Abstract
Primary open-angle glaucoma (POAG) is the second leading cause of blindness worldwide. Although a number of genetic loci have shown association or genetic linkage to monogenic forms of POAG, the identified genes and loci do not appear to have a major role in the common POAG phenotype. We seek to identify genetic loci that appear to be major risk factors for POAG in the Afro-Caribbean population of Barbados, West Indies. We performed linkage analyses in 146 multiplex families ascertained through the Barbados Family Study of Glaucoma (BFSG) and identified a strong linkage signal on chromosome 2p (logarithm of odds score = 6.64 at θ = 0 with marker D2S2156). We subsequently performed case-control analyses using unrelated affected individuals and unaffected controls. A set of SNPs on chromosome 2p was evaluated in two independent groups of BFSG participants, a discovery group (130 POAG cases, 65 controls) and a replication group (122 POAG cases, 65 controls), and a strong association was identified with POAG and rs12994401 in both groups (P < 3.34 E−09 and P < 1.21E−12, respectively). The associated SNPs form a common disease haplotype. In summary, we have identified a locus with a major impact on susceptibility to the common POAG phenotype in an Afro-Caribbean population in Barbados. Our approach illustrates the merit of using an isolated population enriched with common disease variants as an efficient method to identify genetic underpinning of POAG.
Keywords: association study, linkage analysis, SNP, eye, intraocular pressure
Primary open-angle glaucoma (POAG) is a group of optic neuropathies that share a common slow progressive degeneration of retinal ganglion cells and their axons, resulting in a distinct appearance of the optic disc and concomitant pattern of visual loss (1). The biological basis of the disease is not well understood, and the factors contributing to its progression are not fully characterized. Increasing age, African ancestry, family history, and elevated intraocular pressure (IOP) are leading risk factors for POAG. High IOP is the only proven treatable risk factor known to date.
Over 66 million individuals worldwide are estimated to have glaucoma (2), and POAG is the most prevalent type. In POAG, there is no identifiable cause for abnormal resistance to aqueous humor outflow through the major drainage apparatus of the eye (the trabecular meshwork). POAG is a particular problem in individuals of African descent, with estimates of prevalence ranging from 4% to 9% among African-Americans and Afro-Caribbeans, respectively, compared with 1–2% in groups of European descent (3, 4). Many clinical studies have documented the familial aggregation of POAG. First-degree relatives of individuals with POAG have a risk 7–10 times greater than that of the general population (5, 6), and there is a high concordance between monozygotic twins (7). A growing number of genome-wide linkage scans have been performed to look for glaucoma susceptibility loci. Family-based linkage and sibling-pair studies have shown the association of several loci with POAG in various families presenting a monogenic autosomal dominant trait (1, 8–13), however, no individual genes in any of these regions have been cloned to show a role in the pathogenesis of common POAG. Despite this lack of conclusive findings, the number of loci associated with POAG provides strong evidence for the polygenic nature of the disease.
In addition to these genomic regions, three genes have been found for POAG presenting in a monogenic, autosomal dominant fashion. However, these three genes are likely to account for <10% of all POAG cases. Myocilin (MYOC) was demonstrated to cause a severe form of juvenile-onset open-angle glaucoma associated with very high IOPs (14) and is inherited in an autosomal dominant fashion. MYOC mutations, however, have only been found in 3–5% of adult-onset POAG patients. The MYOC protein is secreted into the trabecular meshwork extracellular matrix (ECM) and interacts with ECM components. Another gene, optineurin (OPTN, GLC1E), has been associated with “normal-tension glaucoma,” that is, POAG without an elevated IOP (15). Finally, mutations in WDR36 have been associated with both “high” and “normal-tension” POAG (GLC1G) (12), however, the function of this gene is not well established.
One factor that might impede progress in the identification of major genetic contributors to POAG may be the presence of genetic heterogeneity within populations. The study of heterogeneous groups may yield spurious associations as a result of population stratification. This concern is minimized, however, in relatively homogeneous groups with low admixture such as the population of Barbados, West Indies. Barbados is a Caribbean nation spanning 21 miles in length and 14 miles in width, with a population of approximately one-quarter million. The inhabitants of this country descended primarily from West Africa. Unlike many other Caribbean islands, Barbados has no indigenous population, remains fairly homogenous and has limited European admixture (16). These characteristics were thought to be advantageous in detecting genetic ancestral variants. For this reason, the Barbados Family Study of Glaucoma (BFSG) was conceived to evaluate the genetic contribution to POAG in this population of African origin, which was known to have high rates of glaucoma (17, 18).
The study population and methods of the BFSG have been described elsewhere (17). The participants include families, isolated cases, and unaffected controls. Approximately one-quarter of relatives within the families were found to have glaucoma and a segregation analysis suggested that POAG in this population could be explained by a major codominant gene (19). A whole genome-wide linkage scan in 146 families with POAG from the BFSG was also performed (20). Dominant, dominant with age adjustment, and codominant models were used in analyzing the genome-wide scan. Suggestive two-point logarithm of odds (LOD) scores were found on chromosomes 1, 2, 9–11, and 14 (20).
As part of ongoing studies related to linkage investigations for previously shown monogenic POAG loci (21), we performed focused genotyping using STR and SNP markers in the candidate regions implicated in a monogenic form of POAG, distinct from the regions identified in the original genome-wide scan by Nemesure et al. (20). Chromosome 2p16 harbors a Mendelian locus for autosomal dominant POAG in a Chinese family (22), and in seven Caucasian families (23). Initially linkage was ascertained with microsatellite markers within this region, and when linkage was established, tagging SNPs covering haplotype blocks in the linked region were assessed for linkage and association studies. We show that chromosome 2q16 harbors a major locus for POAG in the Afro-Caribbean population of Barbados.
Results
Genetic Linkage and Association Studies.
Focused genotyping on the chromosome 2p16 region among 146 BFSG families yielded a maximum LOD score of 6.67 at θ = 0.01 for D2S2156 under a codominant model (Table 1). Since subsequent association results yielded the greatest odds ratio for homozygous risk alleles (Table 2), the analysis was repeated using an autosomal recessive model producing a suggestive maximum LOD score of 2.76 at θ = 0.16 for rs1533428. Further analysis using a dominant model yielded a maximum LOD score of 8.511 at θ = 0.18 for D2S2156. An additional significant positive LOD score was obtained for the codominant model with SNPs rs1533428 (Zmax = 3.38 at θ = 0), whereas the autosomal dominant LOD score obtained with this marker was 2.73 at θ = 0.18. D2S391 and D2S337 set the limits of the linked region.
Table 1.
STR and SNP markers and LOD scores for BFSG glaucoma families (n = 146)
| Marker | bp | cM | 0 | 0.01 | 0.05 | 0.1 | 0.2 | 0.3 | 0.4 | Zmax | θmax |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Codominant model | |||||||||||
| D2S391* | 46,265,004 | 73.8 | −3.0 | −2.8 | −2.0 | −1.3 | −0.5 | −0.2 | −0.1 | 0.00 | 0.5 |
| D2S2156 | 51,111,121 | 77.1 | 6.6 | 6.7 | 6.6 | 6.1 | 4.4 | 2.3 | 0.6 | 6.67 | 0.01 |
| rs1533428 | 51,870,909 | 3.4 | 3.4 | 3.2 | 2.8 | 1.9 | 0.9 | 0.2 | 3.38 | 0.00 | |
| rs12994401 | 51,983,897 | 0.8 | 0.8 | 0.8 | 0.8 | 0.6 | 0.3 | 0.1 | 0.81 | 0.05 | |
| D2S337* | 61,523,435 | 84.1 | −5.7 | −5.2 | −3.5 | −2.2 | −0.8 | −0.3 | −0.1 | 0.00 | 0.5 |
| Autosomal recessive model | |||||||||||
| D2S391* | 46,265,004 | 73.8 | −19.9 | −17.4 | −11.1 | −6.6 | −2.2 | −0.4 | −0.2 | 0.00 | 0.50 |
| D2S2156 | 51,111,121 | 77.1 | −17.7 | −14.5 | −7.5 | −3.1 | 0.5 | 1.2 | 0.8 | 0.30 | 1.19 |
| rs1533428 | 51,870,909 | −1.3 | −0.5 | 1.3 | 2.4 | 2.6 | 1.8 | 0.9 | 2.76 | 0.16 | |
| rs12994401 | 51,983,897 | −7.8 | −6.3 | −2.9 | −0.8 | 0.7 | 0.8 | 0.6 | 0.29 | 0.87 | |
| D2S337* | 61,523,435 | 84.1 | −32.8 | −28.4 | −18.3 | −11.3 | −4.1 | −1.1 | 0.1 | 0.42 | 0.12 |
| Autosomal dominant model | |||||||||||
| D2S391* | 46,265,004 | 73.8 | −26.8 | −22.4 | −13.6 | −8.0 | −2.7 | −0.7 | −0.2 | 0.00 | 0.5 |
| D2S2156 | 51,111,121 | 77.1 | −10.8 | −5.8 | 3.2 | 7.4 | 8.5 | 5.7 | 1.8 | 8.51 | 0.18 |
| rs1533428 | 51,870,909 | −2.2 | −1.0 | 1.3 | 2.5 | 2.7 | 1.7 | 0.5 | 2.73 | 0.18 | |
| rs12994401 | 51,983,897 | −6.9 | −5.7 | −2.9 | −1.3 | 0.0 | 0.2 | 0.1 | 0.23 | 0.33 | |
| D2S337* | 61,523,435 | 84.1 | −37.8 | −31.1 | −17.6 | −9.6 | −2.5 | −0.3 | 0.0 | 0.00 | 0.5 |
Table 2.
Association results of rs1533428 and rs12994401 from POAG cases and controls by Fisher's exact test
| Snps | rs1533428 |
rs12994401 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | Discovery group |
Replication group |
Combined group |
Discovery group |
Replication group |
Combined group |
||||||
| Case | Control | Case | Control | Case | Control | Case | Control | Case | Control | Case | Control | |
| Phenotype (n) | 127 | 64 | 116 | 65 | 243 | 129 | 127 | 64 | 122 | 64 | 249 | 128 |
| Risk allele (T) frequency, % | 59.06 | 30.47 | 57.76 | 33.08 | 58.44 | 31.78 | 41.34 | 12.50 | 41.39 | 7.81 | 41.37 | 10.16 |
| Genotypic P Values | 6.16E−06 | 2.24E−05 | 1.58E−10 | 1.50E−08 | 3.86E−08 | 9.76E−16 | ||||||
| Allelic P values | 1.55E−07 | 6.75E−06 | 4.90E−12 | 3.34E−09 | 1.21E−12 | 1.84E−20 | ||||||
| Recessive P values | 3.32E−06 | 6.24E−06 | 4.07E−11 | 7.22E−09 | 2.75E−08 | 2.39E−16 | ||||||
| Dominant P values | 1.1E−03 | 3.46E−02 | 8.79E−05 | 2.7E−03 | 2.52E−06 | 2.98E−08 | ||||||
| ORhom | 7.86 | 5.74 | 6.7 | 33.15 | 36.67 | 34.93 | ||||||
| [3.11, 19.85] | [2.35, 13.99] | [3.57, 12.73] | [4.42, 248.60] | [4.88, 275.27] | [8.40, 145.24] | |||||||
| ORhet | 1.6 | 0.98 | 1.27 | 0.67 | 1.855 | 1.083 | ||||||
| [0.79, 3.22] | [0.48, 2.00] | [0.77, 2.08] | [0.29, 1.55] | [0.74, 4.63] | [0.59, 1.99] | |||||||
| PAR, % | 41.11 | 36.88 | 39.07 | 32.96 | 36.43 | 34.74 | ||||||
Multipoint analysis for the complex POAG trait was not carried out directly with this heterogeneous mixture of families. However, admixture analysis was performed for D2S2156 under the autosomal dominant and codominant models using the HOMOG program (24), giving maximum hLODs of 10.43 at θ = 0.02 and α = 0.45 and 6.97 at θ = 0 and α = 0.8 respectively. For both of these models five families were estimated to be of the linked type with P > 0.95 (P = 0.947 for family 34076 under the codominant model, supporting information (SI) Fig. S1A). Admixture analysis of D2S2156 under the recessive model did not yield a significant hLOD score. However, analysis of rs1533428 gave a hLOD score of 3.45 at θ = 0 and α = 0.45, although the only family linked with P > 0.95 is family 34066. Results obtained when these families were analyzed under the codominant and autosomal dominant models are shown in Table S1. The maximum LOD score with the linked families for D2S2156 under the codominant model fell to 5.3 at θ = 0, whereas the maximum LOD score under the autosomal dominant model increased to 9.9 at θ = 0. Interestingly, the maximum LOD score for these five families obtained with rs1533428 was 2.4 under both the codominant and autosomal dominant models, whereas the flanking microsatellite markers remained negative (Table S1). A multipoint analysis of these 5 families using the autosomal dominant model yielded a maximum LOD score of 9.6 at D2S2156 (Fig. S1B, Table S1). The LOD score remained positive throughout the linked interval, falling to 0.7 at D2S391 and −2.5 at D2S337. Under the codominant model these families yielded a maximum multipoint LOD score of 5.4 at D2S2156 but remained positive at the flanking markers.
To refine the region of interest, we then performed case and control association analyses using two sets of BFSG participants (a discovery set and a replication set). The discovery set included 130 unrelated affected individuals and 65 unrelated unaffected controls. We chose tagging SNPs centered on the highest linkage signal, D2S2156, for this analysis and alleles at both rs1533428 and rs12994401 demonstrated a significant association by Fisher exact test (P < 1.55E−07 and 3.34E−09, respectively, Table 2) and χ2 test (P < 1.33E−07 and 1.07E−08 respectively, Table S2). We then confirmed these results in a replication set of 122 BFSG participants affected with POAG and 65 unaffected controls ascertained independently from the study. The results for both SNPs were also significant in this second set, with P < 6.75E−06 for rs1533428 and P < 1.21E−12 for rs12994401 by Fisher exact test (Table 1). Overall, the combined sets showed highly significant associations for rs1533428 and rs12994401 for POAG by Fisher exact test (with P < 4.48E−12 for rs1533428 and P < 1.46E−18 for rs12994401, Table 2) and χ2 test (with P < 4.48E−12 for rs1533428 and P < 1.46E−18 for rs12994401, Table S2). Both markers were in Hardy-Weinberg equilibrium (HWE) in the controls of both sets, but affected individuals showed significant deviation from HWE (ranging from P < 8.50E−05 to P < 6.06E−19, Table S2). This may be due to a founder effect and the very high association of the T allele of rs1533428 and rs12994401 with POAG in this population. The combined odds ratio for heterozygotes was 1.27 (0.77–2.08) for rs1533428 and 1.08 (0.59–1.99) for rs12994401. For homozygotes, the combined odds ratio was 6.70 (3.53–12.73) for rs1533428 and 34.93 (8.40–145.24) for rs12994401 (Table 2).
Haplotype Analysis.
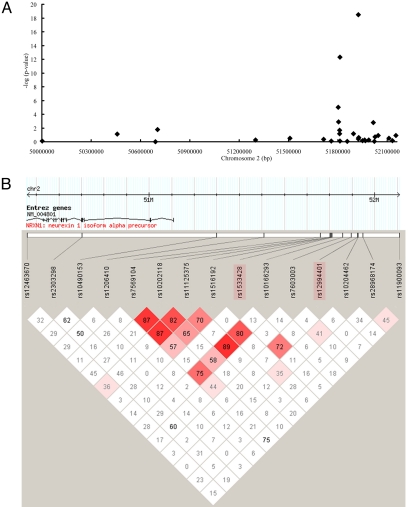
To refine the association and survey the haplotype structures within this region, we genotyped 28 additional SNPs scanning haplotype blocks over a 2-Mb region surrounding D2S2156 in both the BFSG cases and controls (Fig. 1, Fig. S2, Table S3). Among the additional SNPs, rs10202118 (P < 1.5−E05) showed association that remained significant after Bonferroni correction. When testing all 30 SNPs, rs11125375 (P < 2.1−E03), rs10208467 (P < 1.4−E02), rs2303298 (P < 2.6−E02), rs12463670 (P < 4.5−E02), and rs11889995 (P < 5.3−E03) showed associations that, although suggestive, did not withstand correction for multiple testing, and the remainder did not show significant association. Most notably, associated SNPs were clustered around 51.8–51.9 Mbp and SNPs outside this limited region do not show significant association. SNP rs1533428 showed a high linkage disequilibrium (LD) value with rs12994401 (D′ = 0.72, Figure. 1B), but did not show LD values (D′ < 0.3) with other tested markers in the region. SNP rs10202118, which also showed association with POAG (allelic P < 1.50E−05) shows only minor LD with rs1533428 and rs12994401 (D′ = 0.15 and 0.20, respectively). We found a disease-associated haplotype, TT, of SNPs rs1533428 and rs1294401, associated with POAG in the BFSG cases and controls (37.2% in cases, 5.0% in controls, P < 1.088E−21), and also a protective haplotype CC (38.0% in cases, 63.1% in controls, P < 4.1025E−11, Table 3).
Fig. 1.
Association of SNPs in chromosome 2 with POAG. (A) −Log p-values (y axis) from association analyses for 30 SNPs in the chromosome 2 region. Black diamonds represent −logp values of all Barbados case and control SNPs. (B) The pairwise D′ and R2 Hapview plot for SNPs in chromosome 2 using these BFSG samples.
Table 3.
Haplotype analysis of rs1533428 and rs12994401
| Haplotype | Frequency | Case, control ratio counts | Case, control frequencies | χ 2 | P |
|---|---|---|---|---|---|
| CC | 0.465 | 191.4: 312.6, 164.1: 95.9 | 0.380, 0.631 | 43.564 | 4.1025E−11 |
| TT | 0.262 | 187.4: 316.6, 13.1: 246.9 | 0.372, 0.050 | 91.55 | 1.088E−21 |
| TC | 0.229 | 105.2: 398.8, 69.6: 190.4 | 0.209, 0.268 | 3.365 | 0.0666 |
| CT | 0.044 | 20.0: 484.0, 13.2: 246.8 | 0.040, 0.051 | 0.516 | 0.4724 |
Population Stratification Analysis.
Population stratification can induce false-positive associations in case-control studies (25). This effect has been minimized by our choice of case-control groups derived from the limited, distinct geographic area of Barbados. The strong positive signal in the family linkage study in the associated region on chromosome 2, which should be unaffected by a population stratification bias, strongly supports the existence of a gene with a major effect on susceptibility to POAG.
We assessed stratification in our cases and controls using 28 ancestry informative markers (AIMS) shown previously to be effective in detecting population structure in African-Caribbean populations (16). The results indicated that there is no substructure in the cases and controls (P > 0.153, χ2 value is 14.451, 10 d.f., Fst = 0.005, Fig. S3). This is consistent with previous report that the Barbados population is a genetically homogenous (16).
Discussion
Here, we report another locus for common POAG in an Afro-Caribbean population in Barbados. This locus is supported both by linkage analysis with microsatellite and SNP markers, and association studies with specific alleles and haplotypes of SNP markers. It is localized to a region that overlaps a Mendelian locus in a Chinese family with autosomal dominant POAG (22). It is also localized to a region that overlaps with a previous linkage study of seven families (23). Thus, this locus appears to be a significant cause of glaucoma in individuals of African, Chinese, and European origin.
The microsatellite markers D2S391 and D2S337 flanking the linked region show negative linkage to POAG, yielding similar results to the original genome wide linkage scan by Nemesure et al. (20), in which only loci showing evidence of linkage with LOD score >1 were explicitly reported. The negative results for D2S391 and D2S337 at θ = 0.05 initially appear to be inconsistent with the localization of D2S2156 ≈5 cM from one and 7 cM from the other. However, these values are calculated under the codominant model with an assumption that the families form a homogeneous group (that is, although there may be several loci contributing to glaucoma in this population, each locus contributes to POAG equally in all families). This is an approximation to the actual situation, and although the codominant model minimizes this problem at smaller recombination fractions, this is not the case at larger true recombination fractions (e.g., the values of the flanking markers D2S391 and D2S337) in which the positive contributions of linked families are overpowered by negative contributions of unlinked families. Despite its limitations, the codominant model provides a good approximation for POAG in the Barbados population and was used in preference to model free methods for its greater power in both the present and original linkage analyses (20, Table S4).
Our data indicate that a locus for POAG with a complex trait inheritance overlaps with one for monogenic POAG. This suggests that severe mutations in the target gene might cause early-onset high-pressure glaucoma inherited in a Mendelian fashion in some families, whereas milder mutations might increase the risk of glaucoma in the Afro-Caribbean general population. A similar pattern has been seen with GALK1 in cataracts, where complete deficiency results in galactosemia with congenital cataracts. Individuals heterozygous for galactokinase deficiency are at increased risk for presenile cataracts, whereas at least one GALK1 allele results in 20% residual activity due to a shortened protein half-life. This allele, the Osaka variant, increases risk of age-related cataracts in Japanese populations (26, 27).
A survey of the chromosome 2p16 region reveals several genes including neurexin, LOC730100, γG-crystallin pseudogene 1, and LOC129656. Neurexin 1 is a large presynaptic cell-surface protein expressed exclusively in the nervous system. It functions in mediating synaptic Ca2+ channel function (28). Although this is a logical candidate gene, sequencing of exons and the 5′ flanking region in a subset of samples with the risk alleles from the association study revealed no plausible functional changes. LOC730100 is a hypothetical gene encoding a predicted protein without known conserved domains but with some homology to tankyrase (TRF1-interacting ankyrin-related ADP-ribose polymerase), which has a role in the control of telomere length (29). The γG-crystallin pseudogene is a transcriptionally inactive 285 bp ORF, of which 141 bp are similar to part of exon 2 of the γD-crystallin gene. Even though it is preceded by an AG dinucleotide and followed by GT, it is not spliced in test systems and appears to be a silent remnant of a γ-crystallin gene (30). LOC129656 is a hypothetical gene with a predicted protein similar to the CREB regulated transcription coactivator 1, CRTC1. The two associated SNPs are not located within or near a gene, making functional studies challenging. However, they may perform a regulatory role influencing neighboring gene expression. Alternatively, one cannot exclude the possibility that these two SNPs are located within a yet-to-be-annotated gene.
A major strength of the BFSG is its large sample size, including more than 140 Afro-Caribbean families as well as isolated affected POAG cases and unrelated unaffected controls. Although the size of the family set used in this study is adequate, the present investigation is limited by having a relatively small dataset for association analyses, especially with regard to the number of unaffected controls. As a result, the odds ratios are approximate and the confidence intervals are wide. All study participants received a comprehensive and standardized examination and a high degree of data completeness was achieved (17). Glaucoma status was based on conservative, objective and uniformly defined criteria. An inherent limitation of studies of this type is the possibility of misclassification among individuals who have not yet manifested symptoms of glaucoma at the time of examination. In addition, selection biases may be present if affected family members participated disproportionately compared to unaffected relatives. However, because more than one-quarter of family members with POAG were newly detected by the study and association analyses corroborated the linkage results, such biases, if they do exist, are not as likely in this study.
POAG is a blinding condition without known cure. Once damage has occurred, visual impairment cannot be reversed. Therefore, early diagnosis and treatment are essential to prevent further glaucomatous optic neuropathy. Although the variants described here can substantially alter an individual's risk of glaucoma in the Afro-Caribbean population of BFSG, these findings may have broader implications for public health and understanding etiology. Thus, identification of the specific gene or sequence changes responsible for the increased risk may lead to increased benefit in screening and follow-up programs. Additionally, these findings may to provide insight into the pathophysiological cause of glaucoma in this and other populations.
Materials and Methods
Study Population.
This study was approved by the Institutional Review Boards of University of California at San Diego, Stony Brook University Medical Center, the Barbados Ministry of Health, University of Utah, and the National Institutes of Health Combined Neuroscience Institutional Review Boards. All participants provided informed consent before participation in the study. The BFSG is a follow-up to the Barbados Eye Study (BES), 1988–1992 and the Barbados Incidence Study of Eye Diseases (BISED), 1992–1997. All eligible probands were individuals of self-reported African descent who met the study criteria for POAG, as confirmed in BFSG following an established protocol (17). This POAG definition required the presence of specific signs of glaucomatous optic nerve damage plus visual field defects, as well as an ophthalmologic evaluation (Table S5). IOP was not considered in this definition.
Probands were identified from eligible BES and BISED participants with POAG and from the Glaucoma Clinic of the Queen Elizabeth Hospital, Bridgetown, Barbados. Recruitment was targeted to first degree relatives of the proband and was extended to any additional family members of those found to have POAG and their first-degree relatives. A total of 235 families were recruited, of which 146 were used in linkage studies after exclusion of families for small size and unclear paternity. Probands without any eligible family members were recruited for separate genetic analyses. Unaffected spouses and unaffected BES or BISED participants without a family history of glaucoma were recruited to serve as the controls for analyses of association. All participants received a comprehensive examination including anthropometric and blood pressure measurements, best corrected visual acuity based on the ETDRS chart, Humphrey perimetry with the C64 suprathreshold program, C24-2 and C30-2 full-threshold programs, applanation tonometry, pupil dilation, lens gradings with the Lens Opacities Classification System II (LOCSII) (31) at the slit lamp, and color stereo fundus photographs of the disc and macula. All participants additionally received a comprehensive examination by the study's ophthalmologists and an interview including medical, ocular, and family history information. A blood sample of 14 mL was obtained from all available members in each family, including affected and unaffected individuals, and was used for DNA isolation. The recruitment of BFSG families has been detailed in ref. 17.
Association studies were carried out with affected patients and unaffected control individuals drawn from the 235 family linkage studies (16 families had 2 affected individuals unrelated genetically) (251 affected and 37 unaffected spouses) and from a series of unrelated individuals (1 affected and 93 unaffected) ascertained through the Glaucoma Clinic of the Queen Elizabeth Hospital, Bridgetown, Barbados. A single affected individual was ascertained from each family who met the BFSG POAG criteria fully as detailed in table 1 of Nemesure et al. (20) and Table S5). These criteria require an individual to have both visual field and optic disc damage to be considered affected for the purposes of the association study. Unaffected individuals showed none of the signs of glaucoma listed in table 1 of Nemesure et al.
The mean ages of the affected individuals ascertained through the family study (71 ± 12) was about 8–10 years higher than both the unaffected individuals ascertained through the family study (63 ± 11) or the unaffected individuals ascertained through Queen Elizabeth Hospital (61 ± 9). Although elevated IOP is not a criteria in the BFSG, the mean IOP of affected individuals was 26.7 ± 9.6 while the mean IOP of unaffected individuals ascertained from the family study was 17.0 ± 3.7 and the mean age of unaffected individuals ascertained through Queen Elizabeth Hospital was 20.0 ± 3.5 (Table S6).
Genotyping.
A total of 252 BFSG participants with POAG were genotyped, and allele frequencies were compared with 130 BFSG controls without a history of glaucoma by lab personnel masked to case/control status. All SNPs were genotyped by using the SNaPshot method according to the manufacturer's recommendations. In brief, a SNP was amplified by PCR, and the PCR product was purified by Exo I and Shrimp Alkaline Phosphatase (SAP) (New England Biolabs). The purified PCR product and the SNaPshot primer were used to perform the SNaPshot reaction (single base pair extension) with the SNaPshot multiplex mix (ABI). After an additional purification step using SAP, the product was run and analyzed on an ABI 3130xl genetic analyzer (ABI) and the genotyping results were obtained directly. Some SNPs were also additionally assayed using the Taqman PCR SNP genotyping assay (Applied Biosystems, www.appliedbiosystems.com/).
Genotyping results on rs1533428 and rs12994401 were independently genotyped and verified, in two separate laboratories, by using the SNaPshot method, Taqman assay, and direct DNA sequencing. All SNPs (30 total) were genotyped in both the discovery and replication groups. SNPs showing a significant P value for association were genotyped to a minimum successful rate of 95%, whereas SNPs not showing significant association were not brought to a 99% call rate unless genotyping of the remaining ungenotyped individuals could result in a P value <0.05. The AIMS were genotyped by using the SNaPshot method, direct DNA sequencing, conventional agarose gel electrophoresis, and 15% acrylamide gel electrophoresis with ethidium bromide staining.
Linkage Analysis.
For linkage analysis, DNA was extracted directly from blood by standard phenol-chloroform protocols (32). Multiplex PCRs were carried out as previously described (33). Briefly, each reaction was carried out in a 5-μL mixture containing 40 ng of genomic DNA, various combinations of 10 μM dye-labeled primer pairs, 0.5 μL of 10× GeneAmp PCR Buffer II, 0.5 μL of l10 mM Gene Amp dNTP mix, 2.5 mM MgCl2 and 0.2 units of TaqDNA polymerase (AmpliTaq Gold Enzyme; Applied Biosystems. Amplification was performed in a GeneAmp PCR System 9700 (PE; Applied Biosystems). Initial denaturation was carried out for 5 min at 95 °C, followed by 10 cycles of 15 sec at 94 °C, 15 sec at 55 °C, and 30 sec at 72 °C and then 20 cycles of 15 sec at 89 °C, 15 sec at 55 °C, and 30 sec at 72 °C. The final extension was performed for 10 min at 72 °C and followed by a final hold at 4 °C. PCR products from each DNA sample were pooled and mixed with a loading mixture containing HD-400 size standards (PE; Applied Biosystems) and loading dye. The resulting PCR products were separated on an ABI 3130 DNA analyzer and analyzed by using the GENEMAPPER 3.1 software package (PE; Applied Biosystems). Two-point and multipoint linkage analyses were performed with the FASTLINK implementation of the MLINK program of the LINKAGE program package (34, 35). Admixture analysis was performed with the HOMOG program (36).
Statistical Analysis.
The Fisher exact test and the χ2 test for different models over genotypes or alleles were performed to assess evidence for association. Odds ratios and 95% confidence intervals were also calculated to estimate risk size for the heterozygotes and homozygotes for the risk alleles using logistical regression (SPSS v13.0). Linkage disequilibrium (LD), tagging SNPs, and HWE were examined using Haploview v3.32. For the risk genotypes identified, we calculated population attributable risks (PAR) using the Levin formula (37). Additional statistics were calculated using Exemplar v. 4.07 (Sapio Sciences). SNPs were selected by increasing the distance from D2S2156 until the associated region and its limits were identified.
Two-point linkage analysis was performed using the FASTLINK (38) version of MLINK from the LINKAGE Program Package (39). Maximum LOD scores were calculated by using ILINK. POAG was analyzed using the gene frequencies and penetrance values used previously in the BFSG genome wide scan (codominant model) (20). Marker allele frequencies were estimated by counting alleles of 100 unrelated unaffected individuals (200 chromosomes) of Afro-Caribbean Barbadian ethnicity.
Stratification Analysis.
Population stratification in 252 cases and 130 controls was assessed using 28 AIMS previously shown to be effective for identifying population structure and potential admixture in African-Caribbean populations (16). Using the EIGENSTRAT software package, we identified 41 samples (14 cases, 27 controls) as “outliers” along the first 10 principal components. These samples were removed, yielding a dataset with very little stratification between cases and controls due to differences in ancestry (FST = 0.005, Fig. S3). Because of the limited number of markers, EIGENSTRAT was not used to adjust p values in the association analysis.
Supplementary Material
Acknowledgments.
We thank the BFSG (the names of investigators are listed below) and other POAG participants and the staff of the F.H. and K.Z. laboratories. This work was supported by grants from the National Institutes of Health (to M.C.L., J.H., J.F.H., and K.Z.), the Foundation Fighting Blindness (to K.Z.), Research to Prevent Blindness (to K.Z.), and the Burroughs Wellcome Fund Clinical Scientist Award in Translational Research (to K.H.). Members of the BFSG are: M.C.L. (Principal Investigator), B.N., Qimei He, Suh-Yuh Wu, Nancy Mendell, Lixin Jiang, and Koumudi Manthani (Coordinating Center, Stony Brook University); A.H., M. Ann Bannister, Muthu Thangaraj, Rajiv Luthra, Coreen Barrow, and Anthanette Holder (Data Collection Center, Ministry of Health, Bridgetown, Barbados, West Indies; Andrew P. Schachat, Judith A. Alexander, Deborah Phillips, and Reva Ward-Strozykowski (Fundus Photography Reading Center, The Johns Hopkins University, Baltimore, MD); J.F.H. and X.J. (Laboratory Center, National Eye Institute); Trevor Hassell, and Henry Fraser (Faculty of Medicine Sciences, University of the West Indies, Bridgetown, Barbados); and Clive Gibbons (Queen Elizabeth Hospital, Bridgetown, Barbados) (Advisory Group).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907564106/DCSupplemental.
References
- 1.Weinreb RN, Khaw PT. Primary open-angle glaucoma. Lancet. 2004;363(9422):1711–1720. doi: 10.1016/S0140-6736(04)16257-0. [DOI] [PubMed] [Google Scholar]
- 2.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90(3):262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leske MC, Connell AM, Schachat AP, Hyman L. The Barbados Eye Study. Prevalence of open angle glaucoma. Arch Ophthalmol. 1994;112:821–829. doi: 10.1001/archopht.1994.01090180121046. [DOI] [PubMed] [Google Scholar]
- 4.Tielsch JM, et al. Racial variations in the prevalence of primary open-angle glaucoma. The Baltimore Eye Survey [see comments] J Am Med Assoc. 1991;266:369–374. [PubMed] [Google Scholar]
- 5.Wolfs RC, et al. Genetic risk of primary open-angle glaucoma. Population-based familial aggregation study. Arch Ophthalmol. 1998;116(12):1640–1645. doi: 10.1001/archopht.116.12.1640. [DOI] [PubMed] [Google Scholar]
- 6.Drance SM, Schulzer M, Thomas B, Douglas GR. Multivariate analysis in glaucoma. Use of discriminant analysis in predicting glaucomatous visual field damage. Arch Ophthalmol. 1981;99(6):1019–1022. doi: 10.1001/archopht.1981.03930011019007. [DOI] [PubMed] [Google Scholar]
- 7.Teikari JM. Genetic factors in open-angle (simple and capsular) glaucoma. A population-based twin study. Acta Ophthalmol. 1987;65(6):715–720. doi: 10.1111/j.1755-3768.1987.tb07069.x. [DOI] [PubMed] [Google Scholar]
- 8.Allingham RR, et al. Early adult-onset POAG linked to 15q11-13 using ordered subset analysis. Invest Ophthalmol Vis Sci. 2005;46(6):2002–2005. doi: 10.1167/iovs.04-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wirtz MK, et al. Mapping a gene for adult-onset primary open-angle glaucoma to chromosome 3q. Am J Hum Genet. 1997;60(2):296–304. [PMC free article] [PubMed] [Google Scholar]
- 10.Trifan OC, et al. A third locus (GLC1D) for adult-onset primary open-angle glaucoma maps to the 8q23 region. Am J Ophthalmol. 1998;126(1):17–28. doi: 10.1016/s0002-9394(98)00073-7. [DOI] [PubMed] [Google Scholar]
- 11.Wirtz MK, et al. GLC1F, a new primary open-angle glaucoma locus, maps to 7q35–q36. Arch Ophthalmol. 1999;117(2):237–241. doi: 10.1001/archopht.117.2.237. [DOI] [PubMed] [Google Scholar]
- 12.Monemi S, et al. Identification of a novel adult-onset primary open-angle glaucoma (POAG) gene on 5q22.1. Hum Mol Genet. 2005;14(6):725–733. doi: 10.1093/hmg/ddi068. [DOI] [PubMed] [Google Scholar]
- 13.Stoilova D, et al. Localization of a locus (GLC1B) for adult-onset primary open angle glaucoma to the 2cen-q13 region. Genomics. 1996;36(1):142–150. doi: 10.1006/geno.1996.0434. [DOI] [PubMed] [Google Scholar]
- 14.Stone EM, et al. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275(5300):668–670. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- 15.Rezaie T, et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295(5557):1077–1079. doi: 10.1126/science.1066901. [DOI] [PubMed] [Google Scholar]
- 16.Benn-Torres J, et al. Admixture and population stratification in African Caribbean populations. Ann Hum Genet. 2008;72(Pt 1):90–98. doi: 10.1111/j.1469-1809.2007.00398.x. [DOI] [PubMed] [Google Scholar]
- 17.Leske MC, et al. Patterns of open-angle glaucoma in the Barbados Family Study. Ophthalmology. 2001;108(6):1015–1022. doi: 10.1016/s0161-6420(01)00566-8. [DOI] [PubMed] [Google Scholar]
- 18.Leske MC. Open-angle glaucoma—An epidemiologic overview. Ophthalmic Epidemiol. 2007;14(4):166–172. doi: 10.1080/09286580701501931. [DOI] [PubMed] [Google Scholar]
- 19.Nemesure B, et al. Inheritance of open-angle glaucoma in the Barbados family study. Am J Med Genet. 2001;103(1):36–43. doi: 10.1002/ajmg.1498. [DOI] [PubMed] [Google Scholar]
- 20.Nemesure B, et al. A genome-wide scan for primary open-angle glaucoma (POAG): the Barbados Family Study of Open-Angle Glaucoma. Hum Genet. 2003;112(5–6):600–609. doi: 10.1007/s00439-003-0910-z. [DOI] [PubMed] [Google Scholar]
- 21.Yao W, et al. Evaluation of the association between OPA1 polymorphisms and primary open-angle glaucoma in Barbados families. Mol Vis. 2006;12:649–654. [PubMed] [Google Scholar]
- 22.Lin Y, et al. A genome-wide scan maps a novel autosomal dominant juvenile-onset open-angle glaucoma locus to 2p15-16. Mol Vis. 2008;14:739–744. [PMC free article] [PubMed] [Google Scholar]
- 23.Suriyapperuma SP, et al. A new locus (GLC1H) for adult-onset primary open-angle glaucoma maps to the 2p15–p16 region. Arch Ophthalmol. 2007;125(1):86–92. doi: 10.1001/archopht.125.1.86. [DOI] [PubMed] [Google Scholar]
- 24.Gusella JF, et al. A polymorphic DNA marker genetically linked to Huntington's disease. Nature. 1983;306(5940):234–238. doi: 10.1038/306234a0. [DOI] [PubMed] [Google Scholar]
- 25.Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat Genet. 2004;36(5):512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- 26.Stambolian D, Scarpino-Myers V, Eagle RC, Jr, Hodes B, Harris H. Cataracts in patients heterozygous for galactokinase deficiency. Invest Ophthalmol Vis Sci. 1986;27(3):429–433. [PubMed] [Google Scholar]
- 27.Okano Y, et al. A genetic factor for age-related cataract: identification and characterization of a novel galactokinase variant, “Osaka,” in Asians. Am J Hum Genet. 2001;68(4):1036–1042. doi: 10.1086/319512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Missler M, et al. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423(6943):939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- 29.Donigian JR, de Lange T. The role of the poly(ADP-ribose) polymerase tankyrase1 in telomere length control by the TRF1 component of the shelterin complex. J Biol Chem. 2007;282(31):22662–22667. doi: 10.1074/jbc.M702620200. [DOI] [PubMed] [Google Scholar]
- 30.Brakenhoff RH, Aarts HJ, Reek FH, Lubsen NH, Schoenmakers JG. Human gamma-crystallin genes. A gene family on its way to extinction. J Mol Biol. 1990;216(3):519–532. doi: 10.1016/0022-2836(90)90380-5. [DOI] [PubMed] [Google Scholar]
- 31.Chylack LT, et al. Lens opacities classification system II (LOCS II) Arch Ophthalmol. 1989;107:991–997. doi: 10.1001/archopht.1989.01070020053028. [DOI] [PubMed] [Google Scholar]
- 32.Smith GL, Sansone C, Socransky SS. Comparison of two methods for the small-scale extraction of DNA from subgingival microorganisms. Oral Microbiol Immunol. 1989;4(3):135–140. doi: 10.1111/j.1399-302x.1989.tb00240.x. [DOI] [PubMed] [Google Scholar]
- 33.Jiao X, et al. Genetic linkage of Bietti crystallin corneoretinal dystrophy to chromosome 4q35. Am J Hum Genet. 2000;67(5):1309–1313. doi: 10.1016/s0002-9297(07)62960-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cottingham RW, Jr, Idury RM, Schaffer AA. Faster sequential genetic linkage computations. Am J Hum Genet. 1993;53(1):252–263. [PMC free article] [PubMed] [Google Scholar]
- 35.Lathrop GM, Lalouel JM. Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet. 1984;36(2):460–465. [PMC free article] [PubMed] [Google Scholar]
- 36.Ott J. Linkage analysis and family classification under heterogeneity. Ann Hum Genet. 1983;47(Pt 4):311–320. doi: 10.1111/j.1469-1809.1983.tb01001.x. [DOI] [PubMed] [Google Scholar]
- 37.Levin ML. The occurrence of lung cancer in man. Acta Unio Int Contra Cancrum. 1953;9(3):531–541. [PubMed] [Google Scholar]
- 38.Cottingham RW, Idury RM, Schaffer AA. Faster sequential genetic linkage computations. Am J Hum Genet. 1993;53:252–263. [PMC free article] [PubMed] [Google Scholar]
- 39.Lathrop GM, Lalouel JM. Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet. 1984;36:460–465. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.