Abstract
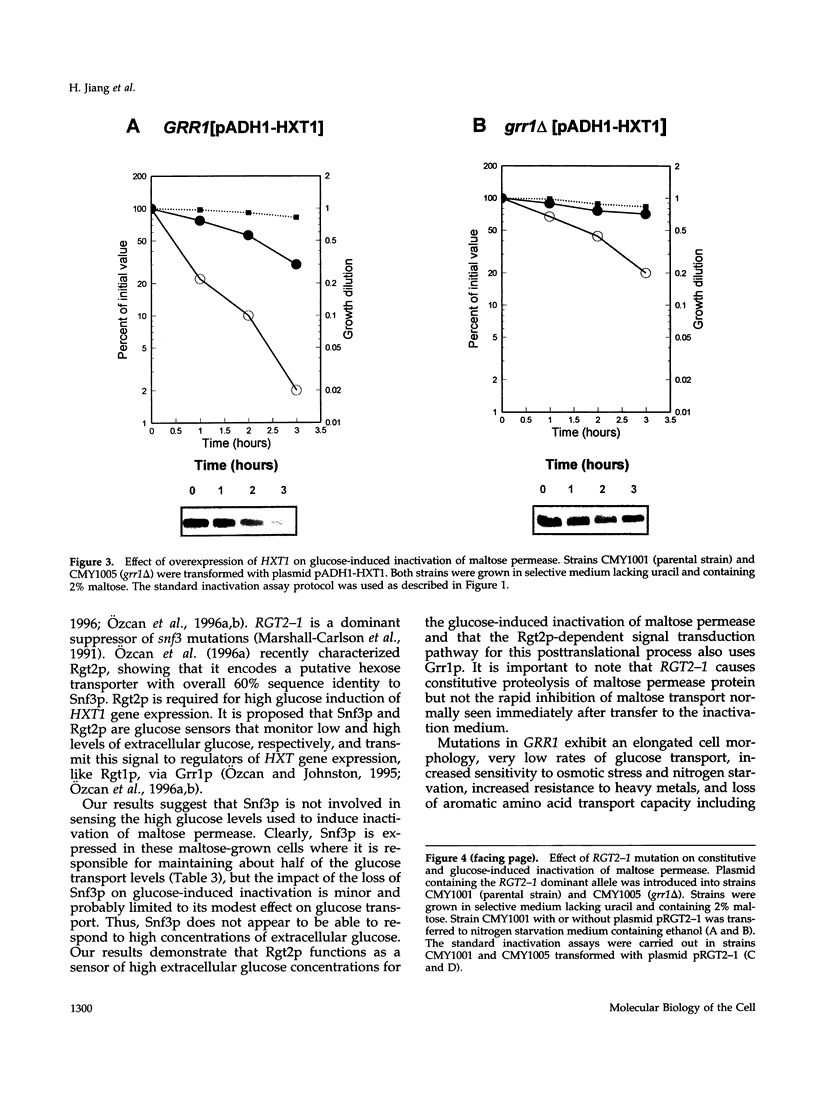
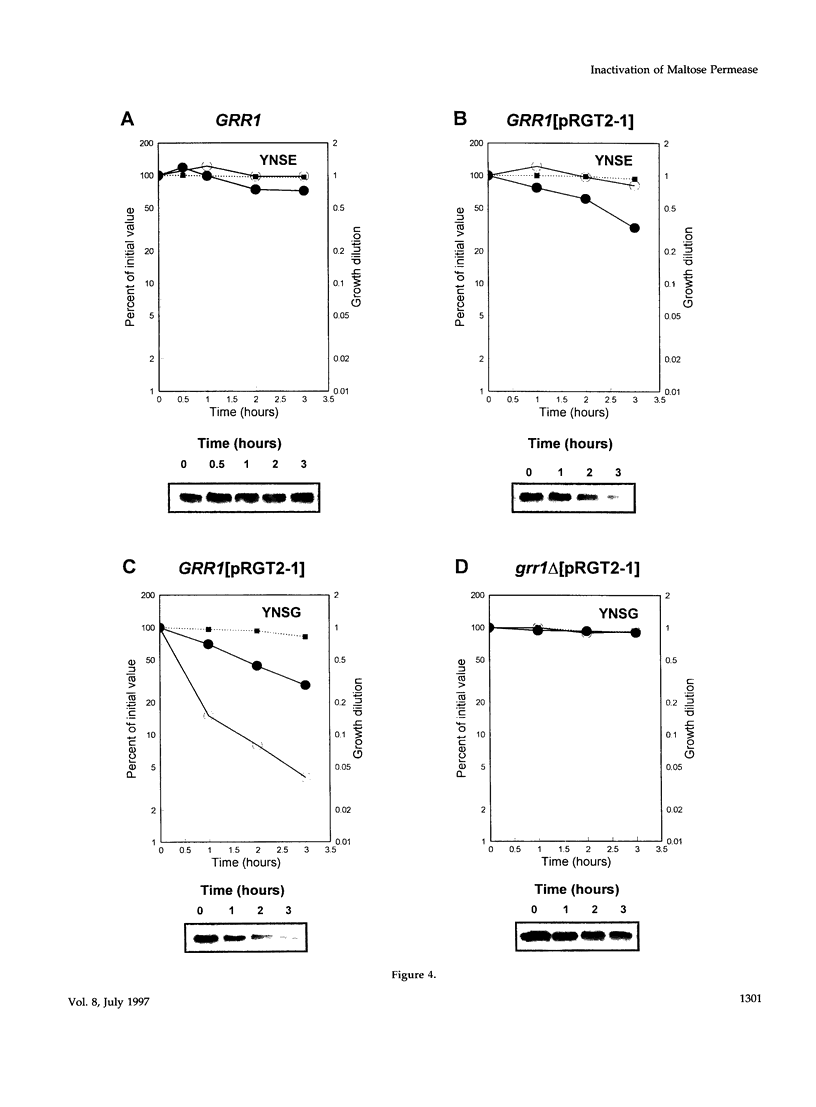
Glucose is a global metabolic regulator in Saccharomyces. It controls the expression of many genes involved in carbohydrate utilization at the level of transcription, and it induces the inactivation of several enzymes by a posttranslational mechanism. SNF3, RGT2, GRR1 and RGT1 are known to be involved in glucose regulation of transcription. We tested the roles of these genes in glucose-induced inactivation of maltose permease. Our results suggest that at least two signaling pathways are used to monitor glucose levels. One pathway requires glucose sensor transcript and the second pathway is independent of glucose transport. Rgt2p, which along with Snf3p monitors extracellular glucose levels, appears to be the glucose sensor for the glucose-transport-independent pathway. Transmission of the Rgt2p-dependent signal requires Grr1p. RGT2 and GRR1 also play a role in regulating the expression of the HXT genes, which appear to be the upstream components of the glucose-transport-dependent pathway regulating maltose permease inactivation. RGT2-1, which was identified as a dominant mutation causing constitutive expression of several HXT genes, causes constitutive proteolysis of maltose permease, that is, in the absence of glucose. A model of these glucose sensing/signaling pathways is presented.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alonso A., Kotyk A. Apparent half-lives of sugar transport proteins in Saccharomyces cerevisiae. Folia Microbiol (Praha) 1978;23(2):118–125. doi: 10.1007/BF02915311. [DOI] [PubMed] [Google Scholar]
- Bai C., Sen P., Hofmann K., Ma L., Goebl M., Harper J. W., Elledge S. J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996 Jul 26;86(2):263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Barral Y., Jentsch S., Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 1995 Feb 15;9(4):399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- Bisson L. F., Coons D. M., Kruckeberg A. L., Lewis D. A. Yeast sugar transporters. Crit Rev Biochem Mol Biol. 1993;28(4):259–308. doi: 10.3109/10409239309078437. [DOI] [PubMed] [Google Scholar]
- Bisson L. F., Neigeborn L., Carlson M., Fraenkel D. G. The SNF3 gene is required for high-affinity glucose transport in Saccharomyces cerevisiae. J Bacteriol. 1987 Apr;169(4):1656–1662. doi: 10.1128/jb.169.4.1656-1662.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R. RAS genes in Saccharomyces cerevisiae: signal transduction in search of a pathway. Trends Genet. 1991 Jan;7(1):28–33. doi: 10.1016/0168-9525(91)90018-l. [DOI] [PubMed] [Google Scholar]
- Cheng Q., Michels C. A. MAL11 and MAL61 encode the inducible high-affinity maltose transporter of Saccharomyces cerevisiae. J Bacteriol. 1991 Mar;173(5):1817–1820. doi: 10.1128/jb.173.5.1817-1820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang H. L., Schekman R. Regulated import and degradation of a cytosolic protein in the yeast vacuole. Nature. 1991 Mar 28;350(6316):313–318. doi: 10.1038/350313a0. [DOI] [PubMed] [Google Scholar]
- Dubin R. A., Needleman R. B., Gossett D., Michels C. A. Identification of the structural gene encoding maltase within the MAL6 locus of Saccharomyces carlsbergensis. J Bacteriol. 1985 Nov;164(2):605–610. doi: 10.1128/jb.164.2.605-610.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J. R., Johnston M. Suppressors reveal two classes of glucose repression genes in the yeast Saccharomyces cerevisiae. Genetics. 1994 Apr;136(4):1271–1278. doi: 10.1093/genetics/136.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick J. S., Johnston M. GRR1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Mol Cell Biol. 1991 Oct;11(10):5101–5112. doi: 10.1128/mcb.11.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo J. M. Carbon catabolite repression in yeast. Eur J Biochem. 1992 Jun 1;206(2):297–313. doi: 10.1111/j.1432-1033.1992.tb16928.x. [DOI] [PubMed] [Google Scholar]
- Görts C. P. Effect of glucose on the activity and the kinetics of the maltose-uptake system and of alpha-glucosidase in Saccharomyces cerevisiae. Biochim Biophys Acta. 1969 Jul 30;184(2):299–305. doi: 10.1016/0304-4165(69)90032-4. [DOI] [PubMed] [Google Scholar]
- Güldener U., Heck S., Fielder T., Beinhauer J., Hegemann J. H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996 Jul 1;24(13):2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko C. H., Liang H., Gaber R. F. Roles of multiple glucose transporters in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Jan;13(1):638–648. doi: 10.1128/mcb.13.1.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage P., Yang X., Carlson M. Yeast SNF1 protein kinase interacts with SIP4, a C6 zinc cluster transcriptional activator: a new role for SNF1 in the glucose response. Mol Cell Biol. 1996 May;16(5):1921–1928. doi: 10.1128/mcb.16.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Gaber R. F. A novel signal transduction pathway in Saccharomyces cerevisiae defined by Snf3-regulated expression of HXT6. Mol Biol Cell. 1996 Dec;7(12):1953–1966. doi: 10.1091/mbc.7.12.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall-Carlson L., Neigeborn L., Coons D., Bisson L., Carlson M. Dominant and recessive suppressors that restore glucose transport in a yeast snf3 mutant. Genetics. 1991 Jul;128(3):505–512. doi: 10.1093/genetics/128.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medintz I., Jiang H., Han E. K., Cui W., Michels C. A. Characterization of the glucose-induced inactivation of maltose permease in Saccharomyces cerevisiae. J Bacteriol. 1996 Apr;178(8):2245–2254. doi: 10.1128/jb.178.8.2245-2254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn L., Schwartzberg P., Reid R., Carlson M. Null mutations in the SNF3 gene of Saccharomyces cerevisiae cause a different phenotype than do previously isolated missense mutations. Mol Cell Biol. 1986 Nov;6(11):3569–3574. doi: 10.1128/mcb.6.11.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S., Dover J., Rosenwald A. G., Wölfl S., Johnston M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci U S A. 1996 Oct 29;93(22):12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S., Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995 Mar;15(3):1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S., Leong T., Johnston M. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol Cell Biol. 1996 Nov;16(11):6419–6426. doi: 10.1128/mcb.16.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riballo E., Herweijer M., Wolf D. H., Lagunas R. Catabolite inactivation of the yeast maltose transporter occurs in the vacuole after internalization by endocytosis. J Bacteriol. 1995 Oct;177(19):5622–5627. doi: 10.1128/jb.177.19.5622-5627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E., Kobayashi H., Harrison D., Hunt T. Destruction of Xenopus cyclins A and B2, but not B1, requires binding to p34cdc2. EMBO J. 1994 Feb 1;13(3):584–594. doi: 10.1002/j.1460-2075.1994.tb06296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein J. M., Hohmann S. Trehalose synthase: guard to the gate of glycolysis in yeast? Trends Biochem Sci. 1995 Jan;20(1):3–10. doi: 10.1016/s0968-0004(00)88938-0. [DOI] [PubMed] [Google Scholar]
- Thevelein J. M. The RAS-adenylate cyclase pathway and cell cycle control in Saccharomyces cerevisiae. Antonie Van Leeuwenhoek. 1992 Aug;62(1-2):109–130. doi: 10.1007/BF00584466. [DOI] [PubMed] [Google Scholar]
- Trumbly R. J. Glucose repression in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1992 Jan;6(1):15–21. doi: 10.1111/j.1365-2958.1992.tb00832.x. [DOI] [PubMed] [Google Scholar]
- Tu J., Carlson M. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 1995 Dec 1;14(23):5939–5946. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L. G., Coons D., Bisson L. F., Carlson M. Altered regulatory responses to glucose are associated with a glucose transport defect in grr1 mutants of Saccharomyces cerevisiae. Genetics. 1994 Apr;136(4):1279–1285. doi: 10.1093/genetics/136.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]











