Abstract
AIM: To confirm the presence of recombination, full-length hepatitis B virus (HBV) from chronic patients was sequenced and analyzed.
METHODS: Full-length HBV genomes from 12 patients were amplified and sequenced in an automated sequencer. Phylogenetic analysis was carried out on full-length, Core and preS2/Surface regions using MEGA software. SimPlot Boot Scanning and amino acid sequence analysis were performed for confirmation of recombination.
RESULTS: Eight patients were infected with genotype D strain; one patient with genotype A and three patients had genotype A and D recombination; two of them had cirrhosis and one had hepatocellular carcinoma. Phylogenetic analysis of core and preS2/surface regions separately showed evidence of genotype A and D recombination. The breakpoints of recombination were found to be at the start of preS2 and at the end of surface coding regions.
CONCLUSION: We identified and characterized recombinant A and D genotype HBV in hepatitis B surface antigen (HBsAg)-positive patients.
Keywords: Hepatitis B virus, Genotype, Variation, Evolution, Recombination
INTRODUCTION
Hepatitis B virus (HBV) is an organ-specific virus causing inflammation of the liver, leading to complications such as chronic liver disease (CLD) and hepatocellular carcinoma (HCC). As compared to Europe and North America, the prevalence of HBV infection in Asia is quite high, with 40 million people harboring chronic HBV infection in India[1].
Two features make HBV unique. First, its way of replication, by which it uses the pregenomic RNA as an intermediate step for reverse transcription. Second, the efficient utilization of its compact genome for production of seven different proteins from four open reading frames (ORFs). Major proteins that are encoded from these four ORFs are the envelope, core the X protein and the polymerase.
Nucleotide substitution, deletion, insertion and recombination are the main factors that results in variation of the HBV genome. HBV genotypes are classified into eight genotypes, from A to H, based on the inter-group divergence of 8% or more in the complete genome nucleotide sequence, or a 4% or greater divergence of the Surface gene[2-4]. Recent studies have reported recombination between the HBV genomes of two genotypes. Two kinds of HBV genotype B have emerged[5-7] i.e. recombinant with genotype C and without recombination with C. Mixed genotype refers to an infection that contains more than one genotype in the same patient and is usually the result of multiple exposures and super-infection, the complete genome of each strain belongs to one genotype. According to Robertson et al[8], recombination can be detected when different genes or different regions within the same gene are placed by phylogenetic analysis into different sequence subtypes.
We and others from India have reported the presence of mixed genotype A and D[9-12]. However, despite the presence of mixed genotypes, there are no reports from India about the presence of recombination, especially using the full-length HBV genome sequencing approach.
In the present study, we have identified recombinant genotype A and D in patients with CLD and HCC due to chronic HBV infection.
MATERIALS AND METHODS
Patients and serological markers
Twelve treatment-naive chronic HBV infected patients [five with cirrhosis, five with chronic hepatitis B (CHB), and two with HCC] were enrolled. The serum from these patients was tested for the presence of hepatitis B surface antigen (HBsAg) by ELISA (Abbot Laboratories, North Chicago, USA and Organon Tecknika, Boxtel, Netherlands). In addition, the serum was tested for hepatitis e antigen (HBeAg), antibody to hepatitis e antigen (anti-HBe), hepatitis B core Antigen (IgG anti-HBc) by ELISA (Organon Tecknika, Boxtel, Netherlands). Assessment of the severity of liver disease was made by Child-Pugh score[13]. Approval of the institutional ethical committee was obtained to undertake this study.
HBV DNA quantitation
HBV DNA was quantified by a commercially available hybrid capture assay (Ultra sensitive kit, Digene, USA) with the lower limit of detection being 4700 copies/mL.
Full-length HBV DNA amplification
HBV DNA was extracted by using 0.5 to 1.0 mL of patient’s plasma using Sera Lysis Buffer (10 mmol/L Tris, 5 mmol/L EDTA, 50 mmol/L NaCl), SDS (1%) and proteinase K (1 mg/mL), followed by extraction with Tris-saturated phenol (pH 7.9) chloroform and then precipitation with ethanol. The obtained pellet was dried and dissolved in 30 μL of 1 × TE buffer (10 mmol/L Tris 1 mmol/L EDTA), a method described previously[12]. Full-length HBV DNA amplification was done by polymerase chain reaction (PCR), as described by Gunther’s method[14]. The Taq polymerase with DNA proof reading activity was used. (Expand high fidelity Taq-Polymerase Roche GmBH Basel, Switzerland). Primers were: P1-CCGGAAAGC TTGAGCTCTTCTTTTTCACCTCTGCCTAATCA (1821-1841), P2-CGGAAAGCTTGAGCTCTTCAAAAAGTTGCA TGGTGCTGG (1823-1806). The reaction conditions for PCR were 94°C for 5 min, 94°C for 1 min, 60°C for 1.5 min; 68°C for 7 min and extension at 72°C for 10 min, 35 cycles were performed. Purified full-length HBV DNA from recombinant vector pCF 80 (Tetramer of 3.2 kb HBV cloned in pBR322) was used as a positive control. DNA extracted from serum samples of healthy individuals and commercially available molecular biology grade water served as the negative control. Every set of PCR amplifications included HBV-positive and-negative controls. Primers were designed using the software Primer Express.
Sequencing full-length HBV genomes
PCR-amplified products were purified using the Qiagen Gel purification kit according to their recommended protocol. Internal primers used for sequencing given in Table 1 were used for sequencing in an automated DNA sequencer (ABI Prism 3730 Applied Biosystems, Foster City, USA). The nucleotide sequence data reported in this paper appears in the GenBank/EMBL/DDBJ nucleotide sequence databases with accession numbers EF103275-EF103285 and AY945305. The genome length has been measured according to Galibert et al[15].
Table 1.
Primers used for sequencing
| Name | Sequence | (nt.) |
| P1_F | 5-TTTTTCACCTCTGCCTAATCA-3 | (1821-1841) |
| SEQ_F1 | 5-AGGCAACTATTGTGGTTTCA-3 | (2194-2212) |
| SEQ_F2 | 5-TCTTTAACCCTCATTGGAAA-3 | (2516-2535) |
| SEQ_F3 | 5-TCACCATATTCTTGGGAACAAGA-3 | (2823-2845) |
| SEQ_F4 | 5-CTTCCTGCTGGTGGCTCCAGTTC-3 | (53-75) |
| SEQ_F5 | 5-CTCGTGGTGGACTTCTCTC-3 | (253-272) |
| SEQ_F6 | 5-ATCCTCAACCACCAGCACG-3 | (492-510) |
| SEQ_F7 | 5-TATTGGGGGCCAAGTCTGTA-3 | (749-768) |
| SEQ_F8 | 5-TTTACCCCGTTGCYAGGCA-3 | (1144-1162) |
| SEQ_F9 | 5-CTCATCTGCCGGACCGTG-3 | (1562-1581) |
| P2_R0 | 5-AAAAAGTTGCATGGTGCTGG-3 | (1825-1841) |
Data analysis
HBV genotyping was done by phylogenetic analysis using full-length sequences, core and preS2 and surface regions. Briefly, sequences were aligned using the CLUSTALW software[16]. Phylogenetic trees were constructed using the Kimura two-parameter matrix and neighbor-joining (NJ) method by MEGA software version 3.1[17]. To confirm the reliability of the phylogenetic tree analysis, bootstrap resampling and reconstruction were carried out 1000 times. Recombination was investigated by SimPlot[18] distributed by the author Ray at (http://www.welch.jhu.edu/). Boot scanning was performed for each of the strains using four sequences at a time[19], i.e. putative recombinant sequence, two consensus sequences of the parental genotype and one consensus sequence as an out-group.
RESULTS
Patients and virological characteristics
Baseline characteristics of the study population are given in Table 2. The majority of the patients were male (M: 10, F: 2). Of the 12 patients, five had cirrhosis, all diagnosed radiologically; [one decompensated with a Child-Turcotte-Pugh (CTP) score of 8, and four compensated with a CPT score of 5], and five with CHB (all biopsy proven), and two had HCC. Of the 12, eight were HBeAg-positive and the remaining four were anti-HBe positive. The EcoRI restriction enzyme site was present in seven of the full-length sequences, whereas it was absent in five. All sequences had a nucleotide (nt.) length of 3182 except genotype A sequence, which had 3221 nt.
Table 2.
Baseline characteristics of patients
| Patient No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| Age (yr) | 8 | 24 | 20 | 20 | 30 | 29 | 88 | 38 | 45 | 40 | 62 | 50 |
| Gender | M | F | M | M | M | M | M | F | M | M | M | M |
| Diagnosis | CHB | CHB | CHB | CHB | CHB | Cirr. | Cirr. | Cirr. | Cirr. | Cirr. | HCC | HCC |
| Bilirubin (mg/dL) | 1.1 | 0.68 | 1.1 | 0.6 | 1.1 | 1 | 0.8 | 0.4 | 2.1 | 1.4 | 0.9 | 1.2 |
| ALT (IU/L) | 46 | 54 | 42 | 59 | 52 | 107 | 78 | 29 | 101 | 98 | 31 | 43 |
| Albumin (g/dL) | 4 | 4.1 | 4.3 | 4.8 | 4.3 | 4 | 2.2 | 3.9 | 3.0 | 3.7 | 4.1 | 4 |
| PT prolongation (s) | 2 | 2 | 1 | 1 | 2 | 4 | 4 | 18 | 8 | 5 | 2 | 2 |
| Ascites | No | No | No | No | No | No | No | No | Yes | No | No | No |
| Encephalopathy | No | No | No | No | No | No | No | No | No | No | No | No |
| CTPscore | 5 | 5 | 5 | 5 | 5 | 6 | 8 | 7 | 9 | 6 | 5 | 5 |
| HBeAg | Pos | Pos | Pos | Pos | Pos | Pos | Pos | Neg | Neg | Pos | Pos | Neg |
| HBV DNA (log copies/mL) | 5.4 | 5.6 | 6.4 | 7.2 | 5.6 | 6.4 | 4.9 | 5.1 | 5.7 | 6.1 | 5.3 | 6.5 |
| HAI | 2 | 5 | 6 | 6 | 5 | - | - | - | - | - | - | - |
| F Score | 1 | 1 | 1 | 3 | 2 | - | - | - | - | - | - | - |
Cirr: Cirrhosis; ALT: Alanine aminotransferase; PT: Prothrombin time; CTP: Child-Turcotte-Pugh; HAI; Histological activity index; F: Fibrosis.
Distribution of genotypes
Phylogenetic analysis using complete HBV genomes of genotypes A to H derived from GenBank revealed the presence of genotype A and D in the study population. Genotype D was predominant, accounting for 92% of the study patients (Figure 1). The nature of genotype D was confirmed by the presence of a 33-bp deletion in the preS1 and a 6-bp deletion in the core terminal regions. Whereas in the genotype A sequence, the 33-bp and 6 bp deletions were absent. Phylogenetic analysis of the core revealed the same results as analysis done with complete HBV genomes as shown in Figure 1A and C.
Figure 1.
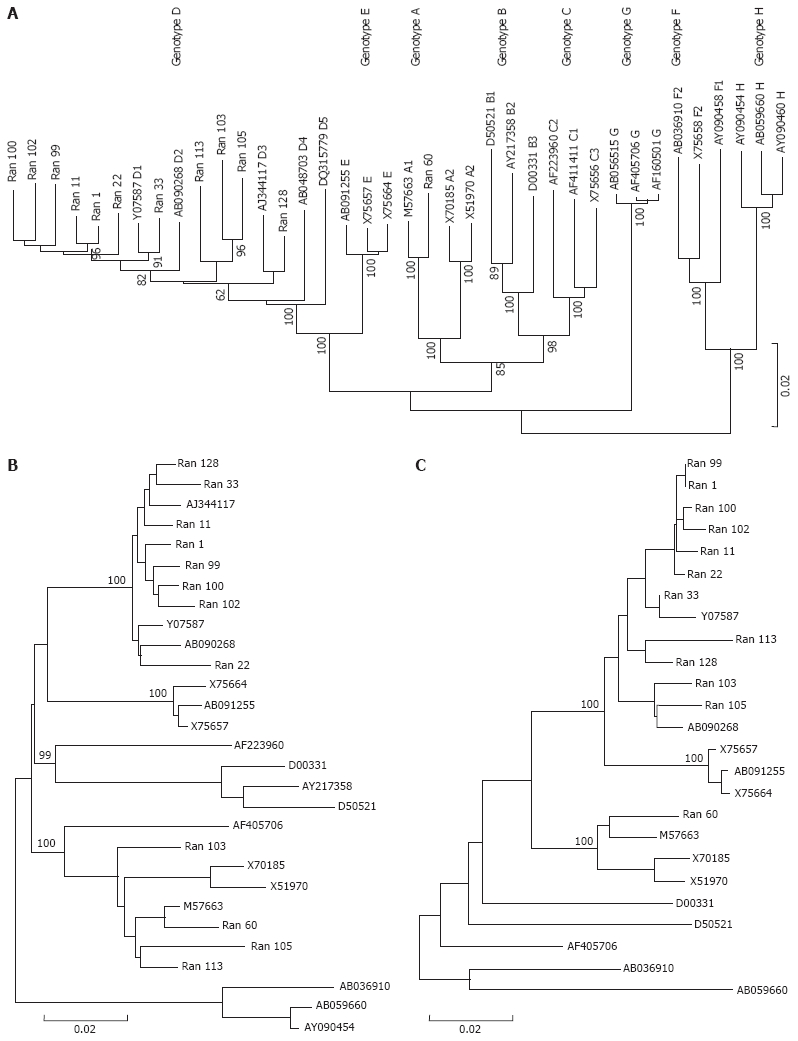
Phylogenetic tree revealed the distribution of genotypes. A: Neighbor joining (NJ) phylogenetic tree was constructed based on the complete genome sequences of 26 HBV reference strains from DDBJ/EMBL/Genbank and 12 Indian HBV isolates in the present study. The bootstrap values obtained from 1000 replicas are shown at the nodes of the main branches. HBV strains from this study are marked as Ran as suffix followed by the isolate number. Reference sequences are denoted by their accession numbers. The capital letter A to H denotes the HBV genotype; B: Phylogenetic analysis done on the PreS2 and surface regions showing three additional (103,105,113) isolates to be grouped in genotype A branch. Phylogenetic trees were constructed by NJ Kimura two-parametric method. Bootstrap values are shown at the nodes of the main branch and at the bottom distance is given; C: Phylogenetic analysis using the Core region. Phylogenetic tree was constructed by NJ Kimura 2-parametric method Bootstrap values are shown at the nodes of the main branch and at the bottom distance given.
Presence of A and D genotype recombination
Phylogenetic analysis of preS2/surface region of 12 isolates revealed clustering of three more sequences in addition to isolate 60 in the genotype A branch (Figure 1B). Presence of recombination was confirmed by boot scanning SimPlot analysis; all the sequences were subjected to analysis using the consensus sequence of genotype A, D and H as the out-group as shown in Figure 2. Recombination break points, of three recombinant strains were detected in preS2 and surface ORFs. Isolate 113 had break points at nt 595-618; isolate 105 had break points at nt 639-659 and 723-737. Isolate 103 had break points at nt 319-359 and 1170-1184. PreS2 and surface regions showed similarity with genotype A at 18 amino acid positions in the recombinant sequences, whereas it was absent in the surface, core and X ORFs. Four of them were identified in the preS2, whereas 13 in the surface region, as shown in Figure 3A and B.
Figure 2.
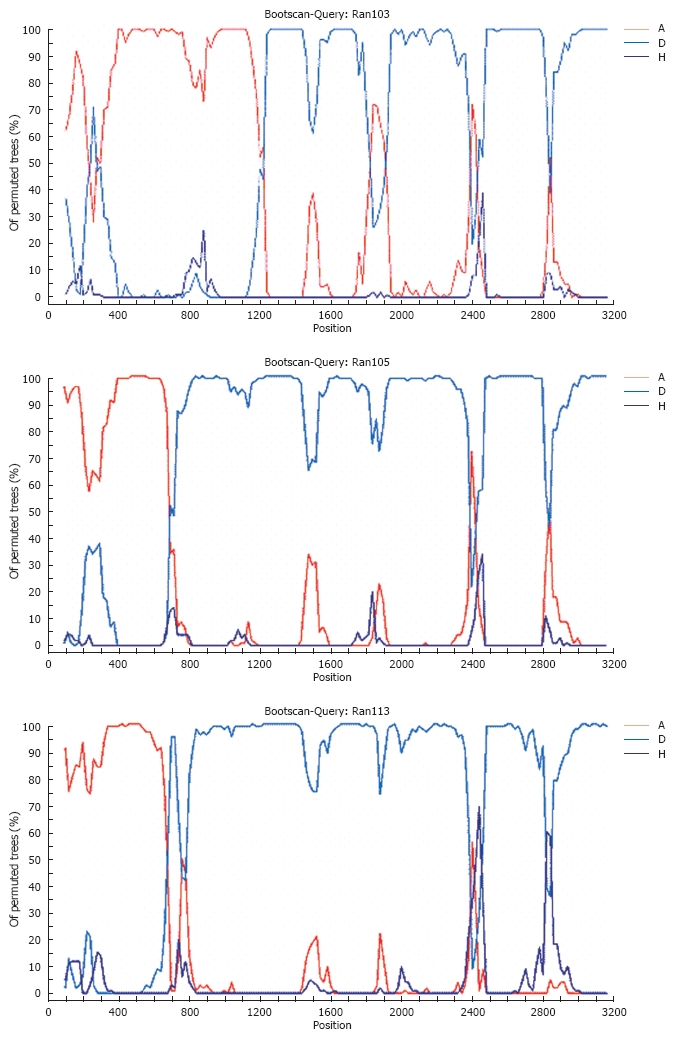
SimPlot analysis demonstrating the recombination in two isolates 103 and105, which were subjected to bootscan analysis over the entire genome using SimPlot program (Lole et al[18]).
Figure 3.
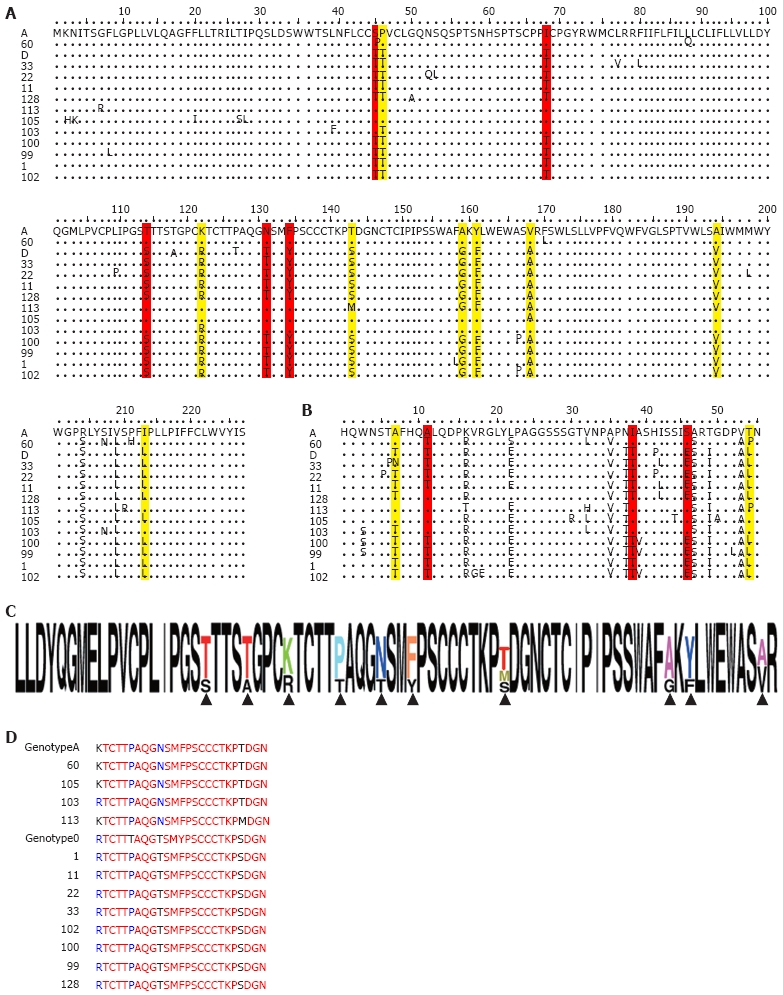
Recombinant sequence similarities with genotype A in PreS2 and Surface region. A: Surface region alignment with genotype A and D amino acids, marked in red shows all three recombinant sequences having similarity with genotype A, whereas yellow marked amino acids shows at least one recombinant sequence similar to genotype A, over all 18 positions; B: Shows same in the preS2 region; C: Shows all the amino acids of MHR of genotype D in black, upward arrow indicates the change of amino acid; D: Shows “a” determinant region and corresponding alignment with 12 isolates.
Major hydrophilic and the “a” determinant regions
As shown in Figure 3C, when analyzed considering only genotype D, the major hydrophilic region (MHR) showed substitutions at 10 amino acid positions. Of the 10 changes, five spanned the “a” determinant region. When similar analysis was done considering genotype A, we could detect a single mutation in isolate 113 at position 144, changing threonine to methionine in the “a” determinant region of the surface region. All the isolates showed the presence of concomitant threonine to proline change at position 131 of the “a” determinant region, which is homologous to the genotype A sequence.
Sequence characteristics of precore/ core and X ORFs
Among the 12 patients, precore stop codon mutation (W28Stop) was found in two patients, and both belonged to the recombinant genotype. We could document the difference in the core nucleotide sequences in the recombinant sequences; however, they were not exactly similar to the typical genotype A pattern. We detected the presence of T1936C nucleotide mutation in the core gene in one of the HCC patients, isolate number 113.
X ORF: In two patients, we detected mutations in X ORF. Both belonged to the recombinant genotype, i.e. isolate 113 mutations were detected at three positions I127T, K130M, V131I, and isolate 105 was harboring a single mutation at position I127L.
DISCUSSION
Phylogenetic analysis based on two different genomic regions, preS2/surface and core, suggested the existence of recombinant strains in Indian isolates of HBV. On examination of the preS2 and the surface region sequences, a close relation with genotype A sequence was detected in three genotype D sequences respective to core, which was genotype D. Further analysis of corresponding genomes allowed us to map the crossover junctions and to confirm the findings of recombination.
HBV recombination is not a new phenomenon, it is important from an evolutionary as well as epidemiological point of view. As increasing number of full-length HBV genome sequences are reported, and a higher frequency of recombinant hybrid genomes is being recognized. Evidence of HBV recombination from different parts of the world suggests the presence of recombination of HBV genotypes involving A/C, A/D, A/E, B/C, B/D, G/A and G/C strains[20-23]. In Asia, recombination of genotype C/D has been reported from Tibet and China[21,23], whereas recombination of B and C has been detected in Japan[24]. HBV strains from Vietnamese patients also show evidence of recombination of C and A genotypes[25]. Genotype A and D recombination has only been documented in CHB patients from South Africa[26]. Although the recombination of A and D genotypes has been detected from Italian and Indian HBV strains, such patients were surface-antigen negative[7]. Furthermore, breakpoints of recombination were different from the presently identified recombinant strains[27,28].
A switch in genotype has been documented during change of HBsAg-positive to serologically negative phase[29]. As we detected recombination in the preS2/surface coding region, we focused on the major hydrophilic region, “a” determinant region of surface ORF, speculating that the changes in the MHR and the “a” determinant region could lead to absence of the surface antigen, using standard serological methods. Our analysis revealed changes at 11 amino acid positions when the analysis was done using genotype D (Figure 3C). When the analysis was carried out considering both A and D genotypes, we could detect changes at only a single position substituting (K) lysine with (M) methionine in the “a” determinant region. However, the amino acid substitutions that are supposed to alter the conformation of “a” determinant region were not found.
Compared to genotype D, genotype A is more prevalent in the HBeAg-positive than in the anti-HBe-positive phase[30,31]. It is known that HBV genotype D virus has a selection advantage to form the precore stop codon mutation, as compared to the genotype A virus, the selection being at the pre-genome encapsidation level. However, in the recombinant sequences identified in the present study, we detected the precore stop codon mutation in two of them. The presence of the stop codon in the precore region and co-infection with other genotypes A, C and H are two important features of HBV genotype G[32]. In presently identified recombinant sequences, we detected the presence of Pc G1896A, and this suggests a possible similar situation and a matter for further investigation.
Recent reports suggest the presence of mixed genotypes in Asia. Europe and Africa in CHB patients, including India[9,11,33]. Moreover, higher levels of HBV replication have been shown to be associated with mixed genotype infection[33].
It is quite possible that the PreS, core, X and P proteins are continually expressed but the preS2 region/protein of genotype D is lost for a short interval during recombination with genotype A sequences, mimicking a molecular window period. It is not yet known whether recombination is advantageous for the virus or the host, but it is quite possible that this phenomenon increases the chances of virus survival and doping the host defense system.
One of the reasons for enhanced HCC development in young African adults could be high HBsAg expression in genotype-A-harboring patients[34] HBV genotype A directs the high level of synthesis of HBsAg in proportion to viral DNA, core protein and HBeAg[35].
The frequency of detection of spliced viral genomes is higher in CHB cases compared to acute and resolved HBV infections. The generation of recombinant HBV could be intracellular, as the ratio of full-length and spliced genomes isolated from the intracellular compartment was significantly higher than from extracellular space. This indicates that, compared to those containing spliced genomes, nucleocapsids containing full-length genomes are preferentially enveloped and released from the cell, and could be one of the reasons for severe liver disease[36]. It would be worth while to study the co-infection of two genotypes, and to establish whether the changes accumulate in one cell or together in the newly infected cells. Genetic exchanges between different viral strains within the infected hepatocytes could be one of the possible reasons for recombination.
HBV infection is the predominant factor for the development of HCC in India[37]. Several reports suggest integration of the preS/S region in cancerous liver tissue[38-40]. Binding of the PreS region with fibronectin and transactivation of TGF α could lead to development of cirrhosis and HCC[41-43]. It is quite unique that HBV uses its strongest promoter preS2/S for expression of the host cellular genes, which are advantageous for the virus itself.
HBV recombinant sequences were analyzed with the orangutan and gibbon monkey hepatitis virus. However, we could not find any association of recombinant sequences with them, and the reason for such transmission was clarified (data not shown). Secondly, phylogenetic analysis was done using the Italian and Indian recombinant sequences reported previously[27,28]. However, they were not clustering in the same region as detected in the present recombinant sequences (data not shown).
There are three theories proposed for evolution of HBV: The new world origin theory[5], co-evolution theory[3,44] and co-speciated theory[45,46]. We postulate a competitive selection theory in which the virus and the host cellular machinery compete, and involvement of various unidentified ways by the virus to combat the host defense mechanisms. A few of these could be the splicing, integration, recombination and down-regulation of MHC I. On the other hand, host APOBEC response to edit the viral genome, CTL proteosome complex and various host genetic factors, taking into consideration ethnicity, may play a part as well. Recombinant detection of mixed genotypes, however, may be the tip of the iceberg as a template switch over, splicing and extensive editing by all APOBEC 3 proteins, which have not been well studied.
It can be argued that, in our study, the HBV sequences were analyzed only at one stage of the disease, the process of sequential changes and the time points were not tracked. This was a preliminary study and such studies are quite cumbersome and expensive.
In summary, we identified new A/D recombinants from Indian CLD and HCC patients. To the best of our knowledge, this is the first report which describes recombinant A and D genotype from HBsAg-positive patients from Asia, and indicates the association of recombinant HBV genotype with HCC. The results of the present study warrants further larger studies to identify populations of recombinant viruses in different clinical categories of HBV patients.
COMMENTS
Background
Recombination is common in retroviruses, especially human immunodeficiency virus (HIV). As the hepatitis B virus (HBV) uses the reverse transcription step using the pre-genomic RNA, the rate of mutation accumulation is at a much higher rate compared to other DNA viruses. India, being highly populated, harbors the second largest pool of HBV carriers. Recombination is also one of the mechanisms of sequence variability and could account for the non-response to antiviral therapy as well as vaccine. Though recombination from the Indian subcontinent has been detected, authors for the first time report recombination of A and D genotype in HBsAg-positive chronic HBV patients.
Research frontiers
Non-response to antiviral drugs and vaccine is one of the hot research related to the article.
Innovations and breakthroughs
This is believed to be the first report describing the recombinant genotype on the Indian subcontinent.
Applications
Large-scale studies are warranted to determine the prevalence and profile of the recombinant genotype on the Indian subcontinent. The affect of antiviral therapy on the recombinant virus is also warranted.
Terminology
Mixed genotype refers to an infection that contains more than one genotype in the same patient, and is usually the result of multiple exposures and super-infection, the complete genome of each strain belongs to one genotype. Recombinant genotype can be detected when different genes or different regions within the same gene are placed by phylogenetic analysis into different sequence subtypes.
Peer review
This is an interesting paper, which confirms the presence of recombination, and full-length HBV from chronic patients were sequenced and analyzed. Authors identified and characterized recombinant A and D genotype HBV in HBsAg-positive patients.
Acknowledgments
Authors thank Indian Council of Medical Research for carrying out this study and providing funds through Advanced Center for Liver Diseases project.
Footnotes
Supported by Indian Council of Medical Research-Advanced Center for Liver Diseases Project (ICMR-ACLD)
Peer reviewer: Vasiliy I Reshetnyak, MD, PhD, Professor, Scientist Secretary of the Scientific Research Institute of General Reanimatology, 25-2, Petrovka str., Moscow 107031, Russia
S- Editor Li DL L- Editor Alpini GD E- Editor Lin YP
References
- 1.Wold Health Organization report. Available from URL: http://www.who.org.
- 2.Kramvis A, Kew M, Francois G. Hepatitis B virus genotypes. Vaccine. 2005;23:2409–2423. doi: 10.1016/j.vaccine.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 3.Norder H, Courouce AM, Magnius LO. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- 4.Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69(Pt 10):2575–2583. doi: 10.1099/0022-1317-69-10-2575. [DOI] [PubMed] [Google Scholar]
- 5.Bollyky PL, Rambaut A, Harvey PH, Holmes EC. Recombination between sequences of hepatitis B virus from different genotypes. J Mol Evol. 1996;42:97–102. doi: 10.1007/BF02198834. [DOI] [PubMed] [Google Scholar]
- 6.Bowyer SM, Sim JG. Relationships within and between genotypes of hepatitis B virus at points across the genome: footprints of recombination in certain isolates. J Gen Virol. 2000;81:379–392. doi: 10.1099/0022-1317-81-2-379. [DOI] [PubMed] [Google Scholar]
- 7.Morozov V, Pisareva M, Groudinin M. Homologous recombination between different genotypes of hepatitis B virus. Gene. 2000;260:55–65. doi: 10.1016/s0378-1119(00)00424-8. [DOI] [PubMed] [Google Scholar]
- 8.Robertson DL, Sharp PM, McCutchan FE, Hahn BH. Recombination in HIV-1. Nature. 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 9.Thakur V, Guptan RC, Kazim SN, Malhotra V, Sarin SK. Profile, spectrum and significance of HBV genotypes in chronic liver disease patients in the Indian subcontinent. J Gastroenterol Hepatol. 2002;17:165–170. doi: 10.1046/j.1440-1746.2002.02605.x. [DOI] [PubMed] [Google Scholar]
- 10.Vivekanandan P, Abraham P, Sridharan G, Chandy G, Daniel D, Raghuraman S, Daniel HD, Subramaniam T. Distribution of hepatitis B virus genotypes in blood donors and chronically infected patients in a tertiary care hospital in southern India. Clin Infect Dis. 2004;38:e81–e86. doi: 10.1086/383144. [DOI] [PubMed] [Google Scholar]
- 11.Chattopadhyay S, Das BC, Kar P. Hepatitis B virus genotypes in chronic liver disease patients from New Delhi, India. World J Gastroenterol. 2006;12:6702–6706. doi: 10.3748/wjg.v12.i41.6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chauhan R, Kazim SN, Bhattacharjee J, Sakhuja P, Sarin SK. Basal core promoter, precore region mutations of HBV and their association with e antigen, genotype, and severity of liver disease in patients with chronic hepatitis B in India. J Med Virol. 2006;78:1047–1054. doi: 10.1002/jmv.20661. [DOI] [PubMed] [Google Scholar]
- 13.Pugh RN, Murray-Lyon IM, Dawson JL, Pietroni MC, Williams R. Transection of the oesophagus for bleeding oesophageal varices. Br J Surg. 1973;60:646–649. doi: 10.1002/bjs.1800600817. [DOI] [PubMed] [Google Scholar]
- 14.Gunther S, Li BC, Miska S, Kruger DH, Meisel H, Will H. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J Virol. 1995;69:5437–5444. doi: 10.1128/jvi.69.9.5437-5444.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 16.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput Appl Biosci. 1994;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- 18.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salminen MO, Carr JK, Burke DS, McCutchan FE. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retroviruses. 1995;11:1423–1425. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- 20.Kurbanov F, Tanaka Y, Fujiwara K, Sugauchi F, Mbanya D, Zekeng L, Ndembi N, Ngansop C, Kaptue L, Miura T, et al. A new subtype (subgenotype) Ac (A3) of hepatitis B virus and recombination between genotypes A and E in Cameroon. J Gen Virol. 2005;86:2047–2056. doi: 10.1099/vir.0.80922-0. [DOI] [PubMed] [Google Scholar]
- 21.Cui C, Shi J, Hui L, Xi H, Zhuoma , Quni , Tsedan , Hu G. The dominant hepatitis B virus genotype identified in Tibet is a C/D hybrid. J Gen Virol. 2002;83:2773–2777. doi: 10.1099/0022-1317-83-11-2773. [DOI] [PubMed] [Google Scholar]
- 22.Suwannakarn K, Tangkijvanich P, Theamboonlers A, Abe Suwannakarn K, Tangkijvanich P, Theamboonlers A, Abe K, Poovorawan Y. A novel recombinant of Hepatitis B virus genotypes G and C isolated from a Thai patient with hepatocellular carcinoma. J Gen Virol. 2005;86:3027–3030. doi: 10.1099/vir.0.81241-0. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Liu Z, Zeng G, Wen S, Qi Y, Ma S, Naoumov NV, Hou J. A new intertype recombinant between genotypes C and D of hepatitis B virus identified in China. J Gen Virol. 2005;86:985–990. doi: 10.1099/vir.0.80771-0. [DOI] [PubMed] [Google Scholar]
- 24.Sugauchi F, Orito E, Ichida T, Kato H, Sakugawa H, Kakumu S, Ishida T, Chutaputti A, Lai CL, Ueda R, et al. Hepatitis B virus of genotype B with or without recombination with genotype C over the precore region plus the core gene. J Virol. 2002;76:5985–5992. doi: 10.1128/JVI.76.12.5985-5992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannoun C, Norder H, Lindh M. An aberrant genotype revealed in recombinant hepatitis B virus strains from Vietnam. J Gen Virol. 2000;81:2267–2272. doi: 10.1099/0022-1317-81-9-2267. [DOI] [PubMed] [Google Scholar]
- 26.Owiredu WK, Kramvis A, Kew MC. Hepatitis B virus DNA in serum of healthy black African adults positive for hepatitis B surface antibody alone: possible association with recombination between genotypes A and D. J Med Virol. 2001;64:441–454. doi: 10.1002/jmv.1070. [DOI] [PubMed] [Google Scholar]
- 27.Simmonds P, Midgley S. Recombination in the genesis and evolution of hepatitis B virus genotypes. J Virol. 2005;79:15467–15476. doi: 10.1128/JVI.79.24.15467-15476.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Xing K, Deng R, Wang J, Wang X. Identification of Hepatitis B virus putative intergenotype recombinants by using fragment typing. J Gen Virol. 2006;87:2203–2215. doi: 10.1099/vir.0.81752-0. [DOI] [PubMed] [Google Scholar]
- 29.Bahn A, Gerner P, Martine U, Bortolotti F, Wirth S. Detection of different viral strains of hepatitis B virus in chronically infected children after seroconversion from HBsAg to anti-HBs indicating viral persistence. J Hepatol. 1997;27:973–978. doi: 10.1016/s0168-8278(97)80139-0. [DOI] [PubMed] [Google Scholar]
- 30.Li JS, Tong SP, Wen YM, Vitvitski L, Zhang Q, Trepo C. Hepatitis B virus genotype A rarely circulates as an HBe-minus mutant: possible contribution of a single nucleotide in the precore region. J Virol. 1993;67:5402–5410. doi: 10.1128/jvi.67.9.5402-5410.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mangia A, Chung YH, Hoofnagle JH, Birkenmeyer L, Mushahwar I, Di Bisceglie AM. Pathogenesis of chronic liver disease in patients with chronic hepatitis B virus infection without serum HBeAg. Dig Dis Sci. 1996;41:2447–2452. doi: 10.1007/BF02100141. [DOI] [PubMed] [Google Scholar]
- 32.Kato H, Orito E, Gish RG, Bzowej N, Newsom M, Sugauchi F, Suzuki S, Ueda R, Miyakawa Y, Mizokami M. Hepatitis B e antigen in sera from individuals infected with hepatitis B virus of genotype G. Hepatology. 2002;35:922–929. doi: 10.1053/jhep.2002.32096. [DOI] [PubMed] [Google Scholar]
- 33.Toan NL, Song le H, Kremsner PG, Duy DN, Binh VQ, Koeberlein B, Kaiser S, Kandolf R, Torresi J, Bock CT. Impact of the hepatitis B virus genotype and genotype mixtures on the course of liver disease in Vietnam. Hepatology. 2006;43:1375–1384. doi: 10.1002/hep.21188. [DOI] [PubMed] [Google Scholar]
- 34.Kew MC, Kramvis A, Yu MC, Arakawa K, Hodkinson J. Increased hepatocarcinogenic potential of hepatitis B virus genotype A in Bantu-speaking sub-saharan Africans. J Med Virol. 2005;75:513–521. doi: 10.1002/jmv.20311. [DOI] [PubMed] [Google Scholar]
- 35.Sugiyama M, Tanaka Y, Kato T, Orito E, Ito K, Acharya SK, Gish RG, Kramvis A, Shimada T, Izumi N, et al. Influence of hepatitis B virus genotypes on the intra- and extracellular expression of viral DNA and antigens. Hepatology. 2006;44:915–924. doi: 10.1002/hep.21345. [DOI] [PubMed] [Google Scholar]
- 36.Sommer G, van Bommel F, Will H. Genotype-specific synthesis and secretion of spliced hepatitis B virus genomes in hepatoma cells. Virology. 2000;271:371–381. doi: 10.1006/viro.2000.0331. [DOI] [PubMed] [Google Scholar]
- 37.Sarin SK, Thakur V, Guptan RC, Saigal S, Malhotra V, Thyagarajan SP, Das BC. Profile of hepatocellular carcinoma in India: an insight into the possible etiologic associations. J Gastroenterol Hepatol. 2001;16:666–673. doi: 10.1046/j.1440-1746.2001.02476.x. [DOI] [PubMed] [Google Scholar]
- 38.Caselmann WH, Meyer M, Kekule AS, Lauer U, Hofschneider PH, Koshy R. A trans-activator function is generated by integration of hepatitis B virus preS/S sequences in human hepatocellular carcinoma DNA. Proc Natl Acad Sci USA. 1990;87:2970–2974. doi: 10.1073/pnas.87.8.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pineau P, Marchio A, Mattei MG, Kim WH, Youn JK, Tiollais P, Dejean A. Extensive analysis of duplicated-inverted hepatitis B virus integrations in human hepatocellular carcinoma. J Gen Virol. 1998;79(Pt 3):591–600. doi: 10.1099/0022-1317-79-3-591. [DOI] [PubMed] [Google Scholar]
- 40.Zhong S, Chan JY, Yeo W, Tam JS, Johnson PJ. Frequent integration of precore/core mutants of hepatitis B virus in human hepatocellular carcinoma tissues. J Viral Hepat. 2000;7:115–123. doi: 10.1046/j.1365-2893.2000.00209.x. [DOI] [PubMed] [Google Scholar]
- 41.Budkowska A, Bedossa P, Groh F, Louise A, Pillot J. Fibronectin of human liver sinusoids binds hepatitis B virus: identification by an anti-idiotypic antibody bearing the internal image of the pre-S2 domain. J Virol. 1995;69:840–848. doi: 10.1128/jvi.69.2.840-848.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ono M, Morisawa K, Nie J, Ota K, Taniguchi T, Saibara T, Onishi S. Transactivation of transforming growth factor alpha gene by hepatitis B virus preS1. Cancer Res. 1998;58:1813–1816. [PubMed] [Google Scholar]
- 43.Yang J, Ding X, Zhang Y, Bo X, Zhang M, Wang S. Fibronectin is essential for hepatitis B virus propagation in vitro: may be a potential cellular target? Biochem Biophys Res Commun. 2006;344:757–764. doi: 10.1016/j.bbrc.2006.03.204. [DOI] [PubMed] [Google Scholar]
- 44.Magnius LO, Norder H. Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene. Intervirology. 1995;38:24–34. doi: 10.1159/000150411. [DOI] [PubMed] [Google Scholar]
- 45.MacDonald DM, Holmes EC, Lewis JC, Simmonds P. Detection of hepatitis B virus infection in wild-born chimpanzees (Pan troglodytes verus): phylogenetic relationships with human and other primate genotypes. J Virol. 2000;74:4253–4257. doi: 10.1128/jvi.74.9.4253-4257.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simmonds P. The origin and evolution of hepatitis viruses in humans. J Gen Virol. 2001;82:693–712. doi: 10.1099/0022-1317-82-4-693. [DOI] [PubMed] [Google Scholar]


