Abstract
Ten complete mammalian genome sequences were compared by using the “feature frequency profile” (FFP) method of alignment-free comparison. This comparison technique reveals that the whole nongenic portion of mammalian genomes contains evolutionary information that is similar to their genic counterparts—the intron and exon regions. We partitioned the complete genomes of mammals (such as human, chimp, horse, and mouse) into their constituent nongenic, intronic, and exonic components. Phylogenic species trees were constructed for each individual component class of genome sequence data as well as the whole genomes by using standard tree-building algorithms with FFP distances. The phylogenies of the whole genomes and each of the component classes (exonic, intronic, and nongenic regions) have similar topologies, within the optimal feature length range, and all agree well with the evolutionary phylogeny based on a recent large dataset, multispecies, and multigene-based alignment. In the strictest sense, the FFP-based trees are genome phylogenies, not species phylogenies. However, the species phylogeny is highly related to the whole-genome phylogeny. Furthermore, our results reveal that the footprints of evolutionary history are spread throughout the entire length of the whole genome of an organism and are not limited to genes, introns, or short, highly conserved, nongenic sequences that can be adversely affected by factors (such as a choice of sequences, homoplasy, and different mutation rates) resulting in inconsistent species phylogenies.
Keywords: alignment-free genome comparison, feature frequency profile (FFP), mammalian phylogeny, noncoding DNA, nongenic regions of the genome
The current understanding of mammalian genomes (and of higher order eukaryotes in general) is primarily a “gene centric” view. As a result, genome comparisons among mammals have been gene based, and highly conserved genes are preferentially used to infer species divergence. However, the coding (coding for proteins, ribosomal RNAs, transfer RNAs, and other functional RNAs) portions of mammalian genomes can amount to as little as 1–3% of the whole genomic sequence, and it is debatable whether species phylogenies derived from a small, alignable subfraction of the whole genome are reliable. As for the noncoding sequence (the other 99%), much of its function is unknown, yet much of this portion is indeed transcribed. Recently, the ENCODE project showed that at least 93% of analyzed human genome nucleotides were transcribed into RNA when all various cell types were considered (1). Similarly, transcriptional analysis of human chromosomes demonstrated that transcripts originating from the nongenic regions comprise the largest fraction of the transcriptional output of the human genome (2). We have operationally defined a nongenic region to be those regions that have not been annotated to contain a gene in the GenBank records. Some known features in the nongenic sequence include transposable elements and sequences whose transcripts are long noncoding RNAs (ncRNAs) or short microRNAs (miRNA). The importance of these nongenic components is just now being realized, and their functions are a matter of current debate. A subject that deserves further investigation is the information embedded in the noncoding regions and the relationship that noncoding genomes share among the mammalian clades. Most of the noncoding sequences are not well conserved among mammals with the exclusion of a tiny fraction which are “ultraconserved”. In this work, we discuss the phylogenic relationship among four partitions of the whole genomic sequence: exonic (all protein-coding exons), intronic (all introns), nongenic (all intergenic-sequence), and whole (entire-sequence) genomes.
Recent observations suggest that large portions of the nongenic genome may in fact be functionally active and under some selective pressure. A very small fraction of the human nongenic genome (0.3–1%) is even “ultraconserved” among mammals (4), and some of them have been implicated to have evolutionary information. For example, rare transposon insertions were shown by Kriegs et al. (5) to be a useful marker for tracing mammalian evolution and the phylogenic relationship between humans and rodents. Also, a selected set of conserved noncoding sequences were shown by Nikolaev et al. (6) to contain an equivalent level of phylogenic information as found in a small portion of genic sequences. They created two separate mammalian phylogenies from 204 kbps of coding sequence and 429 kbps of conserved noncoding sequence and both had identical topologies. Thus, there is strong evidence that a traceable evolutionary history lies embedded in some selected highly conserved nongenic regions as well as genic regions. These and other previous works have focused on studying and inferring phylogenies from highly conserved noncoding sequences, which represent only a small fraction of the genome (1–2%). However, phylogenic inferences based on small fractions of the genome may be incorrect because of tree-building artifacts; in the case of genic sequences, the effects of limited sequence selection have been shown to give incorrect tree topologies. The method we discuss here can be used to compare entire nongenic sequences, including both rare ultraconserved nongenic sequences and less-conserved regions, because the rigors of alignment are not required in our method.
Early constructions of mammal gene-based phylogenies exclusively used multiple alignments of mitochondrially encoded sequences (e.g., ref. 7), arriving at topologies supporting a basal position for rodents (glires) among Boreoeutherians (primates, glires, and Laurasiatherians). We refer to this mitogenomic tree as the type-II topology. However, subsequent analysis with a concatenated set of nuclear genes (8) indicated a different tree topology—a sister relationship between rodents and primates, forming another infraorder, Euarchontaglires (type I). Gene-selection bias always remains a possibility because the choice of gene set plays a critical role in the ultimate species tree obtained, as illustrated by Huerta-Cepas et al. (9, 10). They investigated the human “phylome”—the individual evolutionary history of each of the genes encoded in the human genome. Among a set of 21,588 individual gene trees, the three dominant topologies in order of abundance were type II (44%), type I (32%), and a third type (23%) with rodents and Laurasiatherians grouped together as a clade. There is a wide range of topological variation among individual gene trees and, thus, species trees based on a limited gene set are highly suspect. Likewise, we would expect the same situation to be true for phylogenies derived from one or a limited set of highly conserved nongenic sequences.
In all cases, a larger dataset tends to provide more support for species-level phylogenies. A recent genome comparison by Prasad et al. (11) using the 28-species University of California–Santa Cruz (UCSC) genome browser alignment (12) and the largest number of nucleic acid characters to date confirms the early results (type I) of Murphy et al. (8). This study, like a number of recent large-scale approaches combines the information obtained from many genes to resolve evolutionary relationships. Prasad et al. use a reduced purine–pyrimidine (RY) two-letter code space, which reduces base composition bias and bias caused by differential evolutionary rates among organisms (heterotachy). Clearly, the more genomic data used for each organism in the analysis, the more stable and reliable the tree topology will become in revealing the “true” species tree. All of the above methods rely on multiple sequence alignment (MSA) and the gene set needs to be present in all of the species. Furthermore, the evolutionary phylogenies derived from MSA measures substitutional differences at the local level only for well-conserved regions. Also, errors in MSA can propagate to errors in phylogenetic inference (13), especially when applied in an uncurated manner—as must necessarily occur when applied on a genomic scale. As mentioned earlier, the phylogenetic signal in noncoding regions has been found before by using highly conserved/ultraconserved sequences, i.e., those regions that can be aligned, which comprise only a tiny fraction across all mammalian genomes. Also, because one can observe different topologies depending on the gene that one selects, the same is expected to be true for different conserved noncoding regions.
Any phylogenic method uses variation in conserved features such as variation in aligned base/amino acid positions, variation in gene content, or variation in gene order to derive phylogenies. The feature frequency profile (FFP) method of alignment-free genome comparison (3) derives phylogenic information from the variations in FFPs. In this paper, we use the FFP method to investigate the grouping of the whole-genome features and the extent of inferred evolutionary relationships embedded within nongenic and genic genome partitions. With this method, it is critical to select the feature length optimal for inferring evolutionary phylogeny (see Materials and Methods). The alignment-free FFP method has several principal advantages over MSA-based methods. (i) Whole genomes (genic and nongenic regions) can be compared. (ii) Genomes do not need to share a common set of genes to be effectively compared. (iii) Nonalignable portions of genomes can be compared. It is therefore possible to compare entire nongenic portions of mammalian genomes (not just easily aligned, highly conserved portions, which are a subset of “conserved features”), where a major portion of the sequence is not well conserved, but may have conserved features, such as those detectable by FFP. (iv) The FFP method is significantly faster than MSA-based methods, especially for large genomes. (v) The FFP can incorporate a wide variety of genomic features into each comparison. Thus, our method can account for large-scale genomic changes such as rare genomic changes (14), intron deletions (15, 16), exon sequence indels (17), and transposable element insertions (18–20), as well as small-scale changes such as base transversions in coding sequences. In particular, rare genomic changes, such as short interspersed element/long interspersed element (SINE/LINE) insertions, are thought to be exceptionally useful markers because they provide unambiguous evolutionary information and are thought to be homoplasy-resistant (21, 22).
We show in this work that the phylogenies obtained with the FFP method, whether we use the whole, intronic, exonic, or nongenic genomes, are all topologically equivalent to the current consensus view of the evolutionary relationships between mammalian clades. Irrespective of the type of genomic region, evolutionary footprints are present in all parts of the genome.
Results
In this section, we show that whole-genome comparison, which includes nongenic, intronic and exonic sequence, best represents whole-genome divergence. Several examples are given where selected genes may lead to biased results supporting a specific gene phylogeny rather than organism phylogeny. We show that noncoding sequences such as intergenic regions and introns contain an evolutionary phylogenic signal, which is comparable with exons by comparing tree topologies obtained by using the FFP method. The FFP-based, alignment-free, whole-genome topology is similar to large-scale-coding, MSA-based trees.
Genome Partitions: Intronic, Exonic, and Nongenic Regions.
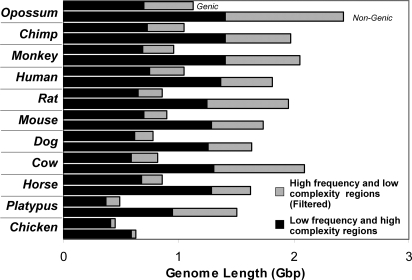
To investigate the conservation of evolutionary information contained within genic and nongenic genome sequences, we partitioned the complete reference genomes of human (Homo sapiens), chimpanzee (Pan troglodytes), rhesus monkey (Macaca mulatta), mouse (Mus musculus), rat (Rattus norvegicus), dog (Canis lupus familiaris), horse (Equus caballus), cow (Bos Taurus), opossum (Monodelphis domesticus), and platypus (Ornithorhynchus anatinus) into their constituent intronic, exonic, and nongenic components. These genomes have the deepest (at present) sequencing coverage (>10×) among sequenced mammals. Exonic sequences were extracted from the genbank assembly records found at the National Center for Biotechnology Information (ftp://ftp.ncbi.nlm.nih.gov/genomes) by using the base-pair positions specified by each genbank coding sequence field. All exons from a species were concatenated together in one exon genome file with an “x”-delimiting character separating exons. The delimiter prevents extracted features (“words”) from spanning two exons. All intervening intron sequences were also concatenated into an intronic genome. Nongenic sequences were extracted from those regions lying outside the range of an annotated gene. It is worth noting that the genbank annotations are known to be incomplete. Therefore, our genome partitions will necessarily misallocate a number of unannotated genes to the nongenic partition, but they will have a negligible effect on FFP construction. The relative sizes (in base pairs) of the mammalian genic (annotated gene regions) and nongenic genome partitions by species are shown in Fig. 1.
Fig. 1.
Sizes of mammalian genome partitions. Sizes of the genomes used in this study after being divided into genic (exons, introns, promoters), and nongenic (no annotated gene) partitions. The sizes of each genomic partition are also indicated after low-complexity and high-frequency features (from l = 18) are filtered out.
Feature Reduction via Filtering, and Feature Redundancy.
Two kinds of feature filtering were applied: high-frequency filtering and low-complexity filtering. High-frequency features are duplicated many times in the genome, and low-complexity features are composed of redundant or highly repetitive sequences. In our analysis, feature complexity and frequency filtering removed ≈35% of the features for each of the four classes of genome partitions. The FFP features at an optimal feature length of l = 18 are highly redundant, ranging between 42% (for exonic features) and 48% redundant (for intronic features). In single-gene analyses, extensive filtering is quite often impractical because too few positions remain available. However given sufficient data, positions in a MSA may be removed because they are suspected for homoplasy (multiple mutations or reversion back to an ancestral state). Additionally, in phylogenetic reconstruction each character in an alignment is assumed to behave independently.* In the FFP method, filtering is safe and justifiable because the features are not independent of each other, and in fact are highly redundant. We can estimate with fairly high confidence that the full set of 18-mer features will be between 40% and 50% redundant based on rank correlation (see SI Materials and Methods). Thus, the elimination of 30–40% of the features (Fig. 1) is not a drastic measure for l = 18, especially because our strict jackknife criteria (10% random selection) demonstrate robust consensus tree topologies. Our rationale for applying filtering was to reduce noise from the comparisons. We observed that the two most-common sources for noise were repetitive, low-complexity sequences such as GC-rich heterochromatin and very high-frequency features. Both kinds of features have a tendency to dominate the Jensen–Shannon (JS) divergence score because they tend to be the largest component of the FFP distribution. Also, the heterochromatin—the tightly packed low-complexity regions—tends to be the least completely assembled portion of the genome.
Whole, Exonic, Intronic, and Nongenic Genome FFP Trees Are in Agreement.
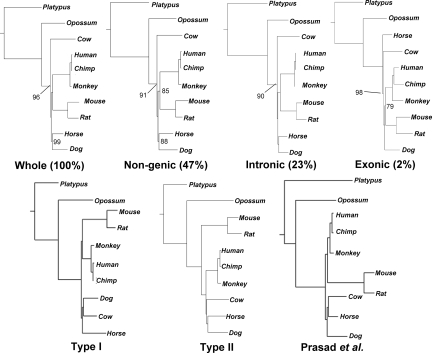
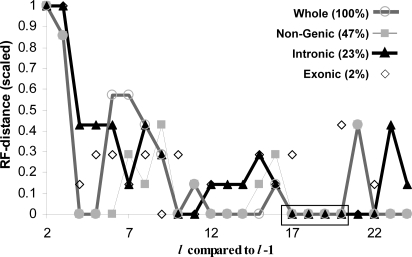
FFP neighbor-joining phylogenies of the whole, nongenic, and intronic genomes yield identical tree topologies at an optimal feature length of l = 18 (see Materials and Methods) as shown in Fig. 2. The exonic genome also yields a very similar topology, with the only difference being the order of divergence within Laurasiatheria. The exonic genomes show more support for a closer relationship between cow and dog (Euungulata), rather than between dog and horse (Zooamata). Analysis of SINE insertions by Nishihara et al. (23) indicated support for the Zooamata clade, whereas analysis by others does not (11, 16, 24). The optimal range of l was found from a tree-convergence plot (Fig. 3). The whole-genome topology converges to the topology shown in Fig. 2 for feature lengths of l = 17–20. The largest noncoding partitions, the intronic and nongenic genomes, converge to the same topology for lengths of l = 16–21 for the intronic partition and l = 16–20 for the nongenic partition. The exonic partition converges for lengths of l = 17–19. All of the branches have reasonably high (>75) jackknife support values for l > 16–21. Beyond l = 21 (l = 20 for the exonic partition), the type-II (rodent basal) topology becomes dominant. The type-II topology is an artifact that is caused by the high mutation rate of murids. When using longer feature lengths, there are fewer common features among species with which to reliably establish evolutionary relationships. In the FFP trees, Laurasiatherians are the first to diverge, but they do not strictly form a monophyletic grouping. However, if we extend the analysis of whole genomes to include the lower coverage (survey) mammalian genomes from other taxa, such as those of the Broad Institute and Washington University, Laurasiatheria form a monoclade. This suggests that the grouping by the FFP method is likely to improve as more whole-genome sequences become available and are included in the analysis.
Fig. 2.
Similar evolutionary information in genic and nongenic genome partitions. (Upper) FFP mammalian species trees created from whole, nongenic, intronic, and exonic genome partitions have identical tree topologies with slight differences in internal branch lengths. For each neighbor-joining FFP tree, the optimal feature length is l = 18. Clade frequencies <100% (from 1,000 replicates) are indicated. (Bottom) For comparison, the two major types (I and II) of individual gene-tree topologies are shown. A tree from Prasad et al. (11) based on a large-genome-scale gene alignment is also shown for comparison. Note that only species common among the three methods are used for comparison (some taxa were pruned from their tree for consistent comparison. (% values indicates the average fraction of the whole genome. Note that 28% are genic regions which are neither intron nor exon).
Fig. 3.
Tree-topology convergence. The tree generated with features of length l is compared with the tree from l−1 by using the RF distance. Largest genome partitions are indicated by lines. The topologies tend to converge (RF = 0) for the largest genome fractions (whole, nongenic, and intronic) in the range from l = 16–21. This convergence range is indicated by the boxed region. The topologies are identical for all fractions for l = 17–19. Percentage values indicate the average fraction of the whole genome represented by each partition. The remaining 28% represents genic regions that are neither introns nor exons.
Comparisons with Phylogenies Based on Single-Gene Alignments.
Of 32 nuclear and mitochondrially encoded mammalian genes analyzed (see Materials and Methods), 12 were observed to have a type-II topology and 11 had a type-I topology (see Table S1). The two tree topologies are shown in Fig. 2. Nine genes flip topologies from type I to type II or vice versa when converted to RY coding. As discussed in Effect of Evolutionary Rate on Phylogeny, this switch may be due to differential evolutionary rates between rodents and other mammals. These examples illustrate that gene-selection bias can alter the resultant phylogeny. The phylogenies of all our genome partitions in the convergence region (l = 18 and 19) match the type-I tree topology, which in turn matches the phylogeny from the large-scale gene [1.9 mega base pairs (Mbp) or roughly half the size of the exonic genome] alignments of Prasad et al. (also shown in Fig. 2). Our FFP whole-genome tree agrees with the comprehensive multigene, alignment-based tree constructed by Prasad et al. (11) and others (6, 8, 27). Only a minority of all of the individual gene alignments yield the topologies of type I. However, the FFP comparisons are effectively made without any knowledge of gene boundaries and without sequence alignment. Thus, although each class of genome partitions reveals evolutionary footprints, we suggest that whole-genome comparison, including both the genic and nongenic sequence, is the proper representation of the whole genome divergence as represented by type I.
Although, we may not fully understand the function of the nongenic genome, our results reveal that the noncoding sequence is under some form of evolutionary constraint even if not at a level which is as understandable as in exons. Although a neutrally evolving (i.e., drifting) genome sequence could also contain evolutionary information, it is likely that the sequence signal within alignable regions would become saturated and obliterated over longer periods of time in the absence of some form of selection, which still preserves feature signals such as those in FFPs. Even if informative, a multigene alignment can only reveal the evolutionary history of the specified multigene set. Topological variations among phylogenies from different genes or gene sets should be expected (25). A particular multigene set is not always a proper proxy for the species or whole genome itself, which highlights the danger of gene-sampling effects in phylogenomic analysis (26).
FFP Methods Yield Bush-Like Tree Topologies.
Like MSA-based trees, FFP also yields mammalian phylogenies that are bush-like, which provides a view consistent with a mammalian radiation characterized by rapid cladogenesis events. The trees for all different partitions are essentially bush-like, and in this respect they are also similar to gene-based reconstructions (42). All of our trees, multigene-based trees of Prasad et al. (11) and Murphy et al. (27), and single or small gene set-based trees have low F values, would be characterized as bush-like. This kind of tree is created by a radiation where serial cladogenesis events occur in a short time span, creating short internal branch lengths. Subsequently, after radiation the external branches lengthen, creating a bush-like topology. Internal branch distances between mammal clades are estimated to be only as short as 1 to 10 million years (24). However, it remains difficult to distinguish between a tree with the incorrect topology and one with several cladogenesis events compressed into a short span of time.
Discussion
Phylogeny of Nongenic and Intronic Regions.
We have shown earlier (3) that the intronic genome contains an evolutionary footprint. It is particularly remarkable that the FFP alignment-free phylogenies from the nongenic genomes yield the type-I topology, the same as that from whole genome. This indicates that coding sequences are not an absolute requirement for tracing the true species tree. This view was also tested by Nikolaev et al. (6), whose results show that a small, select fraction of the noncoding region called “conserved noncoding sequences” (CNCs) or conserved coding sequences can serve equally well as phylogenetic markers. The CNCs are present in intronic and nongenic regions and account for roughly <3% of the entire genome (4, 28). Our alignment-free method reveals that whole, noncoding genomes accurately reconstruct the same phylogenomic topology, suggesting that CNCs are not the only noncoding sequences that contain evolutionary footprints. Furthermore, because as few as 10% of the features can be randomly sampled from either the intronic or nongenic partitions and used to build a highly supported consensus tree (see Jackknife Validation Tests with FFP), the phylogenetic signal must be fairly evenly distributed throughout the whole genome.
Effect of Evolution Rate on Phylogeny.
The earliest phylogenies of mammals based on mitochondrial genes yielded a type-II topology, which is also the most common topology observed among individual gene-tree phylogenies. The prevalence of rodent-basal type-II trees in the literature (29, 30) may be due to the limited and preferential selection of genes where the murid lineage has acquired saturating mutations more quickly than Laurasiatherian mammals. However, the rodent–carnivore controversy is still a matter of debate. For example, a recent studying using a different method based on breakpoint graphs showed a type-II topology (31). Differences among species or gene nucleotide substitution rates can cause the faster-evolving lineage to migrate toward the outgroup [i.e., long branch attraction (32)]. Rodents have been shown to have the highest rates of coding-sequence substitutions when compared with primates and Laurasiatherians (33). Note, the murid speed-up directly conflicts with the concept of a universal mammalian molecular clock (34). The speed-up may be partially explained by the large difference between murid and primate generation times. Murid rodents reach sexual maturity in 5–6 weeks, female chimpanzees at ≈11 years (35) and female rhesus monkeys at between 2.5 and 4 years (36). Ideally, branch lengths should be normalized by generation times.
RY Coding Reduces Compositional Bias.
The two letter RY scheme we employ in the FFP method might at first glance appear to be an overreduction in the complexity of the sequence. However, it has been shown to improve results in phylogenomic analysis. There are three principle advantages to using RY coding with the FFP method. (i) RY coding provides a means of reducing the greater part of the computer resource burden. Longer feature lengths may be used because of the reduction in the size of the feature space and the feature frequencies can be tallied for large mammalian genomes very quickly. (ii) In cases where compositional bias (a known form of systematic error) is present, RY coding has been found to be very useful for increasing the ratio of the evolutionary/nonevolutionary signal (37, 38). For example, RY coding suffices to remove the murid compositional bias in individual gene trees (Table S1), and this coding scheme seems reasonable because of the characterized differences in murid DNA-repair processes (e.g., re. 38). (iii) The rates of transition to transversion can often be two to one in vertebrate genomes, and, also, transition rates can vary highly between species, more so than transversion rates (40). Furthermore, RY coding for whole-genome analysis is also justifiable, especially because the overabundance of evolutionary information in the whole-genome sequence more than overcomes the reduction in the complexity of the sequence by RY coding.
Rare Genomic Changes in FFP.
Poux et al. (17) and Nishihara et al. (23) have both used evidence from rare genomic insertions and deletions to support the existence of unified clades consisting of Archonta + Glires and Perrisodactyla + Carnivora. The FFP method also can analyze rare genomic changes, but on a global, whole-genome scale. The insertion/deletion (indel) events are handled passively in FFP, without special consideration or even prior knowledge of the location of each feature within the genome. These events are accounted for merely by the sliding frame implementation of feature counting. The FFP method is able to characterize changes such as indel events because the original features present in the ancestor and the new features formed by an indel event are reflected in the frequency profile and the JS divergence score. In MSA-based methods of comparison, indels require special treatment, both in tuning of gap penalties and in how alignment gaps will be weighted in the tree reconstruction. By default, some MSA methods ignore gaps in the alignment, (i.e., the gap is treated as an unknown nucleotide). In the Phylip implementation of parsimony, gaps are considered as a “fifth” nucleotide state, so large gaps are heavily weighted in the parsimony method. Unfortunately, different weightings can lead to different tree topologies. So with MSA we must decide, arbitrarily, how important the gap is relative to other characters in the ultimate phylogeny. Citing a limited number of rare genomic changes as phylogenetic evidence does, however, come with a caveat. It is possible that incomplete lineage sorting can give support for a false topology. If speciation occurs before fixation of the allele containing the insertion in the population, derived species may lack the feature. A more robust approach is to consider the FFP profiles collected from all of the insertions through whole-genome comparison. A whole-genome comparison contains a signal derived from all of the insertion events.
Interpreting Feature Changes in Evolutionary Distance.
It is difficult to associate branch lengths in our alignment-free trees with specific divergence times. The JS divergences in our model are not a formal evolutionary distance. However, the two concepts are clearly related. Although JS divergences cannot be directly correlated to evolutionary time, they can be used in the ranking of evolutionary events. Advocates of the use of SINE/LINE insertions as evolutionary phylogenic markers encounter a similar dilemma (22). SINE/LINE insertion analysis cannot currently be applied to branch-length estimation because insertions are most likely episodic events rather than clock-like (21), and the statistical framework for these events has yet to be developed. In the case of FFP, further work would be necessary to develop a model that links feature substitution rates with evolutionary distances. Work by Dermitzakis (28) indicates that conservation patterns associated with conserved nongenic sequences are more like protein-binding sites than coding sequences. As l-mer models have been successfully implemented to classify and compare transcription-factor binding sites (43), it may be possible in the future to develop an evolutionary model, based on features, that is specifically suited to a noncoding sequence.
Conclusion
To summarize, we emphasize the following key points:
A whole genome comparison, including both the genic and nongenic sequence, is representative of the whole genome divergence, which may reflect the divergence of an organism better than methods based on selected genes. The latter account for a very small fraction of the mammalian whole genome and are subject to sampling effects which can lead to biased results supporting a specific gene phylogeny rather than an organism phylogeny.
The entire collection of noncoding (nonexonic) sequences, such as intergenic regions and introns, contain an evolutionary phylogenic signal.
The signal from nongenic (the whole genome minus the exonic, intronic, and regulatory regions) sequences of mammals on a whole-genome scale is very similar to the evolutionary signal present in exonic and genic regions.
Rare genomic changes, such as indels and retroposon insertions, are represented in FFP. These events constitute a significant portion of the evolutionary signal present in mammalian genomes.
The trees reconstructed by using FFP are bush-like, which is consistent with the hypothesis of a rapid mammalian radiation.
Materials and Methods
Single-Gene Phylogenies and Alignment-Based Phylogenies.
We compared the phylogeny obtained by the FFP method with the established mammalian evolutionary phylogenies and a number of single-gene phylogenies. Thirty-two highly conserved mammalian genes were selected, some of which have been used previously in phylogenies by Madsen et al. (44) and Murphy et al. (8) (Table S1). By using the UCSC genome browser, multiple alignments of coding sequences were obtained for each of these genes. Phylogenic analysis was performed with the Phylip package (45). Sequences in each gene MSA were compared with the Kimura-2 (46) distance (by using dnadist); the phylogenies were constructed with neighbor joining (47), and Platypus serves as an outgroup. The tree topologies were examined before and after translation to two-letter alphabet, RY bases (see FFP Alignment-Free Genome Comparison), and placed into one of two dominant tree topologies (Table S1). A tree from Prasad et al. (see figure 1 of ref. 11) was also used for comparison after the species not used in this work were pruned from the tree. Prasad's tree, as well as representatives of type I and type II, is shown in Fig. 2.
FFP Alignment-Free Genome Comparison.
All of the genomic partitions were compared with one another by using the FFP alignment-free method. We have described this method elsewhere (3), but we give a brief description in the supplementary method section that is more relevant for this work.
FFP Tree Building, Optimal Feature Length (l), and Tree Convergence.
The different forms of genome partitions were compared between species by using the FFP method, and NJ trees were constructed from the JS divergence matrix from each type of genome partition (Fig. 2). We have determined from previous research (3) that there is an optimal range of l for features for mammalian genome comparison, which can be estimated by (i) the length of the genomes and (ii) the relative sequence conservation among genomes. An empirical method for finding the optimal l range is to observe when tree topologies begin to converge on a single topology as l is increased; beyond the optimal length range, topologies again become more divergent. The topological distance between trees is evaluated with the Robinson–Foulds (RF) distance (48). Fig. 3 shows a topological convergence plot for each of the genome partitions. Here the RF distance is calculated between tree topologies for l and l−1.
Jackknife Validation Tests with FFP.
A form of jackknife validation test was used to assess the robustness of each tree topology for lengths l = 11–24. We also use this test to determine how uniformly the phylogenetic signal is distributed throughout each genome partition. In the case of MSA, characters within an alignment are sampled without replacement to form a number of replicate alignments. Sampling for a single replicate continues until the number of characters sampled is some fraction of the total alignment, and then all of the sampled characters are replaced. For the FFP method, we have applied a form where each feature (after low-complexity filtering) has a fixed 10% probability of being sampled for each replicate. High-frequency filtering is applied individually to each replicate and then normalized to form an FFP. A JS divergence matrix, D, is calculated for each subset of features, and then a neighbor-joining tree is constructed. A consensus tree was then built from the forest of trees by using Consense from the Phylip package applying extended majority rule. The support values <100% are indicated in the internal nodes of Fig. 2 for l = 18. Many of the features are redundant, by virtue of the sliding window frame used in the method. Although each replicate is randomized, many of the features are not entirely independent of each other. An assessment of feature correlation is described in the SI Materials and Methods.
Supplementary Material
Acknowledgments.
We are grateful to Drs. Kevin Rowe and Susan P. Holmes for their expert advice and discussion. This work was supported by National Institutes of Health Grant GM62412 and the Korean Ministry of Science and Technology (World Class University Project R31–2008-000–10086–0).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909377106/DCSupplemental.
Note: This assumption is debatable. Several well-known examples of character dependence exist. See Dixon and Hillis (40) for an example.
References
- 1.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng J, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 3.Sims GE, Jun SR, Wu GA, Kim SH. Alignment-free genome comparison with feature frequency profiles (FFP) and optimal resolutions. Proc Natl Acad Sci USA. 2009;106:2677–2682. doi: 10.1073/pnas.0813249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dermitzakis ET, et al. Evolutionary discrimination of mammalian conserved nongenic sequences (CNGs) Science. 2003;302:1033–1035. doi: 10.1126/science.1087047. [DOI] [PubMed] [Google Scholar]
- 5.Kriegs JO, et al. Retroposed elements as archives for the evolutionary history of placental mammals. PLoS Biol. 2006;4:e91. doi: 10.1371/journal.pbio.0040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nikolaev S, et al. Early history of mammals is elucidated with the encode multiple species sequencing data. PloS Genetics. 2007;5:e2. doi: 10.1371/journal.pgen.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nikaido M, et al. Maximum likelihood analysis of the complete mitochondrial genomes of eutherians and a reevalution of the phylogeny of bats and insectivores. J Mol Evol. 2001;53:508–516. doi: 10.1007/s002390010241. [DOI] [PubMed] [Google Scholar]
- 8.Murphy WJ, et al. Molecular phylogenetics and the origins of placental mammals. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- 9.Huerta-Cepas J, Dopazo H, Dopazo J, Gabaldon T. The human phylome. Genome Biol. 2007;8:R109. doi: 10.1186/gb-2007-8-6-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huerta-Cepas J, Bueno A, Dopazo J, Gabaldon T. PhylomeDB: A database for genome-wide collections of gene phylogenies. Nucleic Acids Res. 2008;36:D491–D496. doi: 10.1093/nar/gkm899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prasad AB, Allard MW. Confirming the phylogeny of mammals by the use of large comparative sequence data sets. Mol Biol Evol. 2008;25:1795–1808. doi: 10.1093/molbev/msn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhn RM, et al. The UCSC genome browser database: 2008 update. Nucleic Acids Res. 2008;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorne JL, Kishino H. Freeing phylogenies from artifacts of alignment. Mol Biol Evol. 1992;9:1148–1162. doi: 10.1093/oxfordjournals.molbev.a040783. [DOI] [PubMed] [Google Scholar]
- 14.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venkatesh B, Ning Y, Brenner S. Late changes in splicesomal introns define clades in vertebrate evolution. Proc Natl Acad Sci USA. 1999;96:10267–10271. doi: 10.1073/pnas.96.18.10267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matthee CA, et al. Indel evolution of mammalian introns and the utility of noncoding nuclear markers in eutherian phylogenetics. Mol Phylogenet Evol. 2007;42:827–837. doi: 10.1016/j.ympev.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Poux C, van Rheede T, Madsen O, de Jong WW. Sequence gaps join mice and men: Phylogenetic evidence from deletions in two proteins. Mol Biol Evol. 2002;19:2035–2037. doi: 10.1093/oxfordjournals.molbev.a004028. [DOI] [PubMed] [Google Scholar]
- 18.Giordano J, et al. Evolutionary history of mammalian transposons determined by genome-wide defragmentations. PLoS Comp Biol. 2007;3:e137. doi: 10.1371/journal.pcbi.0030137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas JW, et al. Comparative analyses of multi-species sequence from targeted genomic regions. Nature. 2003;424:788–793. doi: 10.1038/nature01858. [DOI] [PubMed] [Google Scholar]
- 20.Nishihara H, et al. A retroposon analysis of Afrotherian phylogeny. Mol Biol Evol. 2005;22:1823–1833. doi: 10.1093/molbev/msi179. [DOI] [PubMed] [Google Scholar]
- 21.Shedlock AM, Okada N. SINE insertions: Powerful tools for molecular systematics. Bioessays. 2000;22:148–160. doi: 10.1002/(SICI)1521-1878(200002)22:2<148::AID-BIES6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 22.Rokas A, Holland PWH. Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol. 2000;11:454–459. doi: 10.1016/s0169-5347(00)01967-4. [DOI] [PubMed] [Google Scholar]
- 23.Nishihara H, Hasegawa M, Okada N. Pegasoferae an unexpected mammalian clade revealed by tracking ancient retroposon insertions. Proc Natl Acad Sci USA. 2006;103:9929–9934. doi: 10.1073/pnas.0603797103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Springer MS, Stanhope MJ, Madsen O, de Jong WW. Molecules consolidate the placental mammal tree. Trends Ecol Evol. 2004;19:430–438. doi: 10.1016/j.tree.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Penny D, Foulds LR, Hendy MD. Testing the theory of evolution by comparing phylogenetic trees constructed from five different protein sequences. Nature. 1982;297:197–200. doi: 10.1038/297197a0. [DOI] [PubMed] [Google Scholar]
- 26.Jeffroy O, Brinkmann H, Delsuc F, Philippe H. Phylogenomics: The beginning of incongruence? Trends Genet. 2006;22:225–231. doi: 10.1016/j.tig.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 27.Murphy WJ, Pevzner PA, O'Brien SJ. Mammalian phylogenomics comes of age. Trends Genet. 2004;20:631–639. doi: 10.1016/j.tig.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Dermitzakis ET, Reymond A, Antonarakis SE. Conserved nongenic sequences: An unexpected feature of mammalian genomes. Nat Rev Genet. 2005;6:151–157. doi: 10.1038/nrg1527. [DOI] [PubMed] [Google Scholar]
- 29.Kullberg M, Nilsson MA, Arnason U, Harley EH, Janke A. Houskeeping genes for phylogenetic analysis of eutherian relationships. Mol Biol Evol. 2006;23:1493–1503. doi: 10.1093/molbev/msl027. [DOI] [PubMed] [Google Scholar]
- 30.Misawa K, Janke A. Revisting the Glires concept—phylogenetic analysis of nuclear sequences. Mol Phylogenet Evol. 2003;28:320–327. doi: 10.1016/s1055-7903(03)00079-4. [DOI] [PubMed] [Google Scholar]
- 31.Alekseyev Max A., Pevzner Pavel A. Breakpoint graphs and ancestral genome reconstructions. Genome Res. 2009;19:943–957. doi: 10.1101/gr.082784.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Syst Zool. 1978;27:401–410. [Google Scholar]
- 33.Zhang J. Rates of conservation and radical nonsynonymous nucleotide substitutions in mammalian nuclear genes. J Mol Evol. 2000;50:56–68. doi: 10.1007/s002399910007. [DOI] [PubMed] [Google Scholar]
- 34.Kumar S, Subramanian S. Mutation rates in mammalian genomes. Proc Natl Acad Sci USA. 2002;99:803–808. doi: 10.1073/pnas.022629899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodall J. The Chimpanzees of Gombe: Patterns of Behavior. Cambridge, MA: Harvard Univ Press; 1986. [Google Scholar]
- 36.Nowak RM. Walker's Mammals of the World. 5th Ed. Vol 1. Baltimore: Johns Hopkins Univ Press; 1990. [Google Scholar]
- 37.Phillips MJ, Delsuc F, Penny D. Genome-scale phylogeny and the detection of systematic biases. Mol Biol Evol. 2004;21:1455–1458. doi: 10.1093/molbev/msh137. [DOI] [PubMed] [Google Scholar]
- 38.Woese CR Achenbach L, Rouviere P, Madelco L. Archael phylogeny: Reexamination of the phylogenetic position of archaeoglobus fulgidus in light of certain composition-induced artifacts. Syst Appl Microbiol. 1991;14:364–371. doi: 10.1016/s0723-2020(11)80311-5. [DOI] [PubMed] [Google Scholar]
- 39.Op het Veld CW, Van Hees-Stuivenberg S, van Zeeland AA, Jansen JG. Effect of nucleotides excision repair on hprt gene mutations in rodent cells exposed to DNA ethylating agents. Mutagenesis. 1997;12:417–424. doi: 10.1093/mutage/12.6.417. [DOI] [PubMed] [Google Scholar]
- 40.Collins DW, Jukes TH. Rate of transition and transversion in coding sequences since the human-rodent divergence. Genomics. 1994;20:386–396. doi: 10.1006/geno.1994.1192. [DOI] [PubMed] [Google Scholar]
- 41.Dixon MT, Hillis DM. Ribosomal RNA secondary structure: Compensatory mutations and implications for phylogenetic analysis. Mol Biol Evol. 1993;10:256–267. doi: 10.1093/oxfordjournals.molbev.a039998. [DOI] [PubMed] [Google Scholar]
- 42.Rokas A, Carroll SB. Bushes in the tree of life. PLoS Biol. 2006;4:e352. doi: 10.1371/journal.pbio.0040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu J, Luo L, Zhang Y. Distance conservation of transcription regulatory motifs in human promoters. Comp Biol Chem. 2008;23:433–437. doi: 10.1016/j.compbiolchem.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 44.Madsen O, et al. Parallel adaptive radiations in two major clades of placental mammals. Nature. 2001;409:610–614. doi: 10.1038/35054544. [DOI] [PubMed] [Google Scholar]
- 45.Felsenstein J. PHYLIP—Phylogeny inference package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 46.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 47.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 48.Robinson DR, Foulds LR. Comparison of phylogenetic trees. Math Biosci. 1981;53:131–147. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.