivo. J Control Release. 2000;65:133–148. doi: 10.1016/s0168-3659(99)00246-1. [DOI] [PubMed] [Google Scholar]
- 11.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Quantum dots versus organic dyes as fluorescent labels. Nat Methods. 2008;5:763–775. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- 12.Qi L, Gao X. Quantum dot-amphipol nanocomplex for intracellular delivery and real-time imaging of siRNA. ACS Nano. 2008;2:1403–1410. doi: 10.1021/nn800280r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bünzli J-CG. Benefiting from the unique properties of lanthanide ions. Acc Chem Res. 2006;39:53–61. doi: 10.1021/ar0400894. [DOI] [PubMed] [Google Scholar]
- 14.Marriott G, Heidecker M, Diamandis EP, Yan-Marriott Y. Time-resolved delayed luminescence image microscopy using an europium ion chelate complex. Biophys J. 1994;67:957–965. doi: 10.1016/S0006-3495(94)80597-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caravan P, Ellison JJ, McMurray TJ, Lauffer RB. Gadolinium(III) chelates as MRI contrast agents: Structure, dynamics, and applications. Chem Rev. 1999;99:2293–2352. doi: 10.1021/cr980440x. [DOI] [PubMed] [Google Scholar]
- 16.Bryant HL, et al. Synthesis and relaxometry of high-generation (G = 5, 7, 9, and 10) PAMAM dendrimer-DOTA-gadolinium chelates. J Magn Reson Imaging. 1999;9:348–352. doi: 10.1002/(sici)1522-2586(199902)9:2<348::aid-jmri30>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 17.Chen HH, et al. MR imaging of biodegradable polymeric microparticles: A potential method of monitoring local drug delivery. Magn Reson Med. 2005;53:614–620. doi: 10.1002/mrm.20395. [DOI] [PubMed] [Google Scholar]
- 18.Chollet P, Favrot MC, Hurbin A, Coll J-L. Side-effects of a systemic injection of linear polyethylenimine-DNA complexes. J Gene Med. 2002;4:84–91. doi: 10.1002/jgm.237. [DOI] [PubMed] [Google Scholar]
- 19.Wightman L, et al. Different behavior of branched and linear polyethylenimine for gene delivery in vitro and in vivo. J Gene Med. 2001;3:362–372. doi: 10.1002/jgm.187. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Reineke TM. Poly(glycoamidoamine)s for gene delivery: Stability of polyplexes and efficacy with cardiomyoblast cells. Bioconjugate Chem. 2006;17:101–108. doi: 10.1021/bc050275+. [DOI] [PubMed] [Google Scholar]
- 21.Rasband WS. IMAGE J. Bethesda: National Institutes of Health; 1997–2007. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.