Abstract
The unique biology of a neoplasm is reflected by its distinct molecular profile compared with normal tissue. To understand tumor development better, we have undertaken a quantitative proteomic search for abnormally expressed proteins in colonic tumors from ApcMin/+ (Min) mice. By raising pairs of Min and wild-type mice on diets derived from natural-abundance or 15N-labeled algae, we used metabolic labeling to compare protein levels in colonic tumor versus normal tissue. Because metabolic labeling allows internal control throughout sample preparation and analysis, technical error is minimized as compared with in vitro labeling. Several proteins displayed altered expression, and a subset was validated via stable isotopic dilution using synthetic peptide standards. We also compared gene and protein expression among tumor and nontumor tissue, revealing limited correlation. This divergence was especially pronounced for species showing biological change, highlighting the complementary perspectives provided by transcriptomics and proteomics. Our work demonstrates the power of metabolic labeling combined with stable isotopic dilution as an integrated strategy for the identification and validation of differentially expressed proteins using rodent models of human disease.
Keywords: colon cancer, min mouse, proteomics, differential mass spectrometry, transcriptomics
Colon cancer is the third most prevalent cancer in the United States, causing nearly 100,000 diagnoses and 50,000 deaths in 2009 (1). Although early stages are treatable, existing screening methods lack specificity or have poor patient compliance. Thus, many cases are undiagnosed until late stages when the prognosis is poor (2). Given its prevalence, research is needed to uncover the molecular basis of colon cancer and reveal biochemical markers of disease.
To address this challenge, animal models have been developed, including several mouse strains (3). The ApcMin/+ (Min) mouse carries a premature stop codon within the adenomatous polyposis coli (Apc) gene, preventing normal expression (4, 5). Min mice develop tumors in the colon and small intestine and are moribund within 4 months of birth. Loss of APC function in humans causes familial adenomatous polyposis, a rare genetic form of colon cancer (6). More generally, the APC gene is the most frequently mutated gene in a genome-wide survey of spontaneous human colonic tumors (7). Thus the Min mouse is relevant for probing the biological basis of colon cancer. We have begun using quantitative proteomics to identify abnormally expressed proteins in colonic tumors from Min mice. Study of tumors should reveal proteins whose abnormal expression is a direct effect of tumorigenesis rather than a secondary sign of ill health.
In a typical quantitative proteomics experiment, sample pairs are compared using isotopic labeling: 1 of the pair is labeled with multiple light isotopes, and the other is labeled with a matching tag containing heavy isotopes. Samples then are combined and analyzed, and the mass spectrometer distinguishes peptides from each sample based on mass. Usually, isotopic tags are introduced in vitro via chemical (8, 9) or enzymatic (10) means. However, an isotopic label may be introduced instead by growth on isotopically enriched food or media. This process, metabolic labeling, often has been used with bacteria, yeast (11), cultured cells (12), and plants (13, 14). Less frequently, it also has been applied in mammals (15–17). Although it can involve increased initial expense and effort, metabolic labeling is desirable because it allows differentially labeled samples to be combined at the outset of an experiment. The resulting internal control during sample preparation and analysis minimizes technical error as compared with in vitro labeling techniques.
Here we describe the use of metabolic labeling to compare protein expression in colonic tumors of Min mice versus normal epithelium from their wild-type counterparts. We subsequently verify abnormal expression of several proteins within colonic tumors via stable isotopic dilution (18), demonstrating the power of untargeted proteomic profiling combined with targeted stable isotopic dilution for discovery and validation of disease-related proteins. We also compare proteomic and transcriptomic profiles and explore the complementary insights these strategies provide.
Results
15N Metabolic Labeling of Mice.
The key challenge in metabolic labeling is introducing an isotopic label via the medium or diet of the organism. Previously, Wu and MacCoss and colleagues demonstrated metabolic labeling of rats using a diet derived from 15N-labeled Spirulina, supplemented with fats, vitamins, and a nitrogen-free carbohydrate base (15). Spirulina is a protein-rich alga that can be purchased in >98% 15N-labeled form. We adapted their protocol for use with mice.
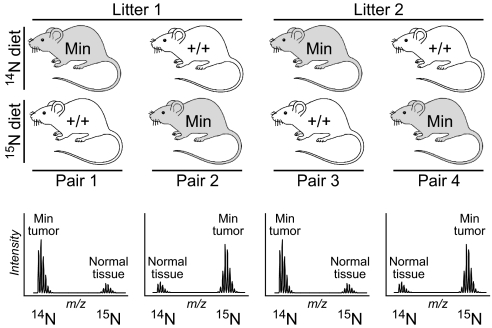
We raised 8 female mice, 4 Min and 4 wild type, from weaning on the Spirulina-based diet. Each mouse was paired with a sister of the opposite genotype. In each pair, 1 mouse was fed the 15N-labeled diet, and the other was fed the natural-abundance form. The isotopic label was switched between Min and wild-type mice from 1 pair to the next. This reciprocal-labeling scheme distinguishes relevant differences in protein expression from experimental artifacts (Fig. 1).
Fig. 1.
Reciprocal-labeling scheme. (Top) Mice (4 Min and 4 wild type) were raised on Spirulina-based food. Each mouse was paired with a sibling bearing the opposite genotype. In each pair, 1 mouse was raised on the 14N diet, and the other was raised on the 15N diet. (Bottom) The isotopic label was exchanged across genotypes from 1 mouse pair to the next, allowing genotype-dependent differences to be distinguished from various artifacts based on the pattern of ratio inversions across the mouse pairs.
Mice were killed at 68 days of age, and organs were frozen in liquid nitrogen. Labeled and unlabeled tissues from each pair were combined, and protein samples were analyzed via liquid chromatography (LC)-MS following extraction and tryptic digestion. Levels of 15N enrichment were determined for peptides from each tissue by matching observed and theoretical isotopic distributions. Average 15N enrichments are summarized in Table 1.
Table 1.
15N enrichment across multiple tissues
| Organ | % 15N Incorporation, Mean (SD)* |
|---|---|
| Brain | 82.8 (1.5) |
| Skeletal muscle | 87.8 (1.7) |
| Heart | 90.3 (1.3) |
| Colon | 91.3 (1.7) |
| Lung | 92.0 (1.6) |
| Kidney | 92.3 (1.7) |
| Mammary gland | 92.5 (1.3) |
| Liver | 92.8 (1.3) |
| Small intestine | 93.0 (0.8) |
| Intestinal tumors† | 93.3 (1.3) |
*Mean 15N incorporation levels are listed for multiple tissues. Standard deviations reflect biological variation across 4 15N-labeled mice.
†Tumors from the colon and small intestine.
We observed considerable variation in 15N enrichment across tissues. As in rats, tissues with high protein turnover (e.g., intestinal tract) showed high levels of enrichment; 15N levels were lower in tissues with slower turnover tissues, such as muscle and brain. Differences in enrichment across proteins within each sample were small (≈1.0%); comparable variation was seen among mouse pairs. All tissues were sufficiently enriched for quantitation, with corrections for incomplete incorporation.
Proteomic Comparison of Colonic Tumors and Normal Epithelium.
Colonic tumors from each Min mouse were combined with normal epithelium from its wild-type sister bearing the opposite isotopic label. After protein extraction and digestion, samples were analyzed via LC-MALDI-TOF/TOF MS. Peptides were identified via Mascot (19) and filtered to a 1% false-discovery rate (FDR) via decoy database searching (20). The protein-level FDR was below 1% for species identified and quantified in multiple replicates. Peptide and protein identities are summarized in Table S1, Table S2, and Table S3.
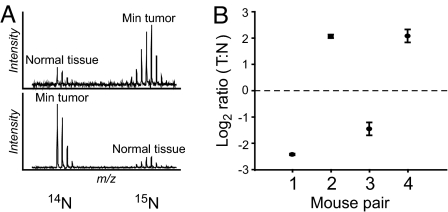
Abundance ratios for 15N-labeled and natural-abundance forms of all peptides were determined via an algorithm designed for this purpose (SI Methods). Each protein's ratio was estimated by averaging the ratios of its constituent peptides within each replicate. Following normalization and log transformation, ratios for each protein were averaged across mouse pairs (Fig. S1). Proteins whose ratios differed from the normalized median by more than 2-fold and whose relative standard errors across replicates were less than 50% were considered differentially expressed (Table 2) (see ref. 21). We identified 10 differentially expressed proteins that met these criteria; other differentially expressed proteins with greater biological variability were seen also (Table S4). Fig. 2 summarizes the quantitation of 1 protein, coronin, across all replicates, demonstrating its upregulation in colonic tumors. Reciprocal labeling confirms that its differential expression reflects tissue type rather than labeling or analytic effects.
Table 2.
Differentially expressed proteins
| Protein Name | Proteomics Abundance Ratio | Microarray Expression Ratio |
|---|---|---|
| ATPase, H+/K+ transporting, non-gastric, α polypeptide | −1.72 | −3.79 |
| SCFP-25 | −1.56 | −0.49 |
| HMG-CoA synthase 2 | −1.13 | 1.27 |
| Actinin α 2 | −1.03 | −0.35 |
| Carboxylesterase 2 | −1.02 | −3.88 |
| Coronin, actin-binding protein 1A | 2.01 | 0.60 |
| Transglutaminase 3, E polypeptide | −1.74 | −3.14 |
| Lymphocyte cytosolic protein 1 | 1.24 | 1.61 |
| Rho, GDP dissociation inhibitor beta | 1.70 | 0.40 |
| RNA binding motif protein 3 | −1.41 | −0.44 |
Each protein was quantified in multiple mouse pairs, with a mean ratio exceeding a 2-fold change and a relative standard error of less than 50% (see ref. 21). Mean microarray expression ratios also are listed for each protein. Ratios are in log2 form and are expressed as the ratio of tumor to normal tissue.
Fig. 2.
Consistent differential expression of coronin. The protein coronin was shown via reciprocal isotopic labeling to be elevated in tumor tissue across all mouse pairs, regardless of isotopic label (A). The mean ratio (±SE) for each mouse pair is plotted in B, showing low variability within and across biological replicates.
Validation of Selected Protein Observations via Stable Isotope Dilution.
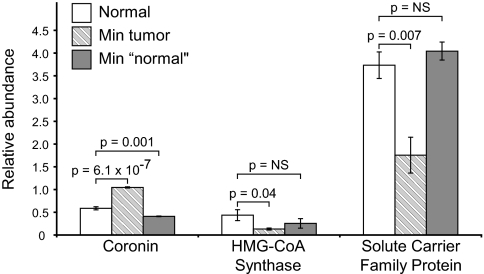
Although we identified several proteins with abnormal abundances, our survey includes a small biological sample. Furthermore, because of biological variability among the mouse pairs, we have chosen liberal criteria to identify differentially expressed proteins. Thus these preliminary observations must be validated in larger populations. Although some observations are confirmed in the literature, we sought to confirm others experimentally. We selected 3 differentially expressed proteins—coronin, 3-hydroxy-3-methylglutaryl (HMG)-CoA synthase 2, and solute carrier family protein 25 (SCFP-25)—for validation via stable isotope dilution (SID) as proposed by Gerber et al. (18).
Synthetic peptides were made to match unique tryptic peptides from each protein. Heavy isotopes were added to each for use as internal standards for quantitation. Labeled and unlabeled peptide forms were assayed via multiple-reaction monitoring (Table S5). These peptides were spiked into tryptic digests from tumors and normal epithelia and were used to quantify each protein (Fig. 3).
Fig. 3.
Confirmation of protein expression via SID. We quantified 3 differentially expressed proteins in tumor and nontumor tissue from Min mice and normal tissue from wild-type controls via SID using synthetic, isotopically labeled peptide standards. Differential expression in tumor relative to wild-type tissue was confirmed for each protein and was significant (P < 0.05) via unpaired 2-sample t-tests, assuming unequal variance. When nontumor tissue from Min mice was compared with normal colonic tissue, no significant difference was observed for HMG CoA synthase or SCFP-25, whereas a small but significant decrease in coronin was observed (P = 0.001). All cases of differential expression remain statistically significant at a 5% FDR. NS, not significant. Error bars represent ±1 SE.
Overall, SID results confirm differential expression of each protein, with additional replicates and greater precision from multiple-reaction monitoring. These changes are statistically significant (P < 0.05) at an FDR below 5% (22). Compared with our initial survey, the observed abundance ratios were reduced in the SID experiments. This difference may arise from biological variability or may reflect slightly different ages of mice used for validation.
These altered protein abundances could be specific to tumors, or they could be specific to the Min mutation and independent of the tumor phenotype. To distinguish these possibilities, we also used SID to quantify each protein in nontumor tissue from the same Min mice. We observed no significant difference in the abundance of HMG CoA synthase or SCFP-25 relative to normal tissue from wild-type controls. In contrast, although we observed increased coronin expression in Min tumor tissue, we detected a slight but significant decrease in nontumor tissue from Min colon relative to normal tissue (P = 0.001). Thus the altered expression of these proteins is specific to tumor tissue.
Colonic Tumor Proteome Versus Transcriptome.
To complement proteomic profiling, we compared gene expression in tumor versus normal epithelial tissue. Although complete analysis of these data is beyond the scope of this report, here we compare protein and transcript expression for the 613 species identified in our proteomic survey (Table S6).
RNA samples were extracted from 4 colonic tumor samples, were fluorescently labeled, and were mixed with a single-labeled control RNA sample from wild-type colonic epithelia. Samples were analyzed via dual-label microarray technology. Protein and RNA ratios then were aligned, and the combined dataset was filtered to include only molecules whose RNA and protein forms were quantified in multiple replicates.
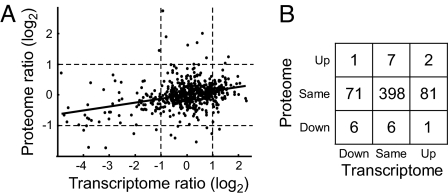
Interestingly, only limited agreement is observed between protein and mRNA ratios for differentially expressed proteins (Table 2). Although 4 show similar changes at protein and mRNA levels, 5 change little at the mRNA level, and 1 displays inverse protein and mRNA ratios. Three proteins whose protein and mRNA ratios are at odds were confirmed via SID, including HMG CoA synthase, whose protein and mRNA ratios are reversed. More generally, when all proteins are considered, only a modest correlation between protein and mRNA ratios is observed (see Fig. 4A). This result is consistent with other reports comparing protein and mRNA abundances (23) and comparing relative changes in protein and mRNA levels after biological perturbation (24).
Fig. 4.
Protein versus mRNA ratios. (A) Mean ratios are plotted for each protein as a function of mean mRNA ratio, with the best-fit line determined by linear regression. (B) Bootstrap analysis of Pearson product moment correlation coefficients among pairs of microarray and proteomics replicates suggests a correlation of only 0.33 (asymmetric 95% confidence interval: 0.05–0.37). The Spearman rank correlation coefficient also was 0.33. Dotted lines indicate 2-fold changes in expression; these thresholds were used to classify protein and mRNA ratio measurements qualitatively as increased (“up”), decreased (“down”), or unchanged (“same”) in B. The correlation between mRNA and protein ratios is limited, and most agreement is among the species showing no significant change at either protein or mRNA levels.
Clearly the quantitative relationship between mRNA and protein ratios is limited. However, requiring quantitative precision may be overly stringent, masking qualitative similarities that are biologically meaningful. Thus we classified protein and mRNA ratios as “increased,” “decreased,” or “unchanged” and determined the frequency at which transcriptome and proteome analyses agreed (Fig. 4B and Table S7). Although the concordance is higher than correlation suggests, it is dominated by species that are unchanged between tumor and normal tissue. Those showing biological response vary, including cases with opposing protein and mRNA ratios. Overall, ≈30% are classified differently at the protein and RNA levels, indicating that transcriptome and proteome data provide complementary views of a tumor's unique molecular profile.
Discussion
A primary goal of our work has been identification of abnormally expressed proteins in colonic tumors from Min mice. Several such proteins have been revealed by the use of 15N metabolic labeling for proteomic comparison of colonic tumors versus normal epithelia. Of these proteins, coronin, HMG CoA synthase, and SCFP-25 have been confirmed via SID experiments. In addition, carboxylesterase 2 has been shown by others via Western blotting to be downregulated in human colonic tumors (25). These results lend support to our approach.
We observed decreased transglutaminase 3 expression in tumors. This protein normally is expressed in mature epithelial cells. Its decreased protein and mRNA expression has been observed previously in oral and esophageal cancer (26, 27).
Interestingly, we observed reduced non-gastric H+/K+ ATPase α in tumors. Although annotated as an H+/K+ ATPase, the rat form resembles a Na+/K+ ATPase (75% sequence identity) and possesses significant Na+/K+ activity (28, 29). The mouse variant also is similar to mouse Na+/K+ ATPase (NP_659149; 65% sequence identity; Blast score: 1373; E-value < 10−100) (30), suggesting it may transport sodium. This finding is significant, because Na+/K+ ATPase activity is reduced in rat distal colon before tumor development (31). Furthermore, this activity has localization and pharmacological properties that match those reported for this H+/K+ ATPase with Na+/K+ activity (28, 29, 31). Notably, the Na+/K+ ATPase α and β subunits suppress cell motility through activation of PI3 kinase and interaction with annexin II (32); thus decreased abundance could signal increasing cell motility.
Continuing this theme of increasing cell motility, our analysis revealed downregulation of α-actinin 2 and upregulation of coronin and lymphocyte cytosolic protein 1 (also called “L-plastin”). α-Actinin interacts with E-cadherin, a structural protein within focal adhesions (33) whose expression is lost as epithelial cells assume mesenchymal properties in Min mice (34). An associated drop in α-actinin also has been noted in tumors (35). The protein L-plastin also is associated with focal adhesions, and its elevation induces proliferation, tissue invasion, and loss of E-cadherin in colorectal tumors (36). In addition, because coronin inactivates the actin-related protein 2/3 complex, its increase promotes cell motility (37). Expression of coronin 1C correlates with malignancy in glioma cells (38). Furthermore, coronin increases in breast cancer cell lines (39). Intriguingly, all these proteins play roles in cellular adhesion, and their observed changes imply increased cell motility that can signal transition of a tumor from relatively benign to invasive stages (34, 40).
This trend toward increasing malignancy is noteworthy, because the tumors we studied are adenomas: tumors in this early stage are not yet capable of tissue invasion and metastasis. Other observations oppose cell motility. Rho-GTPases activate cytoskeletal rearrangement and have been implicated in a variety of cancers. We observed an increase in Rho-GDP dissociation inhibitor β, which inactivates Rho-GTPases, opposing cell motility (41). Interestingly, activation of Rho-GTPases involves isoprenylation, and inhibition of this modification reduces cell motility and cancer progression. This inhibition has been achieved by blocking activity of HMG CoA reductase, an enzyme that provides precursors for this posttranslational modification (41). We observed decreased HMG CoA synthase, which immediately precedes HMG CoA reductase in this biosynthetic pathway. Although not previously implicated in tumor resistance, a decrease in HMG CoA synthase could reduce isoprenylation. Together, our proteomic observations capture the dynamic and conflicted nature of tissue in transition toward malignancy.
Although our observations generally are consistent with the literature, the decreased expression of RNA-binding motif protein 3 (RBM-3) has not previously been reported. Previously, researchers identified RBM-3 as an oncogene whose expression in human colonic adenocarcinomas and colon-derived cell lines is elevated and increases as disease progresses (42). Our observation of decreased RBM-3 expression in adenomas suggests that expression of this protein varies during tumor development, and that high levels of expression may be specific to advanced stages of disease.
Importantly, several preceding observations are novel for colonic tumors, and neither HMG CoA synthase nor SCFP 25 have previously been implicated in tumor development. Differential expression of these proteins was confirmed via SID. Although further investigation will be required to understand the role, if any, of their diminished expression in early Min tumors, their discovery highlights the value of untargeted discovery approaches.
In principle, differences in protein expression could be effects of the Min mutation in normal tissue as well as in tumor tissue. This possibility is important, because biochemical abnormalities have been reported in normal colonic tissue from humans lacking APC function (43). However, we have observed that levels of coronin are decreased in nontumor colonic tissues of Min mice, in stark contrast to the increased levels in tumor tissue. Furthermore, we demonstrated via SID that our observations of the downregulation of HMG CoA synthase and SCFP 25 are specific to tumors. Additionally, many of the other changes we found were observed previously in tumors from independent disease models. Thus most of the differentially expressed proteins we observed are likely to be tumor specific. Nonetheless, the differentially expressed proteins identified by our strategy of differential MS can be biologically significant, whether they arise in normal tissue heterozygous for the Min mutation or only in tumors.
This study has yielded intriguing biological observations, and, importantly, it has confirmed the utility of metabolic labeling for quantitative proteomics in mammals. Although metabolic labeling is often used in yeast, plants, bacteria, and cells in culture, its use in mammals has been relatively limited. Previously, metabolic labeling of rats for 1 and 2 generations had been reported (15, 17), as well as stable isotope labeling by amino acids in cell culture (SILAC) labeling of mice (16). We demonstrated sufficient levels of 15N incorporation across many tissues and subsequently used metabolic labeling to identify differentially expressed proteins in colonic tumors.
The reciprocal-labeling design of our experiment has been important for ensuring high-quality peptide quantitation and identification. Requiring inversion of 15N:14N abundance ratios across replicates as isotopic labels are exchanged allows the exclusion of quantitation artifacts arising from inconsistent changes, coincidentally co-eluting species, and differences in diets (see ref. 44). Furthermore, exchanging isotopic labels minimizes bias arising from different identification rates for labeled and unlabeled peptides, ensuring that all peptides have comparable likelihood of identification. Although the bias against identification of 15N-labeled peptides is most pronounced with incomplete incorporation (45), subtle differences have been observed for peptides labeled to ≈98% incorporation (13), especially as length increases (45).
In addition to metabolic labeling, many in vitro labeling techniques are available, including chemical labeling approaches, such as isotope-coded affinity tags (8), isobaric tagging for relative and absolute quantitation (9), and acrylamide labeling (46), and enzymatic approaches, such as 18O labeling (10). For detailed comparisons of in vitro and in vivo labeling techniques see reviews by Beynon and Pratt (47) and Ong and Mann (48). Because these methods introduce an isotopic label late during sample preparation, they are applicable to any protein sample. However, labeling via these techniques sometimes is incomplete: only peptides with certain functional groups are labeled (8), or labeling reactions may be reversible (10). Additionally, in vitro labeling techniques cannot control error introduced during early steps such as tissue homogenization and protein extraction before labeling and mixing of samples. In contrast, because metabolic labeling introduces an isotopic label into all proteins early, it provides internal control through all experimental steps. Metabolic labeling thus enables extensive fractionation of the mixed, differentially labeled samples, particularly before tryptic digestion: techniques such as subcellular fractionation, immunoprecipitation (49), and SDS/PAGE may be used without increasing error. This internal control makes metabolic labeling a powerful tool to address the challenges of dynamic range.
Although metabolic labeling has been used in many multicellular organisms, it could be problematic in larger organisms or in experiments imposing dietary constraints. Another approach in such circumstances is partial metabolic labeling (50, 51). Rather than requiring nearly complete substitution of 15N for 14N, partial metabolic labeling employs 6% 15N labeling. Through deconvolution of overlapping labeled and unlabeled peptide isotopic envelopes, ratios are derived with accuracy comparable to full metabolic labeling (51). Much lower isotopic enrichment is required, extending the technique to organisms that cannot be efficiently or economically labeled to ≈100% 15N.
Although metabolic labeling and untargeted quantitative proteomics in general provide a valuable overview of protein expression, they often are not well suited to assessing biological variability. Achieving high proteome coverage usually requires significant fractionation; thus many LC-MS analyses are required for each biological sample. The associated time and expense restrict biological replicates, limiting characterization of biological variability. In contrast, although it requires prior selection of peptide targets, SID is well suited to rapid quantitative analysis of many samples to assess variability. These complementary techniques provide a powerful means of discovering broadly and then validating specifically differentially expressed proteins.
Our experimental design used 4 biological replicates for discovery of differentially expressed proteins, minimizing technical variability while providing some indication of biological variability. This approach revealed 3 altered proteins whose expression was verified via SID. Integration of these strategies was efficient: proteomic survey data aided selection of peptides from each target protein with favorable ionization, fragmentation, and chromatographic properties for use as internal standards. With technological advances allowing higher-throughput peptide design (52–54), synthesis (55), and multiplexing to quantify dozens of peptides in a single run (56), the use of SID as a complement to untargeted proteomics surveys surely will increase.
Previous researchers have used microarrays to probe the transcriptome of intestinal tumors (57, 58). Recently, Hung et al. used tumor transcriptome data to identify the provenance of plasma proteins whose abundance was associated directly with tumor development in Apc-mutant mice (59). Although transcriptome analyses provide insight, our data suggest that protein-level surveys provide an important complementary view. We found weak correlation between protein and mRNA ratios, in accord with previous published work (23, 24). This finding reflects biological processes such as posttranslational modification, protein degradation, and mRNA transcript processing and is especially pronounced among species with altered abundance. Ultimately, both protein and mRNA expression profiles are required for comprehensive characterization of biological systems.
Overall, this study confirms the utility of metabolic labeling for proteomic characterization of mice, identifying abnormally expressed proteins in colonic tumors. Combined with SID for targeted validation, metabolic labeling provides a powerful integrated approach for identifying differentially expressed proteins. This protein-level view provides unique and valuable insight into biological systems, complementing gene expression profiling. Ultimately, this work can serve as the basis for future studies that will offer greater protein-level insight into this and other biological systems.
Materials and Methods
Experimental Procedures.
An overview of experimental procedures is provided here. For complete details, see SI Materials.
Quantitative Proteomic Analysis of Mice via Metabolic Labeling.
We raised 4 pairs of female Min and wild-type mice from weaning on a diet derived from natural-abundance or 15N-labeled Spirulina. When the mice were killed, natural-abundance and 15N-labeled samples were combined for protein extraction and digestion. Tissue digests were analyzed via nano-LC-ElectroSpray Ionization-QuadrupoleTOF MS. Peptides were identified via Mascot and filtered to a 1% FDR via decoy database searching. 15N enrichment levels were determined by fitting observed isotopic envelopes to predicted isotopic distributions. Colonic tumor samples were analyzed via LC-MALDI using a TOF/TOF mass spectrometer, and peptides were quantified (see SI Methods). The peptide FDR for the colonic tumor samples was 1%, and the protein FDR was 5%, although this FDR fell below 0.8% when the dataset was restricted to peptides observed in multiple replicates, as required for quantitative analysis.
Stable Isotopic Dilution Experiments.
Colonic tumor and normal tissue samples were homogenized, and proteins were extracted. During tryptic digestion, synthetic isotopically labeled peptides whose sequences matched unique tryptic peptides within the target proteins were introduced as standards for quantitation. Samples were analyzed via multiple-reaction monitoring on a Q-Trap mass spectrometer (Applied Biosystems, Inc.) with HPLC separation.
Microarray Analysis.
RNA was extracted from 4 colonic tumors obtained from 3 female Min mice and from normal epithelium from a single female wild-type mouse. RNA samples were labeled with fluorescent dyes, and each tumor RNA sample was pooled with a portion of RNA from the single wild-type control. Samples were analyzed via dual-labeling microarray techniques (60). Array data have been deposited on the National Institutes of Health Gene Expression Omnibus Web site (http://www.ncbi.nlm.nih.gov/geo/).
Supplementary Material
Acknowledgments.
We thank Linda Clipson for skilled aid with manuscript preparation and Alexandra Shedlovsky for help with dissections. We also thank Barbara Michelson (Harlan Teklad) for preparation of the Spirulina-based mouse diet, Gary Case for synthesis of peptide standards, and Takis Papoulias (University of Michigan) for use of his software (T2DExtractor). E.L.H. was supported by the University of Wisconsin-Madison Biotechnology Training Program (National Institutes of Health Grants 5 T32 GM08349 and T32 HG002760), the Louis and Elsa Thomsen Wisconsin Distinguished Graduate Fellowship, and the University of Wisconsin-Madison Genome Sciences Training Program. M.R.S. was supported by National Science Foundation Grants DBI-0701846 and MCB-0448369. W.F.D. was supported by National Institutes of Health Grant R37-CA63677 and by the University of Wisconsin-Madison Institutional Clinical and Translational Research Program Grant UL 1RR025011.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909282106/DCSupplemental.
References
- 1.American Cancer Society. Cancer Facts and Figures 2009. Atlanta, GA: American Cancer Society; 2009. [Google Scholar]
- 2.Smith RA, et al. American Cancer Society guidelines for the early detection of cancer: Update of early detection guidelines for prostate, colorectal, and endometrial cancers. Update 2001—testing for early lung cancer detection. CA Cancer J Clin. 2001;51:38–75. doi: 10.3322/canjclin.51.1.38. quiz 77–80. [DOI] [PubMed] [Google Scholar]
- 3.Taketo MM. Mouse models of gastrointestinal tumors. Cancer Science. 2006;97:355–361. doi: 10.1111/j.1349-7006.2006.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 5.Su LK, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 6.Rustgi AK. The genetics of hereditary colon cancer. Genes Dev. 2007;21:2525–2538. doi: 10.1101/gad.1593107. [DOI] [PubMed] [Google Scholar]
- 7.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 8.Gygi SP, et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 9.Ross PL, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Molecular and Cellular Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Proteolytic 18O labeling for comparative proteomics: Model studies with two serotypes of adenovirus. Anal Chem. 2001;73:2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 11.Washburn MP, Ulaszek R, Deciu C, Schieltz DM, Yates JR., III Analysis of quantitative proteomic data generated via multidimensional protein identification technology. Anal Chem. 2002;74:1650–1657. doi: 10.1021/ac015704l. [DOI] [PubMed] [Google Scholar]
- 12.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Molecular and Cellular Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 13.Nelson CJ, Huttlin EL, Hegeman AD, Harms AC, Sussman MR. Implications of 15N-metabolic labeling for automated peptide identification in Arabidopsis thaliana. Proteomics. 2007;7:1279–1292. doi: 10.1002/pmic.200600832. [DOI] [PubMed] [Google Scholar]
- 14.Ippel JH, et al. In vivo uniform (15)N-isotope labelling of plants: Using the greenhouse for structural proteomics. Proteomics. 2004;4:226–234. doi: 10.1002/pmic.200300506. [DOI] [PubMed] [Google Scholar]
- 15.Wu CC, MacCoss MJ, Howell KE, Matthews DE, Yates JR., III Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal Chem. 2004;76:4951–4959. doi: 10.1021/ac049208j. [DOI] [PubMed] [Google Scholar]
- 16.Kruger M, et al. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 17.McClatchy DB, Dong MQ, Wu CC, Venable JD, Yates JR., III 15N metabolic labeling of mammalian tissue with slow protein turnover. Journal of Proteome Research. 2007;6:2005–2010. doi: 10.1021/pr060599n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 20.Huttlin EL, Hegeman AD, Harms AC, Sussman MR. Prediction of error associated with false-positive rate determination for peptide identification in large-scale proteomics experiments using a combined reverse and forward peptide sequence database strategy. Journal of Proteome Research. 2007;6:392–398. doi: 10.1021/pr0603194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong MQ, et al. Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science. 2007;317:660–663. doi: 10.1126/science.1139952. [DOI] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Roy Stat Soc B. 1995;57:289–300. [Google Scholar]
- 23.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Godoy LM, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 25.Tang X, et al. Carboxylesterase 2 is downregulated in colorectal cancer following progression of the disease. Cancer Invest. 2008;26:178–181. doi: 10.1080/07357900701560786. [DOI] [PubMed] [Google Scholar]
- 26.Choi P, et al. Examination of oral cancer biomarkers by tissue microarray analysis. Archives of Otolaryngology—Head and Neck Surgery. 2008;134:539–546. doi: 10.1001/archotol.134.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uemura N, et al. Transglutaminase 3 as a prognostic biomarker in esophageal cancer revealed by proteomics. Int J Cancer. 2009;124:2106–2115. doi: 10.1002/ijc.24194. [DOI] [PubMed] [Google Scholar]
- 28.Codina J, Pressley TA, DuBose TD., Jr The colonic H+,K+-ATPase functions as a Na+-dependent K+(NH4+)-ATPase in apical membranes from rat distal colon. J Biol Chem. 1999;274:19693–19698. doi: 10.1074/jbc.274.28.19693. [DOI] [PubMed] [Google Scholar]
- 29.Cougnon M, Bouyer P, Planelles G, Jaisser F. Does the colonic H,K-ATPase also act as an Na,K-ATPase? Proc Natl Acad Sci USA. 1998;95:6516–6520. doi: 10.1073/pnas.95.11.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 31.Davies RJ, Sandle GI, Thompson SM. Inhibition of the Na+,K(+)-ATPase pump during induction of experimental colon cancer. Cancer Biochemistry Biophysics. 1991;12:81–94. [PubMed] [Google Scholar]
- 32.Barwe SP, et al. Novel role for Na,K-ATPase in phosphatidylinositol 3-kinase signaling and suppression of cell motility. Mol Biol Cell. 2005;16:1082–1094. doi: 10.1091/mbc.E04-05-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Craig DH, Haimovich B, Basson MD. Alpha-actinin-1 phosphorylation modulates pressure-induced colon cancer cell adhesion through regulation of focal adhesion kinase-Src interaction. Am J Physiol Cell Physiol. 2007;293:C1862–1874. doi: 10.1152/ajpcell.00118.2007. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Halberg RB, Burch RP, Dove WF. Intestinal adenomagenesis involves core molecular signatures of the epithelial-mesenchymal transition. Journal of Molecular Histology. 2008;39:283–294. doi: 10.1007/s10735-008-9164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slater M, Cooper M, Murphy CR. The cytoskeletal proteins alpha-actinin, ezrin, and talin are de-expressed in endometriosis and endometrioid carcinoma compared with normal uterine epithelium. Applied Immunohistochemistry and Molecular Morphology. 2007;15:170–174. doi: 10.1097/01.pai.0000194762.78889.26. [DOI] [PubMed] [Google Scholar]
- 36.Foran E, McWilliam P, Kelleher D, Croke DT, Long A. The leukocyte protein L-plastin induces proliferation, invasion and loss of E-cadherin expression in colon cancer cells. Int J Cancer. 2006;118:2098–2104. doi: 10.1002/ijc.21593. [DOI] [PubMed] [Google Scholar]
- 37.Uetrecht AC, Bear JE. Coronins: The return of the crown. Trends Cell Biol. 2006;16:421–426. doi: 10.1016/j.tcb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Thal D, et al. Expression of coronin-3 (coronin-1C) in diffuse gliomas is related to malignancy. J Pathol. 2008;214:415–424. doi: 10.1002/path.2308. [DOI] [PubMed] [Google Scholar]
- 39.Pitteri SJ, et al. Plasma proteome profiling of a mouse model of breast cancer identifies a set of up-regulated proteins in common with human breast cancer cells. Journal of Proteome Research. 2008;7:1481–1489. doi: 10.1021/pr7007994. [DOI] [PubMed] [Google Scholar]
- 40.Halberg RB, et al. Long-lived Min mice develop advanced intestinal cancers through a genetically conservative pathway. Cancer Res. 2009;69:5768–5775. doi: 10.1158/0008-5472.CAN-09-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang Y, Olufemi L, Wang MT, Nie D. Role of Rho GTPases in breast cancer. Frontiers in Bioscience. 2008;13:759–776. doi: 10.2741/2718. [DOI] [PubMed] [Google Scholar]
- 42.Sureban SM, et al. Translation regulatory factor RBM3 is a proto-oncogene that prevents mitotic catastrophe. Oncogene. 2008;27:4544–4556. doi: 10.1038/onc.2008.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeung AT, et al. One-hit effects in cancer: Altered proteome of morphologically normal colon crypts in familial adenomatous polyposis. Cancer Res. 2008;68:7579–7586. doi: 10.1158/0008-5472.CAN-08-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huttlin EL, Hegeman AD, Sussman MR. Metabolic labeling approaches for the relative quantification of proteins. In: Whitelegge JP, editor. Protein Mass Spectrometry, Comprehensive Analytical Chemistry. Vol 52. Amsterdam: Elsevier; [Google Scholar]
- 45.Gouw JW, Tops BB, Mortensen P, Heck AJ, Krijgsveld J. Optimizing identification and quantitation of 15N-labeled proteins in comparative proteomics. Anal Chem. 2008;80:7796–7803. doi: 10.1021/ac801249v. [DOI] [PubMed] [Google Scholar]
- 46.Faca V, et al. Quantitative analysis of acrylamide labeled serum proteins by LC-MS/MS. Journal of Proteome Research. 2006;5:2009–2018. doi: 10.1021/pr060102+. [DOI] [PubMed] [Google Scholar]
- 47.Beynon RJ, Pratt JM. Metabolic labeling of proteins for proteomics. Molecular and Cellular Proteomics. 2005;4:857–872. doi: 10.1074/mcp.R400010-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nature Chemical Biology. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 49.Anderson NL, et al. A human proteome detection and quantitation project. Molecular and Cellular Proteomics. 2009;8:883–886. doi: 10.1074/mcp.R800015-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitelegge JP, et al. Subtle modification of isotope ratio proteomics; an integrated strategy for expression proteomics. Phytochemistry. 2004;65:1507–1515. doi: 10.1016/j.phytochem.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 51.Huttlin EL, Hegeman AD, Harms AC, Sussman MR. Comparison of full versus partial metabolic labeling for quantitative proteomics analysis in Arabidopsis thaliana. Molecular and Cellular Proteomics. 2007;6:860–881. doi: 10.1074/mcp.M600347-MCP200. [DOI] [PubMed] [Google Scholar]
- 52.Fusaro VA, Mani DR, Mesirov JP, Carr SA. Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat Biotechnol. 2009;27:190–198. doi: 10.1038/nbt.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kline KG, Frewen B, Bristow MR, Maccoss MJ, Wu CC. High quality catalog of proteotypic peptides from human heart. J Proteome Res. 2008;7:5055–5061. doi: 10.1021/pr800239e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prakash A, et al. Expediting the development of targeted SRM assays: Using data from shotgun proteomics to automate method development. J Proteome Res. 2009;8:2733–2739. doi: 10.1021/pr801028b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maeji NJ, Bray AM, Valerio RM, Wang W. Larger scale multipin peptide synthesis. Pept Res. 1995;8:33–38. [PubMed] [Google Scholar]
- 56.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Molecular and Cellular Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaiser S, et al. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biology. 2007;8:R131. doi: 10.1186/gb-2007-8-7-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leclerc D, Deng L, Trasler J, Rozen R. ApcMin/+ mouse model of colon cancer: Gene expression profiling in tumors. J Cell Biochem. 2004;93:1242–1254. doi: 10.1002/jcb.20236. [DOI] [PubMed] [Google Scholar]
- 59.Hung KE, et al. Comprehensive proteome analysis of an Apc mouse model uncovers proteins associated with intestinal tumorigenesis. Cancer Prevention Research. 2009;2:224–233. doi: 10.1158/1940-6207.CAPR-08-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiao J, Delenstarr G, Visitacion M, Argonza-Barrett R, Lucas AB. New stringent 2-color gene expression workflow enables more accurate and reproducible microarray data. Agilent Technologies Incorporated. [Accessed April 2009]. Available at: http://www.chem.agilent.com/en-us/Search/Library/_layouts/Agilent/PrimaryDocumentViewer.ashx?whid=45778.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.