Abstract
Simian betaretroviruses, (formerly Type D retroviruses; SRV) are a group of closely related retroviruses for which the natural host species are Asian monkeys of the genus Macaca. Five serotypes have been identified by classical neutralization assays and three additional untyped variants have been reported (SRVTsukuba, SRV-6, SRV-7). These viruses may be significant pathogens in macaque colonies, causing a broad spectrum of clinical disease secondary to viral-induced immune suppression. Undetected SRV infections in research macaques also represent a potential confounding variable in research protocols and a concern for human caretakers. Intensive testing efforts have been implemented to identify infected animals in established colonies. A real-time quantitative generic multiplex PCR assay was developed that is capable of simultaneous detection of proviral DNA of SRV serotypes 1, 2, 3, 4 and 5. This assay incorporates amplification of the oncostatin M (OSM) gene for confirmation of amplifiable DNA and allows quantitation of the number of proviral copies per cell analyzed in each multiplex reaction. Detection of multiple serotypes by PCR increases the efficiency and cost-effectiveness of SRV screening programs. A panel of SRV serotype-specific uniplex real-time PCR assays for discrimination among the 5 recognized serotypes is also described.
Keywords: Simian betaretrovirus, SRV, PCR, SPF
1. Introduction
The exogenous simian type D retroviruses (SRV), now classified in the genus Betaretrovirus, subfamily Orthoretrovirinae comprise a group of closely related viruses that are important pathogens in many species of Asian monkeys of the genus Macaca, that are capable of causing significant morbidity and mortality secondary to viral-induced immunosuppression (Nishida et al., 1986; Chopra, 1970; Jensen et al., 1970; Lerche, 1992; Lerche et al., 1997; Lerche, 2003; Nishida, 1992; Power et al., 1986; Sonigo et al., 1986). SRV is endemic in many populations of wild and captive macaques and to date five distinct serotypes have been identified by conventional virus neutralization assays (Hara et al., 2007; Hara et al., 2005; Lerche, 2005; Lerche et al., 1997; Lerche et al., 1986; Lerche et al., 1994; Lowenstine et al., 1986; Maul et al., 1986; Wolfe-Coote, 2005). Undetected SRV infection has also been recognized as a potential confounding variable in biomedical research protocols using macaques as research subjects (Lerche, 2003; Maul et al., 1986). As a result, SRV is one of several persistent viruses targeted for elimination to establish and maintain specific pathogen-free (SPF) breeding colonies of rhesus macaques (Macaca mulatta) for biomedical research. Antibody testing alone, while effective in identifying the presence of SRV at a population level, is inadequate to identify all infected individuals within a population, as infected macaques may exhibit a long interval between infection and seroconversion, and some infected macaques fail to make a detectable antibody response (Lerche, 2003; Lerche et al., 1994; Liska et al., 1997). The application of SRV testing algorithms incorporating real-time PCR together with sensitive and specific assays for SRV antibody detection have been successful in detecting and eliminating SRV infection in breeding and research colonies in which recognized serotypes are endemic (Lerche et al., 1997; Liska et al., 1997; Lowenstine et al., 1986). The existence of five distinct serotypes of exogenous SRV, as determined by conventional virus neutralization assays, recognition of significant genetic variation within specific serotypes, as well as the presence of an endogenous full-length intact betaretrovirus genome in all Old World monkeys, has complicated diagnostic and screening programs utilizing PCR (Anderson, 1999; Chung et al., 2008; Hara et al., 2005; Lerche, 1992; Lerche et al., 1994; Li et al., 2000; Marracci et al., 1995; Marx et al., 1985; Nishida, 1992). While some generalizations are possible regarding the association of the various serotypes with different macaque species, rhesus macaques (M. mulatta) are most commonly infected with SRV type 1, while cynomolgus (M. fascicularis) and pig-tailed macaques (M. nemestrina) are most often infected with SRV type 2 viruses (Lerche, 1992; Lerche et al., 1987; Lowenstine et al., 1986; Marx et al., 1984; Nishida, 1992), these virus-macaque species associations are not exclusive. In the research colony environment macaques may be infected with any of the five serotypes and infection with more than one serotype has been observed (Lerche, 2005; Wolfe-Coote, 2005). Sensitive and specific real-time PCR assays have been developed for detection of proviral DNA of various SRV serotypes, but to date, real-time multiplex or generic PCR assays for SRV have not been reported (Chung et al., 2008).
A prototype generic SRV multiplex real-time PCR assay (GSRV), incorporating GAPDH as an internal control for the presence of amplifiable DNA, was developed and validated in parallel testing with uniplex end point PCR on over 1000 samples submitted for SRV diagnostic testing from a variety of primate facilities to the pathogen detection laboratory at the California National Primate Research Center (CNPRC) ((Lerche et al., 1997), N. Lerche, unpublished). While the SRV detection capabilities of this multiplex prototype were robust compared to a previously published end point assay (kappa=0.6966), frequent failure to amplify GAPDH, possibly due to competition between the GADH and SRV amplification reactions, rendered many of the assays invalid, requiring repeat testing (Lerche et al., 1994). A generic SRV multiplex real-time PCR assay (OSRV) is described that is capable of simultaneous detection of a conserved sequence region in all 5 SRV serotypes and that incorporates detection of the diploid oncostatin (OSM) gene as an internal control for the presence of amplifiable DNA that reduces competition with SRV amplification in the multiplex reaction, greatly reducing the need for repeat testing. Incorporating OSM into the assay has the added benefit of allowing quantitation of the number of SRV proviral DNA copies. (Biosystems, 2008; Bruce et al., 2005; Lerche et al., 1997; Malik et al., 1989). In addition a panel of SRV serotype-specific uniplex real-time PCR assays is described for discrimination among the 5 different serotypes.
2. Materials and Methods
2.1 Animal Samples
SRV infection status of samples used to generate a test validation panel was determined by previous analysis for SRV-specific antibodies by multiplex microbead immunoassay (MMIA) and confirmatory Western immunoblot (WB) and for SRV proviral DNA by both end point PCR and a prototype generic multiplex real-time PCR. An SRV positive was defined as a sample with PCR amplification in 2 of 2 or 3 of 4 tests in both an endpoint PCR assay and a prototype real-time PCR assay as well as positive antibody results from the MMIA. An SRV indeterminate sample had PCR amplification in either 1 or 2 out of 4 PCR tests on the end point and prototype multiplex PCR assay as well as indeterminate antibody results from the MMIA and WB assays. An SRV negative sample was defined as having no PCR amplification in either the end point or prototype real time PCR assays as well as no antibody detection in either the MMIA or the WB assays. A panel of 44 samples comprising 15 SRV positive, 15 SRV negative, and 14 SRV indeterminate DNA samples were randomly selected from a reference DNA bank derived from >1000 heparinized whole blood samples obtained from various species of macaque and submitted to the pathogen detection laboratory for diagnostic SRV testing in 2007.
2.2 SRV variants
In development of the generic and uniplex assays both sequence data and DNA from specific isolates were used. Full sequence data was available for SRV-1, -2, and -3 serotypes while partial sequence data was used for SRV-5ORE and SRVTsukuba. Two isolates of SRV-5, SRV-5Cal and SRV-5Ore (generously provided by Dr. Michael Axthelm, Oregon NPRC), SRVTsukuba (generously provided by Dr. Ryozaburo Mukai, Tsukuba Primate Research Center, Japan)(Hara et al., 2005) were tested on both the generic multiplex and uniplex assays, as was the prototype SRV-4Cal isolate originally identified by neutralization assays, but which has only been partially sequenced (N. Lerche, unpublished). Variants identified as “SRV-6” and “SRV-7” were not available for analysis in the generic assay (Nandi et al., 2000; Nandi et al., 2003; Nandi et al., 2006).
2.3 DNA Isolation and Sample Preparation
DNA was isolated from heparinized whole blood on a Qiagen M48 Biorobot using the QiaAmp midi kit (cat. # 951336, Valencia, CA, USA). From each sample, 200ul of heparinized blood was removed and placed in an open microfuge tube in the Biorobot for DNA extraction according to the kit protocol. The remaining heparinized blood sample was centrifuged and plasma was removed for SRV specific antibody testing (Khan et al., 2006). Negative control samples from uninfected macaques and positive control samples obtained from SRV-1 and -2 infected macaques were processed as described above or by “bulk” processing of 10ml whole blood using the Gentra Puregene DNA Purification Kit (Qiagen cat.# 158389, Valencia, CA, USA). DNA isolated using the “bulk” method was re-suspended in (∼12ml) DNA hydration solution to make a final concentration of ∼25ng/ul. A negative cell culture control consisting of DNA extracted from uninfected Raji or Sup-T-1 cell lines and positive cell culture controls consisting of proviral DNA of SRV serotypes -3, -4Cal -5Cal, -5Ore and SRVTsukuba obtained from infected Raji or SupT cell cultures were prepared as described. Isolated DNA was re-suspended in 4ml of DNA hydration solution to make a final concentration of ∼25ng/ul.
2.4 Primers and Probes
Probe and primer sequences (Tables 1 and 2) were selected according to parameters defined by the Primer Express software (version 2.0, Applied Biosystems, Foster City, CA) to amplify proviral DNA sequences of exogenous SRV serotypes 1, 2, 3, 4, and 5 but not endogenous betaretrovirus sequences. Full length sequence information from SRV serotypes 1, 2, and 3 were used in primer and probe design in addition to partial sequence data from SRV-5Ore and SRVTsukuba. Primers and probes for the generic PCR assay were derived from highly conserved sequences within the envelope (env) gene. Once primers and probes were designed based on the sequence data available, testing was completed with additional unsequenced SRV isolates. Primers and probes for the uniplex type-specific PCR assays were derived from highly variable regions within the env gene incorporating sequences coding for the neutralization epitope region that define the specific serotypes (Anderson, 1999; Malley et al., 1991; Torres et al., 1991a; Torres et al., 1991b; Werner et al., 1991). Partial sequence data for SRVTsukuba was used in development of an SRV-4 specific assay. Isolates of SRV-4cal and SRVTsukuba were both tested on the SRV-4 specific assay to confirm the sensitivity of the assay. In order to confirm specificity of all the uniplex assays DNA controls for all 5 serotypes were tested on each assay. (Table 4)
Table 1.
SRV-specific env primers and probes used in the generic multiplex real-time PCR assay.
| Primer | Position | Virus/Sequence (5′ to 3′) | Sequence position (bp) |
|---|---|---|---|
| OSRV PCR: | |||
| SRVFor | Sense | CTG GWC AGC CAA TGA CGG G | SRV-2,7585-7603 |
| SRVRev | Antisense | CGC CTG TCT TAG GTT GGA GTG | SRV-2,7715-7695 |
| SRVa | Probe | 6FAM - TCA CTA ACC TAA GAC AGG AGG GTC GTC A – TAMRA | SRV-2,7621-7648 |
| SRVb | Probe | 6FAM-TCC TAA ACC TAA GAC AGG AGG GCT GTC A-TAMRA | SRV-2,7626-7642 |
| OSMFor | Sense | CCT CGG GCT CAG GAA CAA C | (Bruce et al., 2005) |
| OSMRev | Antisense | GGC CTT CGT GGG CTC AG | |
| OSM | Probe | VIC- TAC TGC ATG GCC CAG CTG CTG GAC AA-MGBNFQ | |
SRV 1 Genbank accession no. M11841, SRV 2 Genbank accession no. M16605 and Mason Pfizer monkey virus/SRV 3 (MPMV) Genbank accession no. AF033815 were used in PCR design. (Bruce et al., 2005; Hara et al., 2005)
Table 2.
SRV serotype-specific env primers and probes for uniplex real-time PCR
| Primer | Position | Virus/Sequence (5′ to 3′) | Sequence position (bp) |
|---|---|---|---|
| SRV-1 onlya: | |||
| SRV1 | Probe | FAM-TGC TGG CTG TGC TTG CGG TCA-TAMRA | SRV-1, 6599-6619 |
| SRV1 For | Sense | CCT CCC AAC GCC AAT TAG C | SRV-1, 6573-6591 |
| SRV 1 Rev | Antisense | GCG AGA GGA ACG GGA TCA C | SRV-1, 6639-6621 |
| SRV-2 onlya: | |||
| SRV2 | Probe | VIC-TAC ATA GAG CCA GGA GAG C-MGBNFQ | SRV-2, 6967-6985 |
| SRV2 For | Sense | CCT GGT GAT GCA CCT GTA CCT | SRV-2, 6924-6944 |
| SRV2 Rev | Antisense | GAC CAG CAG TCC CAG TTG AGA | SRV-2, 7053-7033 |
| SRV-3 only: | |||
| SRV3 | Probe | FAM-CCT TTG CTT TGC TTA TTG-TAMRA | MPMV, 7214-7230 |
| SRV3 For | Sense | CAT TTT GGA CCA GTT TTC ATG GA | MPMV, 7161-7183 |
| SRV3 Rev | Antisense | AGG TCA AAT ATG AGC CAC CAC TGT | MPMV, 7373-7350 |
| SRV-4 onlyb: | |||
| SRV4 | Probe | FAM-CCC CCT TTT TAG TGC AAC-MGBNFQ | SRV-TSUKUBA 6699-6716(Hara et al., 2005) |
| SRV4 For | Sense | AAA TTG TTC TTG CCC CAT TGT T | SRV-TSUKUBA 6676-6687 |
| SRV4 Rev | Antisense | GGC AGA CAG ATT CTG TGA AAT TAA AA | SRV-TSUKUBA 6743-6718 |
| SRV-5 only: | |||
| SRV5 | Probe | FAM- CCT TTT TTA CAG AAA TGA AC-MGBNFQ | NOT PUBLISHED |
| SRV5 For | Sense | TCC CTG AGA CTG CCC TTT T | |
| SRV5 Rev | Antisense | CAA GGC CCC AAA TGA GGA T | |
Primers designed by Jason Michaels, Sierra Biomedical, Charles River Laboratories.
Modified from Hara, et. al. 2005.
SRV 1 Genbank accession no. M11841, SRV 2 Genbank accession no. M16605 and Mason Pfizer monkey virus/SRV 3 (MPMV) Genbank accession no. AF033815 were used in PCR design. (Hara et al., 2005)
Table 4.
SRV controls amplified in the multiplex and each of the uniplex real-time PCR assays and the generic multiplex OSRV assay.
| Sample type | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| negative whole blood control |
SRV 1 whole blood control |
SRV 2 whole blood control |
negative Raji cell culture control |
SRV 3 Raji cell culture control |
SRV 4 Raji cell culture control |
SRV 5 CA SupT cell culture control |
SRV 5 OR SupT cell culture control |
Tsukuba SupT cell culture control |
||
| PCR assay | SRV 1 only |
und | 32.1 | und | und | und | und | und | und | |
| SRV 2 only |
und | und | 29.3 | und | und | und | und | und | und | |
| SRV 3 only |
und | und | und | und | 23.2 | und | und | und | und | |
| SRV 4 only | und | und | und | und | und | 26.5 | und | und | 26 | |
| SRV 5 only |
und | und | und | und | und | und | 22.8 | 33.9 | und | |
| OSRV Generic |
und | 30.6 | 23 | und | 22.2 | 27 | 22.7 | 34.8 | 29.5 | |
2.5 Calibration Plasmids
Full-length plasmid constructs for SRV-1, -2, -3 were generously provided by Dr. Paul Luciw, UC Davis. Plasmid constructs for SRV-4 and -5 were prepared commercially (BioClone, San Diego, CA, USA) to specifications based on unpublished sequence. Plasmid constructs for each of the 5 serotypes were prepared at a concentration of 107 copies/ul in 1×TE (Tris EDTA) buffer, pH 8.0. Serial tenfold dilutions from 106 copies/reaction (15ul) to 10−1 copies/reaction were made in a background of DNA extracted from negative macaque buffy coat at a concentration of 25ng/ul. Plasmids for SRV serotypes 1, 3 and 5 were diluted in rhesus macaque DNA while plasmids for serotypes 2 and 4 were diluted in cynomologus macaque DNA to more accurately represent species-serotype associations in test samples (Anderson, 1999; Lerche, 1992; Lerche et al., 1994; Nishida, 1992). (Table 3)
Table 3.
Sensitivity of the cycle threshold (CT) values for 10-fold plasmid dilutions in the generic multiplex real-time PCR performed in triplicate. CT values are displayed where samples are amplified and undetermined (und) is displayed where samples were not amplified.
| Plasmid Concentration |
SRV-1 Plasmid |
SRV-2 Plasmid |
SRV-3 Plasmid |
SRV-4 Plasmid |
SRV-5 Plasmid |
|---|---|---|---|---|---|
| 106copies/15ul | 22.4, 22.7, 22.7 | 21.7, 21.8, 21.2 | 23.8, 24.4, 24.3 | 34.1, 33.3, 33.3 | 38.7, 38.6, 38.2 |
| 105copies/15ul | 25.7, 26.0, 25.9 | 24.9, 25.3, 24.9 | 28.0, 28.0, 27.6 | 37.2, 36.2, 37.1 | 43.1, 43.2, 45.4 |
| 104copies/15ul | 29.1, 29.4, 29.1 | 28.3, 30.1, 28.3 | 31.5, 31.7, 31.6 | 42.5, 42.1, 44.1 | 49.6, 52.1, 48.4 |
| 103copies/15ul | 32.7, 32.7, 32.5 | 32.0, 32.3, 31.6 | 34.9, 35.1, 34.9 | 45.0, 50.4, 48.3 | und, und, 48.0 |
| 102copies/15ul | 36.9, 37.3, 36.2 | 35.0, 36.3, 35.5 | 38.2, 37.4, 40.6 | und, und, 47.3 | und, und, und |
| 101copies/15ul | 37.5, 39.6, und | 38.1, 40.0, und | 40.9, 42.0, 40.6 | und, und, und | und, und, und |
| 100copies/15ul | und, und, und | und, 41.3, 40.0 | und, und, und | und, und, und | und, und, und |
| 10-1copies/15ul | und, 41.1, und | und, und, 40.1 | und, und, und | und, und, und | und, und, und |
2.6 Multiplex Detection and Quantitation of SRV Proviral DNA
The procedure for PCR amplification of OSM gene sequences as described previously (Bruce et al., 2005) was modified slightly for use on the Applied Biosystems platform and was multiplexed with the SRV reaction. The reaction conditions have been optimized for the multiplex detection of OSM and all five SRV serotypes. The multiplex reaction mixture consisted of 50ul containing 1ul of 5uM OSM probe, 0.25ul of each 10uM OSM primers, 0.25ul of 20uM SRV probes (SRVa/SRVb) and 0.5ul of each 20uM SRV primers (SRVFor/ SRVRev), 25ul of TaqMan Universal master mix (cat # 4304437, Applied Biosystems, Foster City, CA), 7ul 1×TE buffer and 15ul of template DNA. Cycling conditions were 2min at 50°C, 10min at 95°C; 55 cycles of amplifications for 15sec at 95°C and 1min at 62°C. All reactions were carried out in a 7900 ABI PRISM Sequence Detector from Applied Biosystems. Positive controls consisted of proviral DNA of SRV-1 -2, -3, -4, and -5 prepared as described above.
2.7 Detection of Specific SRV Serotypes
To further identify specific serotypes in samples testing positive (or indeterminate) in the generic SRV multiplex PCR assay, 5 additional serotype-specific uniplex real-time PCR assays are performed. Probe and primer sequences (Table 2) were selected according to parameters defined by the Primer Express software (version 2.0, Applied Biosystems, Foster City, CA) for serotype specificity and to not amplify the endogenous betaretrovirus sequences of Old World monkeys. Reaction conditions for the individual uniplex assays are summarized in table 5.
Table 5.
Different uniplex reaction mixtures for each of the five serotypes.
| Reaction Contents: | SRV-1 | SRV-2 | SRV-3 | SRV-4 | SRV-5 | |
|---|---|---|---|---|---|---|
| Forward Primer | 0.625ul of 12uM | 0.625ul of 12uM | 0.5ul of 20uM | 0.5ul of 20uM | 1ul of 20uM | |
| Reverse Primer | 0.625 of 12uM | 0.625 of 12uM | 0.5ul of 20uM | 0.5ul of 20uM | 1ul of 20uM | |
| Probe | 0.625ul of 10uM | 0.625ul of 10uM | 0.5ul of 20uM | 0.5ul of 20uM | 0.5ul of 20uM | |
| TaqMan Universal master mixc | 12.5ul | 12.5ul | 12.5ul | 12.5ul | 12.5ul | |
| 1 × TE buffer | 5.625ul | 5.625ul | 1ul | 1ul | none | |
| DNA | 5ul | 5ul | 10ul | 10ul | 10ul | |
| total | 25ul | 25ul | 25ul | 25ul | 25ul | |
cat# 4304437, Applied Biosystems, Foster City, CA, USA
Cycling conditions for each assay consisted of 2min at 50°C, 10min at 95°C; 55 cycles of amplifications for 15sec at 95°C and 1min at 60°C. All reactions were carried out in an ABI 7900 ABI PRISM Sequence Detector (Applied Biosystems). The type-specific plasmid constructs for all 5 SRV serotypes and cell culture controls described above were tested on each individual typing assay in order to determine both the lower limit of detection and for confirmation of serotype specificity.
2.8 Interpretative Criteria
The cycle threshold (CT) value reflects the linear phase in the amplification curve that can be used for quantitation of the template input (Biosystems, 2008). Samples were initially amplified in duplicate on separate reaction plates. Samples were considered SRV negative if no amplification was present in the initial duplicate samples but amplification was observed in the control gene OSM. Samples giving discrepant results on initial testing were repeated in duplicate. Samples were considered SRV positive if amplification occurred in both the initial duplicate tests, or in 3 out of 4 replicates. Samples that showed amplification in 1 or 2 out of 4 replicates were considered “indeterminate”.
2.9 Statistical Analysis
Kappa statistic, a proportion of the amount of agreement observed due to chance alone compared to the maximum amount of agreement possible (Landis, 1977), was calculated to compare the agreement between a previously published end point PCR assay and a previously unpublished generic multiplex real-time PCR (GSRV). A second kappa analysis was performed to compare the infection status (determined by combining GSRV and antibody testing) to the newly described OSRV generic multiplex real-time PCR assay (Table 6). Kappa calculations were carried out using the STATA software package (STATA Statistics/Data Analysis ver. 9.2, Statacorp, College Station, Texas, USA).
Table 6.
Results of generic multiplex real-time PCR assay for a panel of 44 DNA samples from macaques with predetermined SRV infection status. (Kappa = 0.8977).
| OSRV Multiplex PCR assay | ||||
|---|---|---|---|---|
| Negative | Indeterminate | Positive | ||
| Infection statusd |
Negative | 15 | 0 | 0 |
| Indeterminate | 1 | 12 | 0 | |
| Positive | 0 | 2 | 14 | |
| Total | 16 | 14 | 14 | |
Infection status was determined by MMIA, WB, end point PCR and GSRV PCR.
3. Results
3.1 Multiplex Detection of SRV Proviral DNA
Proviral DNA of all 5 known serotypes isolated from the blood of infected macaques or infected cell lines in tissue culture was consistently amplified in the multiplex real-time PCR assay (Table 4). Two different isolates of SRV-5 (SRV-5Ore and SRV-5Cal) were detected in the generic PCR, as was SRVTsukuba, an isolate of undetermined serotype. Results of the panel of 44 DNA test samples by the OSRV assay and the GSRV prototype showed very good agreement between the two assays, with only 2 discrepant samples detected among the “indeterminates” (Table 6, Kappa = 0.8977).
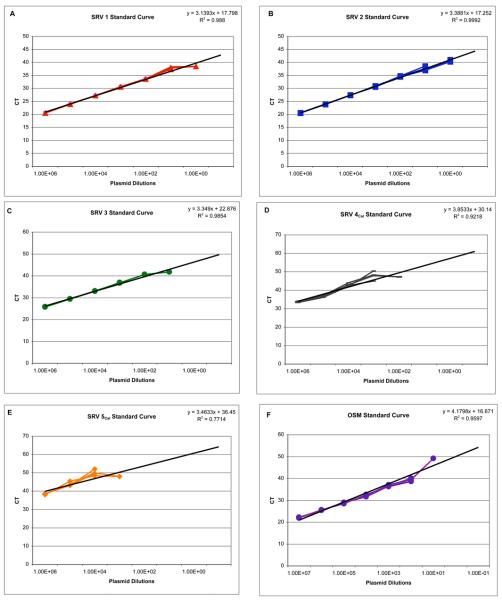
The standard curves generated to determine the detection limits of the multiplex assay for SRV serotypes 1 through 5, and for OSM are presented in Figure 1 A-F. The lower limit of detection in the multiplex assay for SRV serotypes 1 through 3 is from 10-100 copies per reaction, while the limit of detection for serotype 2 in this assay is from 1-10 copies per reaction. The limit of detection for serotypes 4 and 5 are not as sensitive (SRV-4 100-1000, SRV-5 1000-10,000 per reaction) but these standard curves are based on limited sequence data from the CNPRC isolates. The sensitivity of the multiplex PCR assay for each of the 5 serotypes is summarized in Table 3.
Figure 1.
Serial dilutions of SRV plasmids in the generic multiplex real-time PCR assay. Three replicates of each dilution for each plasmid are tested to generate a standard curve for each viral serotype. Dilutions of each plasmid shown on the x-axis of each figure with the corresponding cycle threshold (CT) value displayed on the Y-axis. The CT value reflects the linear phase in the amplification curve that can be used for quantitation of the template input (Biosystems, 2008). Each of the specific standard curves for SRV 1-5 are shown in A-E respectively along with the R2 values for each dilution series. The standard curve for OSM is shown in F.
3.2 Serotype Determination by Uniplex Real-time PCR
Samples positive in the multiplex real-time PCR assay may be further characterized in uniplex typing assays. Specificity of the uniplex PCR reactions is summarized in Table 4. Uniplex PCR amplification was specific only for the matched SRV serotype. Primers and probe generated to amplify sequences of SRVTsukuba, also amplified SRV-4 (for which no sequences have been published), suggesting that SRVTsukuba is a type 4-like variant. No amplification was observed in genomic DNA isolated from SRV negative macaques, demonstrating that none of the type-specific primers amplified endogenous betaretrovirus sequences.
3.3 Quantitation of Proviral DNA copy number
Once the specific serotype has been determined, the equations generated by the standard curves shown in Figure 1 A-F allow for the calculation of the number of proviral DNA sequences detected in the multiplex assay in relation to the number of cells analyzed (Bruce et al., 2005). Once the specific serotype in a given sample is determined by previous information or uniplex PCR assays, the results from the multiplex reaction can be used to perform relative quantitation of proviral load for the specific SRV serotype per number of cells tested in the same reaction well. Using the appropriate SRV standard curve for whichever serotype is present in a sample and inserting the CT value will give the number of proviral copies detected (numerator). Using the equation generated from the OSM standard curve from the multiplex reaction the number of cellular genomes can be calculated (denominator). This allows for a relative quantitation of the number of proviral copies of a specific SRV serotype detected in a sample compared to the number of cellular genomes detected in the same reaction well.
4. Discussion
The OSRV generic multiplex real-time PCR reaction is a sensitive and specific test for routine SRV surveillance that is capable of detecting all 5 SRV serotypes in one PCR reaction. Efficient, accurate and cost-effective testing for SRV in macaque populations is important for maintaining the health of animals maintained in research colonies, as well as the integrity of research data collected utilizing macaques. Incorporation of OSRV generic real-time multiplex PCR into surveillance testing algorithms can significantly increase the efficiency of such programs for detecting the presence or absence of generic SRV in macaque populations. If desired, secondary specific SRV type determinations and quantitation of proviral DNA copy number can be achieved using a second tier of uniplex assays and associated standard curves.
There is also potential for use of the generic SRV multiplex PCR in combination with the specific typing PCR assays as a tool for detection of novel SRV variants. Amplification of proviral DNA sequences in the multiplex assay, but not in any of the 5 type-specific assays could indicate the presence of a previously unrecognized variant of SRV. Recently reported “SRV-6” and “SRV-7” of Indian langurs and rhesus macaques respectively (Nandi et al., 2000; Nandi et al., 2003; Nandi et al., 2006), have not been typed by conventional serum neutralization assays and were not available for testing in the multiplex assay. However, recent detection of SRV by the OSRV multiplex assay in samples obtained from langurs and rhesus macaques in Bangladesh which do not amplify in any of the type-specific uniplex assays (data not shown), suggests the existence of additional novel variants requiring further characterization.
5. Conclusions
A multiplex real-time PCR was developed for the detection of all 5 SRV serotypes. Separate serotype-specific uniplex real-time PCR assays were also developed allowing for differentiation between the different serotypes. These assays allow for accurate, efficient and less expensive screening of macaque colonies.
6. Acknowledgements
The authors would like to thank Jason Michaels and Shannon Flynn for technical assistance. We also thank Paul Luciw for providing reagents, and Masayuki Hara, Ryozaburo Mukai and Mike Axthelm for providing SRV isolates. This study was supported in part by grant number RR00169 National Center for Research Resources (NCRR) of the National Institutes of Health (NIH).
Abbreviations
- SRV
Simian betaretrovirus
- OSM
oncostatin M gene
- GSRV
GAPDH SRV multiplex PCR reaction
- OSRV
oncostatin M gene and SRV multiplex PCR
- env
SRV envelope gene
- MMIA
multiplex microbead immunoassay
- WB
western blot
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson DE, Torres JV. Simian retrovirus receptor and neutralization mechanism by antibodies to the envelope glycoprotein. Viral Immunol. 1999;12:47–56. doi: 10.1089/vim.1999.12.47. [DOI] [PubMed] [Google Scholar]
- Biosystems A. Guide to Performing Relative Quantitation of Gene Expression Using Real-Time Quantitative PCR. Applied Biosystems. 2008;2009 [Google Scholar]
- Bruce AG, Bakke AM, Thouless ME, Rose TM. Development of a real-time QPCR assay for the detection of RV2 lineage-specific rhadinoviruses in macaques and baboons. Virol J. 2005;2:2. doi: 10.1186/1743-422X-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant ML, Gardner MB, Marx PA, Maul DH, Lerche NW, Osborn KG, Lowenstine LJ, Bodgen A, Arthur LO, Hunter E. Immunodeficiency in rhesus monkeys associated with the original Mason-Pfizer monkey virus. J Natl Cancer Inst. 1986;77:957–65. [PubMed] [Google Scholar]
- Chopra HC, Mason MM. A new virus in a spontaneous mammary tumor of a rhesus monkey. Cancer Res. 1970;30:2081–6. [PubMed] [Google Scholar]
- Chung HK, Unangst T, Treece J, Weiss D, Markham P. Development of real-time PCR assays for quantitation of simian betaretrovirus serotype-1, -2, -3, and -5 viral DNA in Asian monkeys. J Virol Methods. 2008;152:91–7. doi: 10.1016/j.jviromet.2008.05.021. [DOI] [PubMed] [Google Scholar]
- Hara M, Kikuchi T, Sata T, Nakajima N, Ami Y, Sato Y, Tanaka K, Narita T, Ono F, Akari H, Terao K, Mukai R. Detection of SRV/D shedding in body fluids of cynomolgus macaques and comparison of partial gp70 sequences in SRV/D-T isolates. Virus Genes. 2007;35:281–8. doi: 10.1007/s11262-007-0076-1. [DOI] [PubMed] [Google Scholar]
- Hara M, Sata T, Kikuchi T, Nakajima N, Uda A, Fujimoto K, Baba T, Mukai R. Isolation and characterization of a new simian retrovirus type D subtype from monkeys at the Tsukuba Primate Center, Japan. Microbes Infect. 2005;7:126–31. doi: 10.1016/j.micinf.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Jensen EM, Zelljadt I, Chopra HC, Mason MM. Isolation and propagation of a virus from a spontaneous mammary carcinoma of a rhesus monkey. Cancer Res. 1970;30:2388–93. [PubMed] [Google Scholar]
- Khan IH, Mendoza S, Yee J, Deane M, Venkateswaran K, Zhou SS, Barry PA, Lerche NW, Luciw PA. Simultaneous detection of antibodies to six nonhuman-primate viruses by multiplex microbead immunoassay. Clin Vaccine Immunol. 2006;13:45–52. doi: 10.1128/CVI.13.1.45-52.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis JR, Koch GG. An application of hierarchical kappa-type statistics in the assessment of majority agreement among multiple observers. Biometrics. 1977;33:363–74. [PubMed] [Google Scholar]
- Lerche NW. Epidemiology and control of type D retrovirus infection in captive macaques. In: T. R, Matano S, Ishida H, Goodman M, editors. Topics in Primatology. Vol. 3. University of Tokyo Press; Tokyo: 1992. pp. 439–448. [Google Scholar]
- Lerche NW. The Laboratory Primate. Elsevier; 2005. Common Viral Infections of Laboratory Primates; p. 11. [Google Scholar]
- Lerche NW, Cotterman RF, Dobson MD, Yee JL, Rosenthal AN, Heneine WM. Screening for simian type-D retrovirus infection in macaques, using nested polymerase chain reaction. Lab Anim Sci. 1997;47:263–8. [PubMed] [Google Scholar]
- Lerche NW, Marx PA, Osborn KG, Maul DH, Lowenstine LJ, Bleviss ML, Moody P, Henrickson RV, Gardner MB. Natural history of endemic type D retrovirus infection and acquired immune deficiency syndrome in group-housed rhesus monkeys. J Natl Cancer Inst. 1987;79:847–54. [PubMed] [Google Scholar]
- Lerche NW, Osborn KG, Marx PA, Prahalada S, Maul DH, Lowenstine LJ, Munn RJ, Bryant ML, Henrickson RV, Arthur LO, et al. Inapparent carriers of simian acquired immune deficiency syndrome type D retrovirus and disease transmission with saliva. J Natl Cancer Inst. 1986;77:489–96. [PubMed] [Google Scholar]
- Lerche NW, Osborn KG. Simian retrovirus infections: potential confounding variables in primate toxicology studies. Toxicol Pathol. 2003;31(Suppl):103–10. doi: 10.1080/01926230390174977. [DOI] [PubMed] [Google Scholar]
- Lerche NW, Yee JL, Jennings MB. Establishing specific retrovirus-free breeding colonies of macaques: an approach to primary screening and surveillance. Lab Anim Sci. 1994;44:217–21. [PubMed] [Google Scholar]
- Li B, Axthelm MK, Machida CA. Simian retrovirus serogroup 5: partial gag-prt sequence and viral RNA distribution in an infected rhesus macaque. Virus Genes. 2000;21:241–8. doi: 10.1023/a:1008104000920. [DOI] [PubMed] [Google Scholar]
- Liska V, Lerche NW, Ruprecht RM. Simultaneous detection of simian retrovirus type D serotypes 1, 2, and 3 by polymerase chain reaction. AIDS Res Hum Retroviruses. 1997;13:433–7. doi: 10.1089/aid.1997.13.433. [DOI] [PubMed] [Google Scholar]
- Lowenstine LJ, Pedersen NC, Higgins J, Pallis KC, Uyeda A, Marx P, Lerche NW, Munn RJ, Gardner MB. Seroepidemiologic survey of captive Old-World primates for antibodies to human and simian retroviruses, and isolation of a lentivirus from sooty mangabeys (Cercocebus atys) Int J Cancer. 1986;38:563–74. doi: 10.1002/ijc.2910380417. [DOI] [PubMed] [Google Scholar]
- Malik N, Kallestad JC, Gunderson NL, Austin SD, Neubauer MG, Ochs V, Marquardt H, Zarling JM, Shoyab M, Wei CM, et al. Molecular cloning, sequence analysis, and functional expression of a novel growth regulator, oncostatin M. Mol Cell Biol. 1989;9:2847–53. doi: 10.1128/mcb.9.7.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malley A, Werner L, Benjamini E, Leung CY, Torres J, Pangares N, Shiigi S, Axthelm M. Characterization of T-and B-cell epitopes of a simian retrovirus (SRV-2) envelope protein. J Med Primatol. 1991;20:177–81. [PubMed] [Google Scholar]
- Marracci GH, Kelley RD, Pilcher KY, Crabtree L, Shiigi SM, Avery N, Leo G, Webb MC, Hallick LM, Axthelm MK, et al. Simian AIDS type D serogroup 2 retrovirus: isolation of an infectious molecular clone and sequence analyses of its envelope glycoprotein gene and 3′ long terminal repeat. J Virol. 1995;69:2621–8. doi: 10.1128/jvi.69.4.2621-2628.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx PA, Bryant ML, Osborn KG, Maul DH, Lerche NW, Lowenstine LJ, Kluge JD, Zaiss CP, Henrickson RV, Shiigi SM, et al. Isolation of a new serotype of simian acquired immune deficiency syndrome type D retrovirus from Celebes black macaques (Macaca nigra) with immune deficiency and retroperitoneal fibromatosis. J Virol. 1985;56:571–8. doi: 10.1128/jvi.56.2.571-578.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx PA, Maul DH, Osborn KG, Lerche NW, Moody P, Lowenstine LJ, Henrickson RV, Arthur LO, Gilden RV, Gravell M, et al. Simian AIDS: isolation of a type D retrovirus and transmission of the disease. Science. 1984;223:1083–6. doi: 10.1126/science.6695196. [DOI] [PubMed] [Google Scholar]
- Maul DH, Lerche NW, Osborn KG, Marx PA, Zaiss C, Spinner A, Kluge JD, MacKenzie MR, Lowenstine LJ, Bryant ML, et al. Pathogenesis of simian AIDS in rhesus macaques inoculated with the SRV-1 strain of type D retrovirus. Am J Vet Res. 1986;47:863–8. [PubMed] [Google Scholar]
- Nandi JS, Bhavalkar-Potdar V, Tikute S, Raut CG. A novel type D simian retrovirus naturally infecting the Indian Hanuman langur (Semnopithecus entellus) Virology. 2000;277:6–13. doi: 10.1006/viro.2000.0567. [DOI] [PubMed] [Google Scholar]
- Nandi JS, Tikute SA, Chhangani AK, Potdar VA, Tiwari-Mishra M, Ashtekar RA, Kumari J, Walimbe A, Mohnot SM. Natural infection by simian retrovirus-6 (SRV-6) in Hanuman langurs (Semnopithecus entellus) from two different geographical regions of India. Virology. 2003;311:192–201. doi: 10.1016/s0042-6822(03)00187-9. [DOI] [PubMed] [Google Scholar]
- Nandi JS, Van Dooren S, Chhangani AK, Mohnot SM. New simian beta retroviruses from rhesus monkeys (Macaca mulatta) and langurs (Semnopithecus entellus) from Rajasthan, India. Virus Genes. 2006;33:107–16. doi: 10.1007/s11262-005-0032-x. [DOI] [PubMed] [Google Scholar]
- Nishida T. Topics in primatology. University of Tokyo Press; Tokyo: 1992. [Google Scholar]
- Power MD, Marx PA, Bryant ML, Gardner MB, Barr PJ, Luciw PA. Nucleotide sequence of SRV-1, a type D simian acquired immune deficiency syndrome retrovirus. Science. 1986;231:1567–72. doi: 10.1126/science.3006247. [DOI] [PubMed] [Google Scholar]
- Sonigo P, Barker C, Hunter E, Wain-Hobson S. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell. 1986;45:375–85. doi: 10.1016/0092-8674(86)90323-5. [DOI] [PubMed] [Google Scholar]
- Torres JV, Werner LL, Malley A, Benjamin E. Neutralization epitope in the envelope glycoprotein of simian retrovirus-1 (SRV-1) and identification of the virus receptor. J Med Primatol. 1991a;20:218–21. [PubMed] [Google Scholar]
- Torres JV, Werner LL, Malley A, Benjamini E. The induction of neutralizing antibodies by synthetic peptides of the envelope protein of type D simian retrovirus-1 (SRV-1) Mol Immunol. 1991b;28:907–13. doi: 10.1016/0161-5890(91)90055-o. [DOI] [PubMed] [Google Scholar]
- Werner LL, Torres JV, Leung CY, Kwang HS, Malley A, Benjamini E. Immunobiological properties of a recombinant simian retrovirus-1 envelope protein and a neutralizing monoclonal antibody directed against it. Mol Immunol. 1991;28:819–26. doi: 10.1016/0161-5890(91)90045-l. [DOI] [PubMed] [Google Scholar]
- Wolfe-Coote S. The laboratory primate. Elsevier Academic Press; Amsterdam ; Boston: 2005. [Google Scholar]