Abstract
The MADS domain homeotic proteins APETALA1 (AP1), APETALA3 (AP3), PISTILLATA (PI), and AGAMOUS (AG) combinatorially specify the identity of Arabidopsis floral organs. AP1/AP1, AG/AG, and AP3/PI dimers bind to similar CArG box sequences; thus, differences in DNA-binding specificity among these proteins do not seem to be the origin of their distinct organ identity properties. To assess the overall contribution that specific DNA binding could make to their biological specificity, we have generated chimeric genes in which the amino-terminal half of the MADS domain of AP1, AP3, PI, and AG was substituted by the corresponding sequences of human SRF and MEF2A proteins. In vitro DNA-binding assays reveal that the chimeric proteins acquired the respective, and distinct, DNA-binding specificity of SRF or MEF2A. However, ectopic expression of the chimeric genes reproduces the dominant gain-of-function phenotypes exhibited by plants ectopically expressing the corresponding Arabidopsis wild-type genes. In addition, both the SRF and MEF2 chimeric genes can complement the pertinent ap1-1, ap3-3, pi-1, or ag-3 mutations to a degree similar to that of AP1, AP3, PI, and AG when expressed under the control of the same promoter. These results indicate that determination of floral organ identity by the MADS domain homeotic proteins AP1, AP3, PI, and AG is independent of their DNA-binding specificity. In addition, the DNA-binding experiments show that either one of the two MADS domains of a dimer can be sufficient to confer a particular DNA-binding specificity to the complex and that sequences outside the amino-terminal basic region of the MADS domain can, in some cases, contribute to the DNA-binding specificity of the proteins.
Full text
PDF





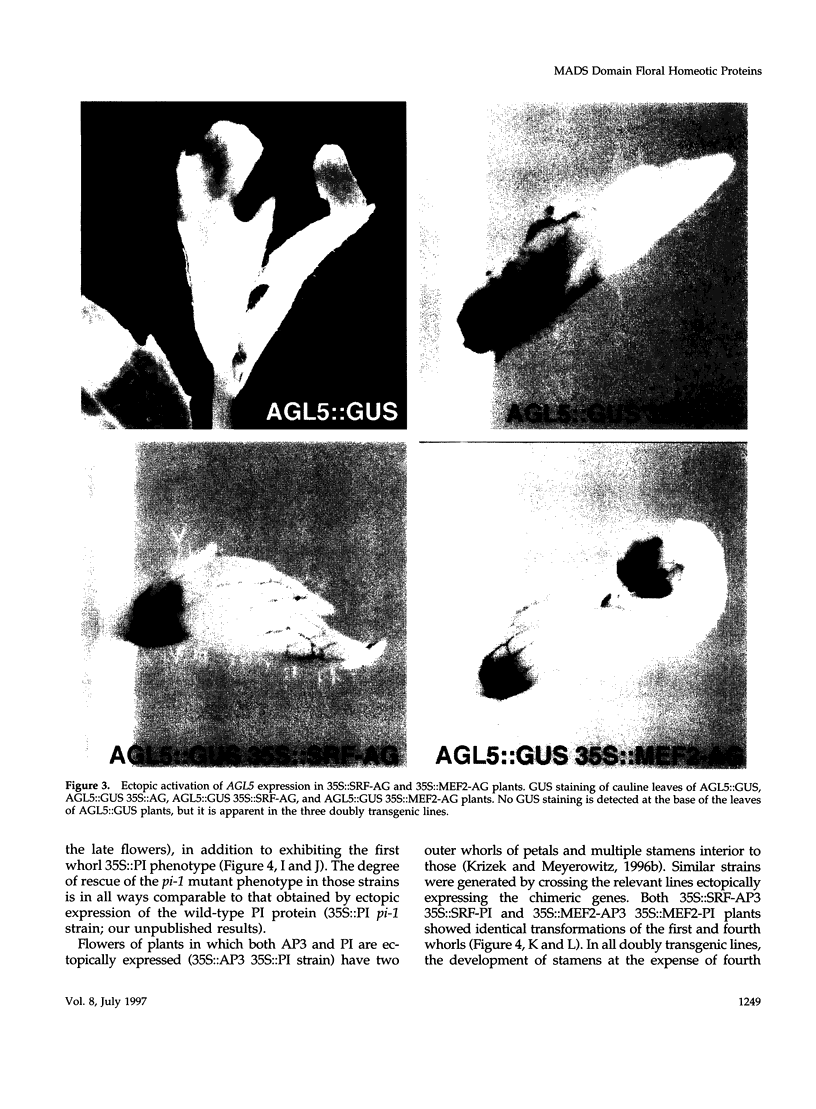
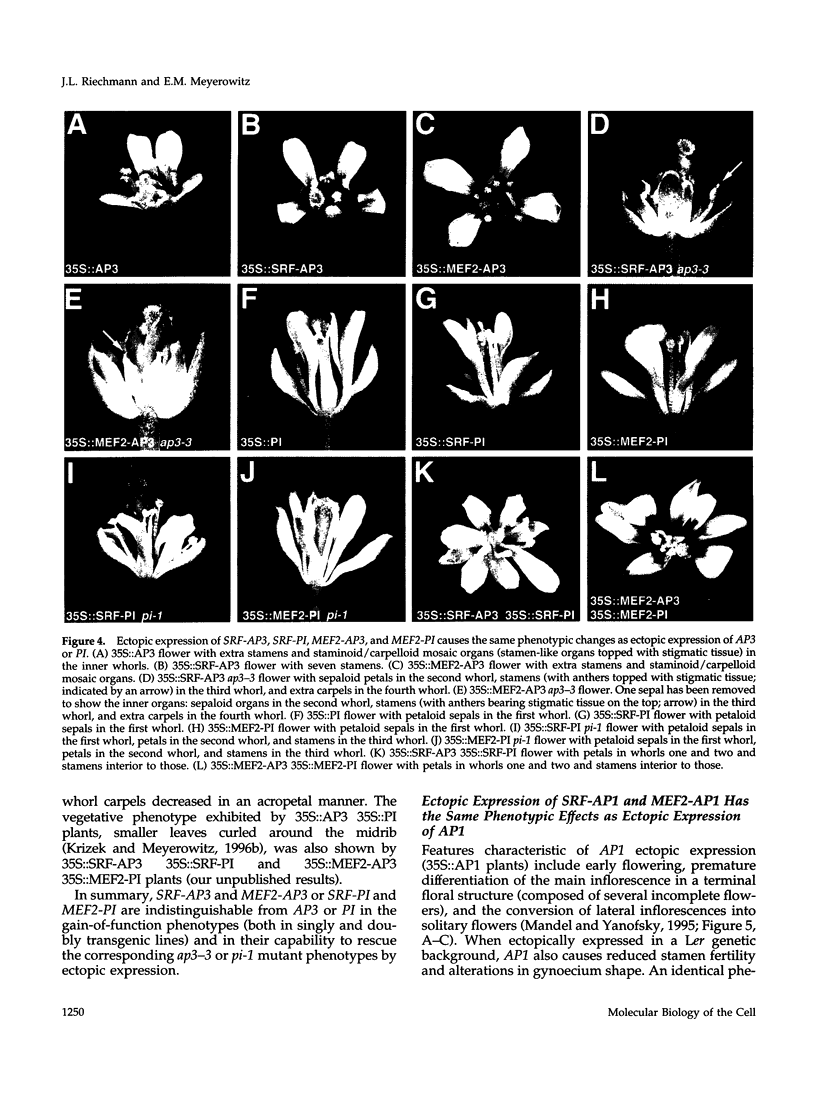
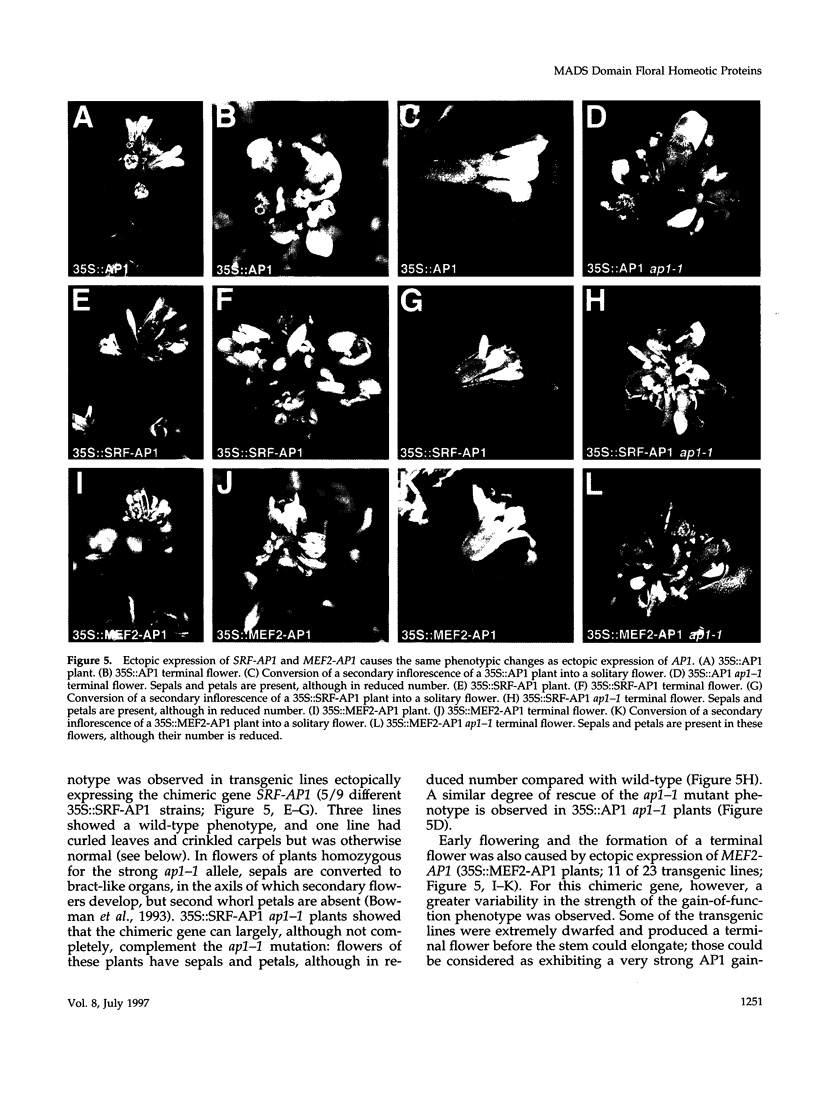







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bowman J. L., Smyth D. R., Meyerowitz E. M. Genes directing flower development in Arabidopsis. Plant Cell. 1989 Jan;1(1):37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. L., Smyth D. R., Meyerowitz E. M. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991 May;112(1):1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- Chan S. K., Jaffe L., Capovilla M., Botas J., Mann R. S. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell. 1994 Aug 26;78(4):603–615. doi: 10.1016/0092-8674(94)90525-8. [DOI] [PubMed] [Google Scholar]
- Chan S. K., Mann R. S. A structural model for a homeotic protein-extradenticle-DNA complex accounts for the choice of HOX protein in the heterodimer. Proc Natl Acad Sci U S A. 1996 May 28;93(11):5223–5228. doi: 10.1073/pnas.93.11.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E. S., Meyerowitz E. M. The war of the whorls: genetic interactions controlling flower development. Nature. 1991 Sep 5;353(6339):31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Davies B., Egea-Cortines M., de Andrade Silva E., Saedler H., Sommer H. Multiple interactions amongst floral homeotic MADS box proteins. EMBO J. 1996 Aug 15;15(16):4330–4343. [PMC free article] [PubMed] [Google Scholar]
- Dessain S., Gross C. T., Kuziora M. A., McGinnis W. Antp-type homeodomains have distinct DNA binding specificities that correlate with their different regulatory functions in embryos. EMBO J. 1992 Mar;11(3):991–1002. doi: 10.1002/j.1460-2075.1992.tb05138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker S. C., von Kessler D. P., Beachy P. A. Differential DNA sequence recognition is a determinant of specificity in homeotic gene action. EMBO J. 1992 Nov;11(11):4059–4072. doi: 10.1002/j.1460-2075.1992.tb05499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell R. B. Inactivation of gene expression in plants as a consequence of specific sequence duplication. Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3490–3496. doi: 10.1073/pnas.91.9.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukubo-Tokunaga K., Müller M., Affolter M., Pick L., Kloter U., Gehring W. J. In vivo analysis of the helix-turn-helix motif of the fushi tarazu homeo domain of Drosophila melanogaster. Genes Dev. 1992 Jun;6(6):1082–1096. doi: 10.1101/gad.6.6.1082. [DOI] [PubMed] [Google Scholar]
- Gentz R., Rauscher F. J., 3rd, Abate C., Curran T. Parallel association of Fos and Jun leucine zippers juxtaposes DNA binding domains. Science. 1989 Mar 31;243(4899):1695–1699. doi: 10.1126/science.2494702. [DOI] [PubMed] [Google Scholar]
- Goto K., Meyerowitz E. M. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 1994 Jul 1;8(13):1548–1560. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- Hai T., Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanes S. D., Riddihough G., Ish-Horowicz D., Brent R. Specific DNA recognition and intersite spacing are critical for action of the bicoid morphogen. Mol Cell Biol. 1994 May;14(5):3364–3375. doi: 10.1128/mcb.14.5.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R. M., Cai Z. L., Ho S. N., Pease L. R. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990 May;8(5):528–535. [PubMed] [Google Scholar]
- Huang H., Mizukami Y., Hu Y., Ma H. Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 1993 Oct 11;21(20):4769–4776. doi: 10.1093/nar/21.20.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Tudor M., Su T., Zhang Y., Hu Y., Ma H. DNA binding properties of two Arabidopsis MADS domain proteins: binding consensus and dimer formation. Plant Cell. 1996 Jan;8(1):81–94. doi: 10.1105/tpc.8.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Tudor M., Weiss C. A., Hu Y., Ma H. The Arabidopsis MADS-box gene AGL3 is widely expressed and encodes a sequence-specific DNA-binding protein. Plant Mol Biol. 1995 Jun;28(3):549–567. doi: 10.1007/BF00020401. [DOI] [PubMed] [Google Scholar]
- Jack T., Brockman L. L., Meyerowitz E. M. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell. 1992 Feb 21;68(4):683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- Jack T., Fox G. L., Meyerowitz E. M. Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. Cell. 1994 Feb 25;76(4):703–716. doi: 10.1016/0092-8674(94)90509-6. [DOI] [PubMed] [Google Scholar]
- Jacobson D. R., Moskovits T. Rapid, nonradioactive screening for activating ras oncogene mutations using PCR-primer introduced restriction analysis (PCR-PIRA) PCR Methods Appl. 1991 Nov;1(2):146–148. doi: 10.1101/gr.1.2.146. [DOI] [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A., Bevan M. W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987 Dec 20;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimyuk V. I., Carroll B. J., Thomas C. M., Jones J. D. Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant J. 1993 Mar;3(3):493–494. doi: 10.1111/j.1365-313x.1993.tb00169.x. [DOI] [PubMed] [Google Scholar]
- Krizek B. A., Meyerowitz E. M. Mapping the protein regions responsible for the functional specificities of the Arabidopsis MADS domain organ-identity proteins. Proc Natl Acad Sci U S A. 1996 Apr 30;93(9):4063–4070. doi: 10.1073/pnas.93.9.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B. A., Meyerowitz E. M. The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development. 1996 Jan;122(1):11–22. doi: 10.1242/dev.122.1.11. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science. 1989 Mar 31;243(4899):1681–1688. doi: 10.1126/science.2494700. [DOI] [PubMed] [Google Scholar]
- Latinkić B. V., Lau L. F. Transcriptional activation of the immediate early gene pip92 by serum growth factors requires both Ets and CArG-like elements. J Biol Chem. 1994 Sep 16;269(37):23163–23170. [PubMed] [Google Scholar]
- Latinkić B. V., Zeremski M., Lau L. F. Elk-1 can recruit SRF to form a ternary complex upon the serum response element. Nucleic Acids Res. 1996 Apr 1;24(7):1345–1351. doi: 10.1093/nar/24.7.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. The unfolding drama of flower development: recent results from genetic and molecular analyses. Genes Dev. 1994 Apr 1;8(7):745–756. doi: 10.1101/gad.8.7.745. [DOI] [PubMed] [Google Scholar]
- Ma H., Yanofsky M. F., Meyerowitz E. M. AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 1991 Mar;5(3):484–495. doi: 10.1101/gad.5.3.484. [DOI] [PubMed] [Google Scholar]
- Mandel M. A., Bowman J. L., Kempin S. A., Ma H., Meyerowitz E. M., Yanofsky M. F. Manipulation of flower structure in transgenic tobacco. Cell. 1992 Oct 2;71(1):133–143. doi: 10.1016/0092-8674(92)90272-e. [DOI] [PubMed] [Google Scholar]
- Mandel M. A., Gustafson-Brown C., Savidge B., Yanofsky M. F. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992 Nov 19;360(6401):273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- Mandel M. A., Yanofsky M. F. A gene triggering flower formation in Arabidopsis. Nature. 1995 Oct 12;377(6549):522–524. doi: 10.1038/377522a0. [DOI] [PubMed] [Google Scholar]
- Mann R. S., Chan S. K. Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 1996 Jul;12(7):258–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- Mann R. S. The specificity of homeotic gene function. Bioessays. 1995 Oct;17(10):855–863. doi: 10.1002/bies.950171007. [DOI] [PubMed] [Google Scholar]
- Matzke M. A., Matzke AJM. How and Why Do Plants Inactivate Homologous (Trans)genes? Plant Physiol. 1995 Mar;107(3):679–685. doi: 10.1104/pp.107.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride K. E., Summerfelt K. R. Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Mol Biol. 1990 Feb;14(2):269–276. doi: 10.1007/BF00018567. [DOI] [PubMed] [Google Scholar]
- Mizukami Y., Ma H. Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell. 1992 Oct 2;71(1):119–131. doi: 10.1016/0092-8674(92)90271-d. [DOI] [PubMed] [Google Scholar]
- Molkentin J. D., Black B. L., Martin J. F., Olson E. N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995 Dec 29;83(7):1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- Norman C., Runswick M., Pollock R., Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988 Dec 23;55(6):989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- Nurrish S. J., Treisman R. DNA binding specificity determinants in MADS-box transcription factors. Mol Cell Biol. 1995 Aug;15(8):4076–4085. doi: 10.1128/mcb.15.8.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Aggarwal A. K., Jordan S. R., Beamer L. J., Obeysekare U. R., Harrison S. C. Conserved residues make similar contacts in two repressor-operator complexes. Science. 1990 Mar 9;247(4947):1210–1213. doi: 10.1126/science.2315694. [DOI] [PubMed] [Google Scholar]
- Pellegrini L., Tan S., Richmond T. J. Structure of serum response factor core bound to DNA. Nature. 1995 Aug 10;376(6540):490–498. doi: 10.1038/376490a0. [DOI] [PubMed] [Google Scholar]
- Percival-Smith A., Müller M., Affolter M., Gehring W. J. The interaction with DNA of wild-type and mutant fushi tarazu homeodomains. EMBO J. 1990 Dec;9(12):3967–3974. doi: 10.1002/j.1460-2075.1990.tb07617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock R., Treisman R. A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res. 1990 Nov 11;18(21):6197–6204. doi: 10.1093/nar/18.21.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock R., Treisman R. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 1991 Dec;5(12A):2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- Purugganan M. D., Rounsley S. D., Schmidt R. J., Yanofsky M. F. Molecular evolution of flower development: diversification of the plant MADS-box regulatory gene family. Genetics. 1995 May;140(1):345–356. doi: 10.1093/genetics/140.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J. L., Krizek B. A., Meyerowitz E. M. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc Natl Acad Sci U S A. 1996 May 14;93(10):4793–4798. doi: 10.1073/pnas.93.10.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J. L., Wang M., Meyerowitz E. M. DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 1996 Aug 15;24(16):3134–3141. doi: 10.1093/nar/24.16.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley S. D., Ditta G. S., Yanofsky M. F. Diverse roles for MADS box genes in Arabidopsis development. Plant Cell. 1995 Aug;7(8):1259–1269. doi: 10.1105/tpc.7.8.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savidge B., Rounsley S. D., Yanofsky M. F. Temporal relationship between the transcription of two Arabidopsis MADS box genes and the floral organ identity genes. Plant Cell. 1995 Jun;7(6):721–733. doi: 10.1105/tpc.7.6.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier A. F., Gehring W. J. Functional specificity of the homeodomain protein fushi tarazu: the role of DNA-binding specificity in vivo. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1450–1454. doi: 10.1073/pnas.90.4.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrocks A. D., Gille H., Shaw P. E. Identification of amino acids essential for DNA binding and dimerization in p67SRF: implications for a novel DNA-binding motif. Mol Cell Biol. 1993 Jan;13(1):123–132. doi: 10.1128/mcb.13.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrocks A. D., von Hesler F., Shaw P. E. The identification of elements determining the different DNA binding specificities of the MADS box proteins p67SRF and RSRFC4. Nucleic Acids Res. 1993 Jan 25;21(2):215–221. doi: 10.1093/nar/21.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi H., Okada K., Shimura Y. Nucleotide sequences recognized by the AGAMOUS MADS domain of Arabidopsis thaliana in vitro. Plant J. 1993 Aug;4(2):385–398. doi: 10.1046/j.1365-313x.1993.04020385.x. [DOI] [PubMed] [Google Scholar]
- Shore P., Sharrocks A. D. The MADS-box family of transcription factors. Eur J Biochem. 1995 Apr 1;229(1):1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- Theissen G., Kim J. T., Saedler H. Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J Mol Evol. 1996 Nov;43(5):484–516. doi: 10.1007/BF02337521. [DOI] [PubMed] [Google Scholar]
- Treisman R. Identification and purification of a polypeptide that binds to the c-fos serum response element. EMBO J. 1987 Sep;6(9):2711–2717. doi: 10.1002/j.1460-2075.1987.tb02564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R., Marais R., Wynne J. Spatial flexibility in ternary complexes between SRF and its accessory proteins. EMBO J. 1992 Dec;11(12):4631–4640. doi: 10.1002/j.1460-2075.1992.tb05565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R. Ternary complex factors: growth factor regulated transcriptional activators. Curr Opin Genet Dev. 1994 Feb;4(1):96–101. doi: 10.1016/0959-437x(94)90097-3. [DOI] [PubMed] [Google Scholar]
- Weigel D., Meyerowitz E. M. The ABCs of floral homeotic genes. Cell. 1994 Jul 29;78(2):203–209. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]
- Williams G. T., Lau L. F. Activation of the inducible orphan receptor gene nur77 by serum growth factors: dissociation of immediate-early and delayed-early responses. Mol Cell Biol. 1993 Oct;13(10):6124–6136. doi: 10.1128/mcb.13.10.6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne J., Treisman R. SRF and MCM1 have related but distinct DNA binding specificities. Nucleic Acids Res. 1992 Jul 11;20(13):3297–3303. doi: 10.1093/nar/20.13.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky M. F., Ma H., Bowman J. L., Drews G. N., Feldmann K. A., Meyerowitz E. M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature. 1990 Jul 5;346(6279):35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]