Abstract
The performance of the Abbott m2000rt RealTime™ HIV-1 assay (RealTime HIV-1) with manual sample preparation was compared against the ROCHE COBAS AmpliPrep/AMPLICOR™ HIV-1 MONITOR Test v1.5 (CAP/CA HIV-1) using samples collected from 100 donors infected with HIV and 20 donors not infected with HIV in northern Tanzania where HIV-1 subtypes A, C, D, and their recombinant forms predominate. The RealTime HIV-1 appeared to have more within-run variability at high HIV-1 RNA concentrations, but total assay variability over the dynamic range tested was within the manufacturer's claim of <0.3 SD copies/mL. Accuracy studies showed 100% concordance for positive and negative values. When continuous values were examined, CAP/CA HIV-1 yielded higher values than the RealTime HIV-1 at higher nominal HIV-1 RNA concentrations. The RealTime-HIV-1 assay showed excellent linearity between 2.5 and 7.0 log copies/mL. Of negative samples, 100% showed negative results, and >95% of samples with nominal concentrations of 40 copies/mL were detected at ≥40 copies/mL by RealTime HIV-1. Manual sample preparation may contribute to higher total assay variability. This study suggests that the Abbott m2000rt RealTime HIV-1 assay with manual sample preparation is an acceptable and feasible alternative to the traditional gold-standard ROCHE COBAS AmpliPrep/AMLICOR HIV-1 Monitor v1.5 assay and that the RealTime HIV-1 assay performs well on samples from East Africa.
Keywords: HIV, reverse transcriptase polymerase chain reaction, viral load, validation studies, Tanzania
1. Introduction
As access to antiretroviral therapy (ART) has expanded in the developing world the detection of treatment failure has become a priority (World Health Organisation, 2006). While treatment failure may be defined using clinical and immunologic criteria, this approach results in detection of treatment failure later than increasing plasma HIV-1 RNA concentration (viral load). Consequently, the World Health Organization (WHO) has indicated that there is a strong argument for moving towards the wider availability of viral load testing in resource constrained settings. In particular, the WHO suggests that simple point-of-care assays are needed (World Health Organization, 2006).
A variety of technologies have been developed and commercialized to measure HIV-1 RNA concentration, including reverse transcriptase polymerase chain reaction (RT-PCR) amplification, isothermal nucleic acid sequence-based amplification (NASBA), and branched-chain DNA signal amplification (bDNA) (Collins et al., 1997; Dyer et al., 1999; Johanson et al., 2001; Sun et al., 1998). The development of real-time RT-PCR assays represents one step towards simpler assays for viral load monitoring. Several recent studies have shown that real-time PCR methods compare favorably with conventional assays for quantitation of HIV-1 RNA (de Mendoza et al., 2005; Rouet et al., 2005; Stevens et al., 2005; Yao et al., 2005).
As in most of East Africa, HIV-1 from patients in Moshi, Tanzania, exhibit considerable subtype diversity with subtypes A, C, and D predominating and the frequent occurrence of HIV-1 recombinant forms (Kiwelu et al., 2003; Osmanov et al., 2002; Ramadhani et al., 2007). By contrast, most HIV-1 from patients in Europe, North and South America, and Australasia belongs to subtype B (Osmanov et al., 2002). A number of studies have shown that HIV-1 diversity can influence the reliability of RNA detection (Barlow et al., 1997; Swanson et al., 2005). Furthermore, inaccurate quantitation can have adverse consequences for patients (Geelen et al., 2003). Therefore, it is essential that as new, simpler assays for measurement of plasma HIV-1 RNA concentration are evaluated in settings where HIV-1 subtype and recombinant form diversity is substantial. The Abbott m2000rt RealTime HIV-1 assay (RealTime HIV-1) (Abbott Laboratories, Abbott Park, IL) offers the potential advantages of the reduced complexity of a real-time assay and, by targeting the pol integrase (IN) region of the HIV-1 genome, may be subject to less variability than assays targeting the gag gene (Geelen et al., 2003; Swanson et al., 2006a).
The RealTime HIV-1 Assay is an RT-PCR assay has been evaluated against the Abbott LCx HIV RNA quantitative assay (Abbot Laboratories, Abbott Park, IL) (Garcia-Diaz et al., 2006; Swanson et al., 2006a; Swanson et al., 2006b), the VERSANT HIV-1 RNA 3.0 (Bayer Diagnostics, Tarrytown, NY) (Swanson et al., 2006a; Swanson et al., 2007), and the ROCHE AMPLICOR HIV-1 MONITOR 1.5 (Roche Molecular Systems, Branchburg, NJ) (Swanson et al., 2006a; Swanson et al., 2007) using patient samples from Brazil (Swanson et al., 2006b), London (Garcia-Diaz et al., 2006; Swanson et al., 2006a), and assorted HIV-1 group M and O isolates (Swanson et al., 2007) and more recently against the NucliSENS HIV-1(Xu et al., 2008). In these evaluations, the Abbott RealTime HIV-1 Assay performed well against other assays, reliably quantifying diverse HIV-1 strains, providing a wide dynamic range and good sensitivity. This study extends the evaluation of the RealTime HIV-1 assay using patient samples from East Africa where subtypes A, C and D, and HIV-1 recombinant forms predominate. Furthermore, the assay was evaluated in a resource poor setting using a simple, manual nucleic acid extraction method. In this study field data were compared directly to data generated in a national reference laboratory using the CAP/CA HIV-1 assay which is commonly used as a reference standard for HIV-1 RNA quantitation.
2. Materials and Methods
2.1 Sample collection and laboratory testing sites
Participants were recruited among consecutive clients at the Kilimanjaro Christian Medical Centre (KCMC) Infectious Diseases Clinic, at a KCMC HIV voluntary counseling and testing clinic, and at the Kikundi cha Wanawake Kilimanjaro Kupambana na UKIMWI (KIWAKKUKI; Women Against AIDS in Kilimanjaro) HIV voluntary counseling and testing clinic, all in Moshi, Tanzania. HIV antibody testing was done according to previously described methods (Mayhood et al., 2008). Forty mL of blood was collected from participants between November 2006 and January 2007 until the goal of 120 samples; 20 (17%) from HIV-uninfected persons and 100 (83%) from HIV-infected persons was reached. Whole blood was collected in tubes containing K3 EDTA. Within 4 hours of collection plasma was separated, divided into four aliquots each of 5mL and frozen at -80°C. Frozen plasma was shipped to the University of Witwatersrand, Johannesburg, South Africa on dry ice. In Johannesburg, plasma was tested using the comparator assay to determine HIV-1 RNA concentration. When necessary to fulfill the requirements of the method validation plan, dilutions of nominal samples were prepared using plasma from donors not infected with HIV. The resulting samples were shipped back to the Kilimanjaro Christian Medical Centre, Moshi, Tanzania, on dry ice and were stored at -80°C before testing.
2.2 HIV-1 RNA concentration determination by RealTime HIV-1 Assay and the CAP/CA HIV-1 comparator assay
The RealTime HIV-1 Assay was performed in the Molecular Section of the Kilimanjaro Christian Medical Centre Biotechnology Laboratory, Moshi, Tanzania according to the manufacturer's specifications. RNA was manually extracted from 1mL plasma samples using the Abbott Sample Preparation System (Abbott Laboratories, Abbott Park, IL). The Tanzania team was blinded to the results of the South Africa testing. The CAP/CA HIV-1 conventional and ultrasensitive assays were performed by the Department of Molecular Medicine and Haematology, University of Witwatersrand, Johannesburg, South Africa, according to the manufacturer's specifications. RNA was extracted from 0.7mL plasma samples using the ROCHE COBAS® AmpliPrep™ System (Roche Molecular Systems, Branchburg, NJ). This laboratory complies with external quality assessment for viral load testing using the external quality assessment programs of both the College of American Pathologists and of the US National Institutes of Health Viral Quality Assessment program (VQA, Rush-Presbyterian-St. Luke's Medical Centre, Chicago, IL).
2.3 Method validation study plan
For within-run precision studies, three runs were performed, each containing replicates of 10 patient samples at high (>5.0 log copies/mL), medium (4.0-5.0 log copies/mL), or low (<4.0 log copies/mL) HIV-1 RNA concentration. Assay controls (low positive, high positive and negative) were included in every run in both Moshi and Johannesburg and results were only accepted if controls were in range according to the standard operating procedures for each assay. For between-run precision studies, runs each containing the same 15 patient samples and 3 control samples were repeated over 3 consecutive days. Accuracy studies used 100 samples from persons infected with HIV run in duplicate. Samples were compiled to represent the range of detection of the RealTime HIV-1 Assay and the patient population in Moshi, Tanzania. For reportable range and linearity experiments, samples were selected from the collection of samples available; one with the closest value to the lower (40 copies/mL) and one with the closest value to the upper (1,000,000 copies/mL) ends of the reportable range for the Abbott m2000rt RealTime™ HIV-1 assay. In addition, four samples representing intermediate values were each run in duplicate. To verify the manufacturer's claims for the limit of blank and reference interval verification studies, plasma samples from 20 patients not infected with HIV were tested. To verify the limit of detection, 20 samples with HIV-1 RNA concentrations of <40 copies/mL were tested over 5 days. The CAP/CA HIV-1 ultrasensitive (reportable range 50-100,000 copies/mL) assay was used to measure these HIV-1 RNA concentrations. Four aliquots were tested per day for 5 days. Operational evaluations included measuring assay time for the Abbott m2000rt RealTime™ HIV-1 assay and for the CAP/CA HIV-1 assay.
2.4 Statistical analyses
For the purpose of this study, results from testing in South Africa were considered to be the reference value. Mean, median, minimum and maximum values were calculated for both assays. Correlation was used to determine the linear relationship between the assays and linear regression to quantitate this relationship. The coefficient of determination (R2) was calculated. Method comparison to determine agreement between the two assays was analyzed by Bland and Altman (Bland and Altman, 1986) and percentage similarity (Scott et al., 2003). All statistical tests were two-sided and all statistical analyses were performed using SAS version 8.2 Enterprise Guide version 2 software (SAS Institute, Inc., Cary, NC) and GraphPad Prism software version 4.02 (GraphPad Software, Inc., San Diego, CA).
2.5 Research ethics
Approval to collect patient samples for this study was granted by the Kilimanjaro Christian Medical Centre Research Ethics Committee.
3. Results
3.1 Instrument familiarization
The RealTime HIV-1 instrument was installed at the Moshi site over a period of one week using existing available bench space. Three laboratory technologists were trained over a subsequent period of two weeks and standard operating procedures were simultaneously developed. Two laboratory technologists each performed successfully three independent training runs before beginning the validation study. One laboratory technologist performed all validation study assays.
3.2 Within-run precision studies
The within-run precision experiment results compared well with the manufacturers' claims. The mean, range, standard deviation and coefficient of variation (CV) of the CAP/CA HIV-1 and of the RealTime HIV-1 Assay are shown in Table 1. Values for the CAP/CA HIV-1 assay were also within the limits of the manufacturer's claims. The manufacturer's claim for the RealTime HIV-1 assay is much tighter and showed some values, especially at log 5copies/mL, with more variability or less precision according to these claims (Table 1). Assay run control values were within limits for both assays. All negative samples gave negative values on all assay runs.
Table 1. Within-run precision studies using patient samples from East Africa, Abbott m2000rt RealTime™ HIV-1 assay with manual sample preparation compared with the ROCHE COBAS® AmpliPrep™/AMPLICOR™ HIV-1 MONITOR® v1.5a.
| Sample run number | ROCHE COBAS® AMPLICOR™ HIV-1 MONITOR® v1.5 | Abbott m2000rt RealTime™ HIV-1 |
|---|---|---|
| 1 (n=10) | ||
| Mean | 5.51 | 5.07 |
| Range (low, high) | (5.43, 5.55) | (4.84, 5.22) |
| SD (manufacturer's claim)b | 0.04 (0.1) | 0.12 (0.04) |
| CV | 0.71% | 2.27% |
| 2 (n=10) | ||
| Mean | 4.32 | 4.20 |
| Range | (4.19, 4.38) | (4.16, 4.24) |
| SD (manufacturer's claim)b | 0.08 (0.14) | 0.03 (0.02) |
| CV | 1.76% | 0.66% |
| 3 (n=10) | ||
| Mean | 3.07 | 3.29 |
| Range | (2.92, 3.31) | (3.15, 3.36) |
| SD (manufacturer's claim)b | 0.13 (0.17) | 0.06 (0.05) |
| CV | 4.31% | 1.85% |
Log transformed HIV-1 RNA values are reported
Standard deviation (manufacturer's claims for total assay variability)
3.3 Between-run precision studies
All samples with continuous values yielded detectable values and no HIV-1 RNA was detected from all negative samples on both assays. After combining the results of between-run studies for each CAP/CA HIV-1 log HIV-1 RNA concentration interval, standard deviations were calculated for the RealTime HIV-1 method and compared with the manufacturer's claims. For CAP/CA HIV-1 log HIV-1 RNA concentration interval 5.0 the RealTime HIV-1 standard deviation (and manufacturer's SD claim) from repeat tests was 0.072 (0.08); for log 4.0 was 0.128 (0.06); for log 3.0 was 0.147 (0.10); and for log 2.0 was 0.07 (0.09). Total variability for CAP/CA HIV-1 log HIV-1 RNA concentration interval 5.0 was 0.192 (0.10); for 4.0 was 0.158 (0.14); and for 3.0 was 0.207 (0.13).
3.4 Accuracy studies
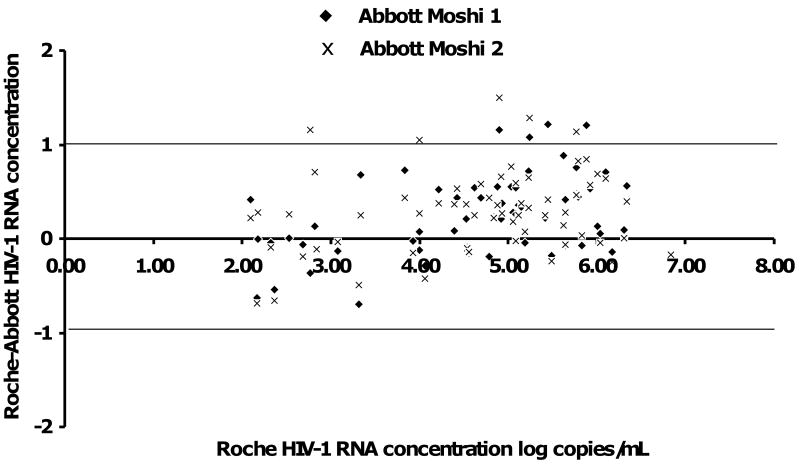
Of 100 patient samples, 39 (39%) were determined to have HIV-1 RNA concentrations <400 copies/mL on the Roche assay. Of the 39 samples in the first Moshi Abbott run, 24 (62%) were not detected, 12 (31%) were detected <40 copies/mL, and 3 (8%) were detected >40 copies/mL. On the second Moshi Abbott run, the only result that was discordant with the first run was that one more sample was detected <40 copies/mL and one less at >40 copies/mL. All methods at all sites showed 100% concordance for discrete values irrespective of the range. Of the 100 patient samples, the remaining 61 (61%) yielded continuous values above 400 copies/mL. Of these 61 samples, the mean (minimum, maximum) log HIV-1 RNA concentration for the CAP/CA HIV-1 method was 4.63 (2.09, 6.83). The mean from the first Moshi RealTime HIV-1 run was 4.39 log copies/mL (1.67, 7.00) and 4.34 log copies/mL (1.60, 7.00) for the second run. The Bland-Altman difference plot for these data shows that the RealTime HIV-1 assay produced higher values than the CAP/CA in the low HIV-1 RNA concentration samples, but lower values on the higher HIV-1 RNA samples (Figure 1). Assay controls passed for each run, with low and high positives within their acceptable ranges and all negative controls yielded negative results on both assays.
Figure 1. Bland-Altman difference plot comparing the Abbott m2000rt RealTime™ HIV-1 assay with manual sample preparation run twice compared with the ROCHE COBAS® AmpliPrep™/AMPLICOR™ HIV-1 MONITOR® v1.5.
3.5 Reportable range and linearity experiments
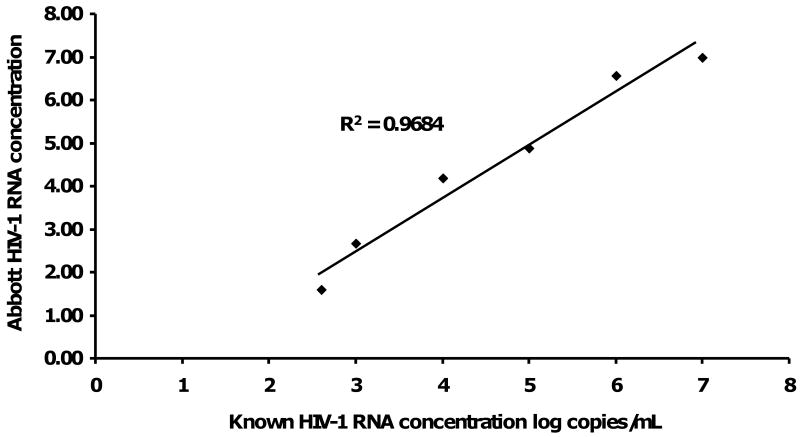
Linearity was determined using known samples at intervals from 2.5 to 7.0 log HIV-1 RNA concentration. HIV-1 RNA levels were established using the CAP/CA HIV-1 assay. The equation of the line for the mean of the two RealTime HIV-1 assay runs was y = -1.09 + 1.21×, (intercept p=0.110); evaluation of correlation with nominal samples yielded an R2 of 0.9684. HIV-1 copy number detected by the RealTime HIV-1 assay is displayed a linear relationship to the known viral concentration over the dynamic range of 2.5 - 7.0 log10 copies/mL (Figure 2).
Figure 2. Linearity plot of mean Abbott m2000rt RealTime™ HIV-1 RNA level against known samples determined by the ROCHE COBAS® AmpliPrep™/AMPLICORTM HIV-1 MONITOR® v1.5.
3.6 Additional verifications
Studies to verify the manufacturer's claims for the limit of blank and reference interval verification studies showed that 20 plasma samples from persons not infected with HIV all were found to have HIV-1 RNA not detected on the ultrasensitive CAP/CA HIV-1 (limit of blank <50 copies/mL). All samples were also HIV-1 RNA not detected on the RealTime HIV-1 Assay. Studies to verify the limit of detection showed that 20 samples with nominal HIV-1 RNA concentrations of 40 copies/mL on the ultrasensitive CAP/CA HIV-1, 19 (95%) had HIV RNA-1 concentrations 40-178 copies/mL and 1 (5%) was <40 copies/mL on the Abbott m2000rt RealTime™ HIV-1 Assay. Total assay time from sample preparation, extraction, amplification and detection was 6 hours for the for the Abbott m2000rt RealTime™ HIV-1 Assay and 8 hours for the CAP/CA HIV-1 assay.
4. Discussion
When compared with the CAP/CA HIV-1, the RealTime HIV-1 assay with manual sample preparation is an acceptable alternative that performs adequately in areas where HIV-1 subtypes and recombinant forms infecting patients in East Africa. Furthermore, this study shows that real-time assays can be established and used for HIV-1 RNA quantitation in resource-constrained settings.
The RealTime HIV-1 assay appeared to have more within-run variability at high HIV-1 RNA concentrations, but total assay variability was within the manufacturer's claim of <0.3 SD copies/mL. Accuracy studies showed 100% concordance for positive and negative values. When continuous values were examined, the CAP/CA HIV-1 assay yielded higher values than the RealTime HIV-1 at higher nominal HIV-1 RNA concentrations. The RealTime HIV-1 assay showed excellent linearity between 2.5 and 7.0 log copies/mL with an R2 value of 0.9684. Of negative samples, 100% showed negative results, and >95% of samples at 40 copies/mL were detected at ≥40 copies/mL.
The manual extraction method and differences between real-time and traditional PCR technologies may contribute to increased variability, as may shipping of samples between Tanzania and South Africa for the validation study. A further contributor to variability may exist due to the differences in detection methodologies; the CAP/CA assay uses endpoint detection whereas the RealTime HIV-1 assay uses real-time detection. The RealTime HIV-1 used a larger input sample volume than the CAP/CA HIV-1 assay which may have contributed further to performance differences. Based on published comparisons of the RealTime HIV-1 with other assays, it is likely that use of the Abbott m2000sp automated sample preparation instrument would reduce the level of total variability observed in this study (Swanson et al., 2006a; Swanson et al., 2007). This question will be addressed through a study currently ongoing in Tanzania.
It was possible to install the RealTime HIV-1 instrument at a site in East Africa in one week and to successfully train staff over two weeks with subsequent independent training runs. The small footprint of the Abbott m2000rt allowed the use of a small amount of existing bench space and manual sample preparation used a similarly small area in another part of the laboratory. Despite the limitations of manual sample preparation, the RealTime HIV-1 assay with manual sample preparation compared favorably to the CAP/CA HIV-1 assay. The lower capital investment and lower complexity of manual sample preparation may make this an attractive option for other laboratories in resource-constrained settings with low to intermediate sample numbers of ≤21 per day (Fiscus et al., 2006).
The RealTime HIV-1 assay offers a number of potential advantages over the CAP/CA HIV-1 assay and other conventional assays. First, the overall turnaround time of real-time assays are shorter than for convention PCR methods. This improves the likelihood that results will be available to clinicians when patients return to clinic, an important consideration in settings where patients travel long distances to receive care. The wider dynamic range of the RealTime HIV-1 assay covers the dynamic range of the CAP/CA ultrasensitive and conventional assays, obviating the need to have more than one assay, and improving cost-effectiveness and work flow. The liquid plasma sample remains a short coming of both real-time and conventional PCR assays and this places constraints on access to the test for patients living in rural and remote areas. The evaluation of more durable dry blood spot samples for viral load monitoring provides a potential solution to this problem and has been the subject of past and ongoing research (Garrido et al., 2009; Leelawiwat et al., 2009).
5. Conclusions
These data suggest that the RealTime HIV-1 assay with manual sample preparation is an acceptable and feasible alternative to the conventional ROCHE COBAS AmpliPrep/AMLICOR HIV-1 Monitor v1.5 assay and that the RealTime HIV-1 assay performs well on samples from East Africa. It is likely that manual sample preparation contributes to higher total assay variability, but this is offset by the lower cost and complexity of manual sample preparation. Further research is needed to compare the RealTime HIV-1 assay with automated sample preparation with non-subtype B samples and evaluating dry blood spot samples as an alternative to liquid plasma samples.
Acknowledgments
The authors thank the staff and patients of the Kilimanjaro Christian Medical Centre Infectious Diseases Clinic and the staff and clients of Kikundi cha Wanawake Kilimanjaro Kupambana na UKIMWI (KIWAKKUKI; Women Against AIDS in Kilimanjaro) for their participation. The authors thank Julitha Kimbi, Devotha Lyimo, and Editha Mushi for assistance with sample collection. This research was supported by the Center for HIV/AIDS Vaccine Immunology, a United States National Institutes of Health (NIH) funded program (U01 AI067854). Authors received additional support from NIH awards International Studies of AIDS-associated Co-infections (ISAAC) (AI 062563 JAC, ABM), and the Duke Clinical Trials Unit and Clinical Research Sites (U01 AI069484-01 JAC). The authors are grateful to Abbott Laboratories for donating reagents for sample testing in Moshi. This publication was made possible by support from the US Agency for International Development (USAID). The contents are the responsibility of the authors and do not necessarily reflect the views of USAID or the US government. The authors acknowledge the National Health Laboratory Service PCR Laboratory staff in the Department of Molecular Medicine and Haematology for performing the routine analysis on the CAP/CA HIV-1.
Footnotes
Potential conflicts of interest. All authors: no conflicts.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
John A. Crump, Email: crump017@mc.duke.edu.
Lesley E Scott, Email: Lesley.Scott@nhls.ac.za.
Emma Msuya, Email: emmafletcher2002@yahoo.com.
Anne B. Morrissey, Email: amorr1023@sbcglobal.net.
Ekyafyose E. Kimaro, Email: ekyafyoose@yahoo.com.
John F. Shao, Email: jshao@kcmc.ac.tz.
Wendy S. Stevens, Email: wendy.stevens@nhls.ac.za.
References
- Barlow KL, Tosswill JHC, Parry JV, Clewley JP. Performance of the Amplicor Human Immunodeficiency Virus Type 1 PCR and analysis of specimens with false-negative results. J Clin Microbiol. 1997;35:2846–53. doi: 10.1128/jcm.35.11.2846-2853.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–10. [PubMed] [Google Scholar]
- Collins ML, Irvine B, Tyner D, Fine E, Zayati C, Chang CA, Horn T, Ahle D, Detmer J, Shen LP, Kolberg J, Bushnell S, Urdea MS, Ho DD. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 moleculs/ml. Nucl Acids Res. 1997;25:2979–84. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mendoza C, Koppelman M, Montes B, Ferre V, Soriano V, Cuypers H, Segondy M, Oosterlaken T. Multicenter evaluation of the NucliSens EasyQ HIV-1 v1.1 assay for the quantitative detection of HIV-1 RNA in plasma. J Virol Methods. 2005;127:54–9. doi: 10.1016/j.jviromet.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Dyer JR, Pilcher CD, Shepard R, Schock J, Eron JJ, Fiscus SA. Comparison of NucliSens and Roche Monitor assays for quantitation of levels of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1999;37:447–9. doi: 10.1128/jcm.37.2.447-449.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiscus SA, Cheng B, Crowe SM, Demeter L, Jennings C, Miller V, Respess R, Stevens W, Forum for Collaborative HIV Research Alternative Viral Load Assay Working Group HIV-1 viral load assays for resource-limited settings. PLoS Med. 2006;3:e550. doi: 10.1371/journal.pmed.0030417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Diaz AG, Clewley GS, Booth CL, Labett W, McAllister N, Geretti AM. Comparative evaluation of the performance of the Abbott real-time human immunodeficincy virus type 1 (HIV-1) assay for the measurement of HIV-1 plasma viral load following automated specimen preparation. J Clin Microbiol. 2006;44:1788–91. doi: 10.1128/JCM.44.5.1788-1791.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido C, Zahonero N, Corral A, Arredondo M, Soriano V, de Mendoza C. Correlation between human immunodeficiency virus type 1 (HIV-1) RNA measurements obtained with dried blood spots and those obtained with plasma by use of Nuclisens EasyQ HIV-1 and Abbott RealTime HIV load tests. J Clin Microbiol. 2009;47:1031–36. doi: 10.1128/JCM.02099-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geelen S, Lange J, Borleffs J, Wolfs T, Weersink A, Schuurman R. Failure to detect a non-B HIV-1 subtype by the HIV-1 Amplicor Monitor test, version 1.5: a case of unexpected vertical transmission. AIDS. 2003;17:781–2. doi: 10.1097/00002030-200303280-00027. [DOI] [PubMed] [Google Scholar]
- Johanson J, Abravaya K, Caminiti W, Erickson D, Flanders R, Leckie G, Marshall E, Mullen C, Ohhashi Y, Perry R, Ricci J, Salituro J, Smith A, Tang N, Vi M, Robinson J. A new ultrasensitive assay for quantitation of HIV-1 RNA in plasma. J Virol Methods. 2001;95:81–92. doi: 10.1016/s0166-0934(01)00297-x. [DOI] [PubMed] [Google Scholar]
- Kiwelu IE, Renjifo B, Chaplin B, Sam N, Nkya WMMM, Shao J, Kapiga S, Essex M. HIV type 1 subtypes among bar and hotel workers in Moshi, Tanzania. AIDS Res Hum Retrovirus. 2003;19:57–64. doi: 10.1089/08892220360473970. [DOI] [PubMed] [Google Scholar]
- Leelawiwat W, Young NL, Chaowanachan T, Ou CY, Culnane M, Vanprapa N, Waranawat N, Wasinrapee P, Mock PA, Tappero J, McNicholl JM. Dried blood spots for the diagnosis and quantitation of HIV-1: stability studies and evaluation of sensitivity and specificity for the diagnosis of infant HIV-1 infection in Thailand. J Virol Methods. 2009;155:109–117. doi: 10.1016/j.jviromet.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Mayhood MK, Afwamba IA, Odhiambo CO, Ndanu E, Thielman NM, Morrissey AB, Shao JF, Pence BW, Crump JA. Validation, performance under field conditions, and cost-effectiveness of Capillus HIV-1/HIV-2 and Determine HIV-1/2 rapid human immunodeficiency virus antibody assays using sequential and parallel testing algorithms in Tanzania. J Clin Microbiol. 2008;46:3946–51. doi: 10.1128/JCM.01045-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmanov S, Pattou C, Walker N, Schwardlander B, Esparza J, WHO-UNAIDS Network for HIV Isolation Charaterization Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J Acquir Immun Defic Syndr. 2002;29:184–90. doi: 10.1097/00042560-200202010-00013. [DOI] [PubMed] [Google Scholar]
- Ramadhani HO, Thielman NM, Landman KZ, Ndosi EM, Gao F, Kirchherr JL, Shah R, Shao HJ, Morpeth SC, McNeill JD, Shao JF, Bartlett JA, Crump JA. Predictors of incomplete adherence, virologic failure, and antiviral drug resistance among HIV-infected adults receiving antiretroviral therapy in Tanzania. Clin Infect Dis. 2007;45:1492–8. doi: 10.1086/522991. [DOI] [PubMed] [Google Scholar]
- Rouet F, Ekouevi DK, Chaix ML, Burgard M, Inwoley A, Tony TD, Danel C, Anglaret X, Leroy V, Msellati P, Dabis F, Rouzioux C. Transfer and evaluation of an automated, low-cost real-time reverse transcription-PCR test for diagnosis and monitoring of human immunodeficiency virus type 1 infection in a West African resource-limited setting. J Clin Microbiol. 2005;43:2709–17. doi: 10.1128/JCM.43.6.2709-2717.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LE, Galpin JS, Glencross DK. Multiple method comparison: statistical model using percentage similarity. Clin Cytometry. 2003;54B:46–53. doi: 10.1002/cyto.b.10016. [DOI] [PubMed] [Google Scholar]
- Stevens W, Wiggill T, Horsfield P, Coetzee L, Scott LE. Evaluation of the NucliSens EasyQ assay in HIV-1 infected individuals in South Africa. J Virol Methods. 2005;124:105–10. doi: 10.1016/j.jviromet.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Sun R, Ku J, Jayakar H, Kuo JC, Brambilla D, Herman S, Rosenstraus M, Spadoro J. Ultrasensitive reverse transcription-PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1998;36:2964–9. doi: 10.1128/jcm.36.10.2964-2969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson P, de Mendoza C, Joshi Y, Golden A, Hodinka RL, Soriano V, Devare SG, Hackett J. Impact of human immunodeficiency virus type 1 (HIV-1) genetic diversity of the performance of four commercial viral load assays: LCx HIV RNA Quantitative, AMPLICOR HIV-1 MONITOR v1.5, VERSANT HIV-1 RNA 3.0, and NucliSens HIV-1 QT. J Clin Microbiol. 2005;43:3860–8. doi: 10.1128/JCM.43.8.3860-3868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson P, Holzmayer V, Huang S, Hay P, Adebiyi A, Rice P, Abravaya K, Thamm S, Devare SG, Hackett J. Performance of the automated Abbott RealTime™ HIV-1 assay on a genetically diverse panel of specimens from London: comparison to VERSANT HIV-1 RNA 3.0, AMPLICOR HIV-1 MONITOR 1.5, and LCx® HIV RNA Quantitative assays. J Virol Methods. 2006a;137:184–92. doi: 10.1016/j.jviromet.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Swanson P, Huang S, Abravaya K, de Mendoza C, Soriano V, Devare SG, Hackett J. Evaluation of performance across the dynamic range of the Abbott RealTime™ HIV-1 assay as compared to VERSANT HIV-1 RNA 3.0 and AMPLICOR HIV-1 MONITOR v1.5 using serial dilutions of 39 group M and O viruses. J Virol Methods. 2007;141:49–57. doi: 10.1016/j.jviromet.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Swanson P, Huang S, Holzmayer V, Bodelle P, Yamaguchi J, Brennan C, Badaro R, Brites C, Abravaya K, Devare SG, Hackett J. Performance of the automated Abbott RealTime™ HIV-1 assay on a genetically diverse panel of specimens from Brazil. J Virol Methods. 2006b;134:237–43. doi: 10.1016/j.jviromet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- World Health Organisation. Progress on global access to HIV antiretroviral therapy: a report on ‘3 by 5’ and beyond. WHO Press (Ed); Geneva, Switzerland: 2006. [Google Scholar]
- World Health Organization. Antiretroviral therapy for HIV infection in adults and adolescents in resource-limited settings: towards a universal approach. World Health Organization (Ed); Geneva: 2006. p. 140. [Google Scholar]
- Xu S, Song A, Li X, Li J, Bao Z, Mao P, Zhao Q, Wang Y. Performance of the Abbott RealTime HIV-1 assay for quantification of HIV-1 clades prevalent in China. J Clin Virol. 2008;41:305–9. doi: 10.1016/j.jcv.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Yao J, Liu Z, Ko LS, Pan G, Jiang Y. Quantitative detection of HIV-1 RNA using NucliSens EasyQ HIV-1 assay. J Virol Methods. 2005;129:40–6. doi: 10.1016/j.jviromet.2005.04.017. [DOI] [PubMed] [Google Scholar]