Abstract
Brain tumors are the most common solid tumors of childhood, accounting for over 20% of cancers in children under 15 years of age. Pilocytic astrocytomas (PAs), World Health Organization grade I, are one of the most frequently occurring childhood brain tumors, yet little is known about genetic changes characterizing this entity. We have used microarray comparative genomic hybridization at 0.97 Mb resolution to study a series of PAs (n = 44). No copy number abnormality was seen in 64% of cases at this resolution. However, whole chromosomal gain (median 5 chromosomes affected) occurred in 32% of tumors. The most frequently affected chromosomes were 5 and 7 (11 of 44 cases each) followed by 6, 11, 15, and 20 (greater than 10% of cases each). Findings were confirmed by fluorescence in situ hybridization and microsatellite analysis in a subset of tumors. Chromosomal gain was significantly more frequent in PAs from patients over 15 years old (p = 0.03, Fisher exact test). The number of chromosomes involved was also significantly greater in the older group (p = 0.02, Mann-Whitney U test). One case (2%) showed a region of gain on chromosome 3 and one (2%) a deletion on 6q as their sole abnormalities. This is the first genomewide study to show this nonrandom pattern of genetic alteration in pilocytic astrocytomas.
Keywords: aCGH, Aneuploidy, Pilocytic astrocytoma
INTRODUCTION
Almost one-fourth of all neoplasia in children under 15 years of age occurs in the brain or spinal cord (1). Pilocytic astrocytomas (PAs), World Health Organization malignancy grade I (2), account for more than 20% of these central nervous system tumors in this age group (3). They occur mainly in children and young adults.
Most cases accessible to gross resection are treated by surgery alone. Prognosis is generally good, with 10-year survival rates of up to 96% (4). PAs are characterized by their relatively circumscribed and often cystic presentation. They show limited infiltrative potential and only very infrequently progress to a more malignant phenotype. When this does occur, it is generally decades after the initial PA diagnosis (5, 6). Reports on the frequency of progression in PAs vary, with one report of <1% in cerebellar PAs, increasing to 1.8% if the patient had received radiotherapy (7). Another report on PAs at all sites suggests progression in 11% of cases in the absence of radiotherapy (8). PAs may also recur (in up to 19% of cases in one study) after apparent gross total resection (8, 9).
The widely varied histology of pilocytic astrocytomas makes their diagnosis challenging, and criteria for a PA diagnosis come with a number of caveats (10). This difficulty is highlighted by a study that found that over half of tumors classified as PAs at review had been misdiagnosed as higher-grade tumors at primary diagnosis (11).
Although the adult diffuse astrocytomas are becoming increasingly well characterized in terms of the molecular mechanisms involved in their tumorigenesis and progression, the mechanisms involved in PA formation remain unknown. Several cytogenetic and molecular genetic studies have looked for common aberrations in PAs (12-24). The majority has found few abnormal results with no consistent pattern. One study using fluorescence in situ hybridization with pericentromeric probes reported chromosomal gains, most frequently of 7 and 8, in one-third of cases but did not investigate all chromosomes (23). Using R-banding and classic cytogenetics, Zattara-Cannoni et al reported chromosomal abnormalities in 13 of 24 cases, including gains of chromosomes 7 and 8 in 3 cases each with a further 6 cases showing random gains or losses (24). A metaphase comparative genomic hybridization (CGH) study reported a pattern of multiple small gains and losses (22).
In an attempt to document genetic copy number changes in PAs, we carried out a microarray CGH analysis of the whole genome at a resolution of 0.97 Mb on a series of well-documented PA samples (n = 44). We report a novel, nonrandom pattern of whole chromosomal gain that shows age-dependent differences in its frequency of occurrence. Smaller regions of gain and loss were also observed in individual cases.
MATERIALS AND METHODS
Patients, Tumor Tissue, and DNA Isolation
Primary tumor samples from 44 patients with pilocytic astrocytoma were included in the analysis. The tumors were resected at the Karolinska Hospital, Stockholm, and the Sahlgrenska University Hospital, Gothenburg, Sweden, between 1987 and 1997. At resection, the median age of the patients was 9 years (range, 0.5–33 years). Seven patients (cases PA8, PA9, PA10, PA27, PA42, PA50, and PA65) had undergone a previous resection for PA. However, in some cases, the second operation may represent removal of residual material after a previous partial resection as opposed to true recurrence. Patients and tumors are described in Table 1. Histopathologic classification was according to the most recent World Health Organization recommendations (2). None of the cases in the series showed features consistent with a diagnosis of pilomyxoid astrocytoma. All tumor pieces were selected for DNA extraction after histologic examination to ensure a minimum of 70% tumor cells. DNA was extracted from tumor pieces and blood lymphocytes as described previously (25). Tumor samples were stored at −135°C and blood samples at −20°C before DNA extraction. Extracted DNA was stored at −80°C.
TABLE 1.
Clinical Data and Summary of Results
| Case | Gender | Age | Tumor Content (%)* | Location | aCGH Result |
|---|---|---|---|---|---|
| Cases from patients under 15 years of age | |||||
| PA2 | M | 5 | 90 | Cerebellum | Normal |
| PA5 | F | 3 | 90 | Cerebellum | Normal |
| PA7 | F | 3 | 85 | Optic nerve | Normal |
| PA8† | F | 8 | 75 | Cerebellum | Normal |
| PA9† | M | 12 | 85 | Hypothalamus | +7 |
| PA14 | M | 5 | 90 | Cerebellum | Normal |
| PA15 | F | 14 | N/A | Cerebellum | +7 |
| PA16 | M | 1 | 85 | Hypothalamus | Normal |
| PA19 | M | 13 | 80 | Third ventricle | Normal |
| PA20 | M | 6 | 90 | Cerebellum | +3p25.2–25.3 |
| PA23 | F | 5 | 90 | Fourth ventricle | Normal |
| PA24 | M | 9 | 90 | Fourth ventricle | Normal |
| PA25 | M | 10 | 85 | Optic nerve | +5, +7, +9, +10, +15 |
| PA27† | F | 13 | 75 | Cerebellum | +7 |
| PA28 | M | <1 yr | 80 | Right temporal lobe | Normal |
| PA32 | M | 4 | N/A | Cerebellum | Normal |
| PA33 | M | 2 | 90 | Third ventricle | Normal |
| PA39 | F | 11 | 90 | Cerebellum | +5 |
| PA40 | M | 4 | 85 | Optic nerve | Normal |
| PA42† | F | 12 | 85 | Left frontal lobe | +4, +5, +7, +12 |
| PA43 | M | 6 m | 90 | Optic nerve | Normal |
| PA44 | M | 1 | 85 | Optic nerve | Normal |
| PA45 | M | 9 | 90 | Cerebellum | Normal |
| PA47 | F | 8 | 90 | Cerebellum | −6q26 |
| PA48 | F | 5 | 85 | Cerebellum | Normal |
| PA49 | M | 3 | N/A | Cerebellum | Normal |
| PA50† | M | N/A | 70 | Fourth ventricle | +5, +6, +11, +15, +20 |
| PA51 | M | 8 | N/A | Cerebellum | Normal |
| PA52 | F | 6 | N/A | Cerebellum | Normal |
| PA57 | F | 9 | 80 | Cerebellum | Normal |
| PA65† | F | 5 | n/a | Brain stem | Normal |
| PA68 | M | 3 | 90 | Cerebellum | Normal |
| Cases from patients over 15 year of age | |||||
| PA1 | F | 33 | 85 | Optic nerve | Normal |
| PA3 | M | 32 | 80 | Cerebellum | Normal |
| PA10† | F | 20 | 85 | Cerebellum | Normal |
| PA12 | F | 17 | 85 | Cerebellum | +5, +6, +7, +10, +15 |
| PA18 | F | 33 | 75 | Pineal region | Normal |
| PA29 | M | 25 | 90 | Cerebellum | +5, +6, +7, +8, +11 |
| PA30 | F | 26 | 85 | Cerebellum | +5, +6, +7, +20 |
| PA35 | F | N/A | 80 | Cerebellum | +3q11.2, +5, +6, +7, +8, +11, +15, +20 |
| PA37 | M | 16 | 85 | Cerebellum | +5, +6, +11, +15, +20 |
| PA41 | F | 23 | 85 | Fourth ventricle | +5, +6, +7, +10, +15 |
| PA60 | M | 21 | N/A | Cerebellum | Normal |
| PA67 | F | 26 | 90 | N/A | +5, +7, +9, +10, +11, +12, +13, +14, +15 |
This figure includes any normal cells present in the tumor sample other than components of the vasculature (see “Discussion”).
This patient had undergone a previous resection for pilocytic astrocytoma (see “Materials and Methods”).
N/A, data not available; aCGH, microarray comparative genomic hybridization.
Microarray Comparative Genomic Hybridization
A genomic microarray at a resolution of 0.97 Mb was constructed with clones obtained from the Wellcome Trust Sanger Institute as described previously (26, 27). Briefly, clone DNA was extracted and amplified with 3 DOP primers. The DOP-polymerase chain reaction (PCR) products were then mixed and amplified with a 5′-amine-modified primer. These PCR products were spotted in duplicate onto amine-binding slides (Amersham Biosciences, Little Chalfont, UK) and stored in the dark at room temperature. Drosophila control clones and clones from each chromosome were distributed evenly through the 24 blocks of the array. Detailed clone information is given in Supplemental Table 1.
Labeling and hybridizations were performed essentially as described previously (28). Briefly, 400 ng of test and reference DNA were labeled using a Bioprime Labeling Kit (Invitrogen, Carlsbad, CA) with a modified dNTP reaction mixture as described (28). Test DNA was hybridized with sex-mismatched reference DNA from samples of pooled blood from 20 normal males or 20 normal females. The labeled and purified test and reference DNA were mixed and coprecipitated with 45 μg Cot1 DNA (Roche Diagnostics, Mannheim, Germany). The precipitated DNA was dissolved in hybridization buffer, incubated at 37°C for 2 hours, and hybridized to the array that had been prehybridized with 480 mg herring sperm DNA (Sigma-Aldrich, St. Louis, MO) and 80 μg Cot1 DNA. Arrays were allowed to hybridize for up to 24 hours at 37°C and then washed as described (28). In one case, a region of alteration was characterized in more detail through use of a tiling path array of chromosome 6. Clones, array construction, and hybridizations were carried out as previously described (29).
Array Analysis
Arrays were scanned and analyzed as described elsewhere (28). Spots were excluded and copy number scored largely as previously described (27). Briefly, clones with log2 ratios falling more than 3 standard deviations (SDs) from the mean log2 ratio of all spots in 6 normal–normal hybridizations were excluded as were those mapped to multiple locations of the NCBI35 assembly (available at www.ensembl.org/Homo_sapiens/) and those displaying inconsistent values compared with neighboring clones in multiple cases. This left 3,038 clones for use in analysis with a mean midclone separation of 0.97 Mb. Spots were excluded for each array if they had a reference intensity of less than quadruple the average intensity of 6 Drosophila bacterial artificial chromosome (BAC) spots. Spots with poor morphology, overlying artefact, or with a difference in ratio of more than 10% between replicates were also excluded. An average of 95.7% of spots passed per array.
Small regions (<10 clones) were assessed as gained or lost if 2 or more consecutive clones showed a log2 ratio outside of 3 SD from the mean of all log2 ratios in normal–normal hybridizations. Larger regions of gain or loss (such as chromosome arms or whole chromosomes) were assessed by subjective judgment in which all clones in a region were noticeably above or below the baseline of log2 ratios in other chromosomes.
Microsatellite Analysis
Microsatellite analysis was performed as described previously (30) using markers D1S243, D1S2667, D1S426, D1S2799, D6S344, D6S276, D6S1657, D6S311, D7S659, D7S494, D7S486, D7S480, D11S1318, D11S926, and D11S915. The primer sequences were those publicly available (www.ensembl.org/Homo_sapiens).
Interphase Fluorescence In Situ Hybridization on a Tissue Microarray
A tissue microarray of a subset of tumors was constructed as follows. Cores (n = 3 for each case) of 0.6-mm diameter were taken from 5 PAs after tumor-rich areas were identified in hematoxylin & eosin-stained sections. These cores were arrayed into a fresh paraffin block using a manual tissue arrayer (Beecher Instruments, Silver Spring, MD). Five-micrometer sections from the array were placed on positively charged slides and dried overnight at 37°C. After dewaxing in xylene and rehydration in an ethanol series, the slides were pretreated with 12 g pretreatment powder (Qbiogene, Inc., Irvine, CA) in 40 mL 2 × SSC at 45°C for 15 minutes and washed briefly in 2 × SSC. Protein digestion in Proteinase K (250 μg/mL, 45°C) was performed for 2 hours. The slides were washed in 2 × SSC and dehydrated. Dual-color fluorescence in situ hybridization (FISH) reactions were performed using commercial centromeric alpha satellite probes for chromosomes 7 (spectrum-Orange) and 17 (spectrumGreen) (Vysis, Downers Grove, IL). Probe mixture (1 μL of each probe, 6 μL buffer, and 2 μL dH2O) was added to the slide and covered with a coverslip. The slide was then heated to 72°C for 5 minutes before being transferred to a 37°C hybridization chamber overnight. Washing of slides was performed as per probe manufacturer’s instructions. Slides were counterstained with VECTASHIELD HardSet Mounting Medium with DAPI (Vector Laboratories, Peterborough, UK). A Zeiss Axiophot epifluorescence microscope equipped with a x63 oil immersion objective and DAPI, spectrumOrange, and FITC filter sets were used to view nuclei. Normal ranges were calculated for all probes by counting the fluorescent signals from 200 nuclei of white and grey matter from normal cerebrum.
Immunohistochemistry
Tissue sections of 4-μm thickness were placed on polyionic slides (Dako, Ely, UK). After rehydration, immunohistochemical analysis was performed using mouse monoclonal antibodies against human CD68 (clone KP1) and CD45 (clones 2B11 and PD7/26) as the primary antibodies (Dako). Detection was by the ChemMate peroxidase/DAB kit and EnVision detection system (Dako). All procedures were carried out according to the manufacturer’s instructions.
RESULTS
The majority of cases in this series (28 of 44 [64%]) showed normal copy number across the genome. Gain of at least one chromosome (median, 5 chromosomes affected; range, 1–9) was seen in 32% of cases (14 of 44). One case (PA35) showed a pericentromeric gain on chromosome 3 in addition to whole chromosomal gains, whereas small regions of loss or gain were the sole abnormality in a further 2 cases (4%). Thus, small regions of copy number change were not a common finding of this study. Potential regions of loss or gain involving only one clone at 0.97 Mb resolution are awaiting further confirmation. A representative microarray comparative genomic hybridization (aCGH) plot of PA29 showing gain of chromosomes 5, 6, 7, 8, and 11 is shown in Figure 1.
FIGURE 1.
Whole genome microarray comparative genomic hybridization (aCGH) plot of DNA copy number ratio for sample PA29 showing gain of chromosomes 5, 6, 7, 8, and 11. Insets show microsatellite data from markers in chromosomes 1, 6, 7, and 11. Note the increase in the intensity of the tumor alleles indicating a gain of genetic material on chromosomes 6, 7, and 11 correlating with the gains shown in the aCGH plot. Microsatellite analysis of chromosome 1 indicates balance, again correlating with the aCGH data.
The most commonly gained chromosomes were chromosomes 5 and 7, which were gained in 11 cases each (25%). Concomitant gain of chromosomes 5 and 7 was seen in 8 cases (18%). Gain of chromosomes 6, 11, 15, and 20 was also seen in greater than 10% of cases each. A summary of all changes is included in Table 1. Raw data and clone information are provided in Supplemental Table 1.
A higher number of female cases with chromosomal gains than males was observed (43% and 22%, respectively), but this difference was not significant (p = 0.20, 2-tailed Fisher exact test).
There was also no significant difference in the frequency of copy number alteration in tumors from different locations. All sites affected in 3 or more cases showed at least one case with chromosomal gain.
Cases with grossly normal copy number at 0.97 Mb resolution were slightly more likely to show histology representative of a “classic” presentation of PA (20 of 30 [66%]), whereas those with whole chromosomal gain were more likely to show unusual features such as marked blood vessel proliferation or necrosis (7 of 14 [50%] showed a classic presentation). However, this difference did not approach significance (p = 0.33, 2-tailed Fisher exact test). The difference in frequency of whole chromosomal gain in cases that had undergone a previous resection (4 of 7 [57%] showed gain) compared with cases undergoing primary surgery (10 of 37 [27%] showed gain) also did not reach statistical significance (p = 0.18, 2-tailed Fisher exact test).
Microarray Comparative Genomic Hybridization of Cases in Patients Under 15 Years of Age (n = 32)
A cutoff age of 15 years was used to divide the cases into childhood and adult groups because this age is used by the American Cancer Society (www.cancer.org) and the United Kingdom Office for National Statistics (www.statistics.gov.uk) to define childhood cancer incidence. The childhood cases had a median age of 6 years with a range of 6 months to 14 years. A normal complement of DNA across the genome was seen in 23 (72%) of these cases.
Seven of the cases (22%) showed gain of between one and 5 chromosomes. Only 3 of these 7 cases showed gain of more than one chromosome. The most commonly gained chromosome in this group was chromosome 7, which was gained in 5 cases. Gain of chromosome 7 was the sole change in 3 cases. The second most commonly involved chromosome was chromosome 5, which was gained in 4 cases.
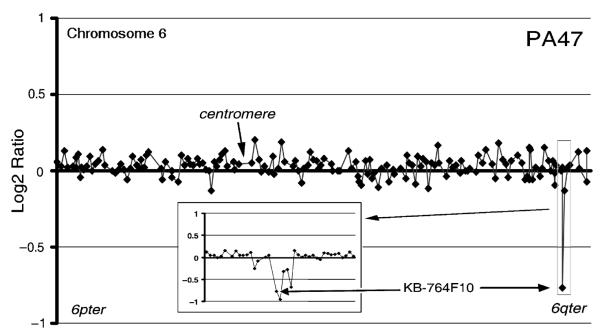
Two cases showed small regions of alteration as the sole abnormality. A region of gain of maximum 4.4 Mb was seen in PA20 at 3p25.2–25.3, encompassing BACs RP11-334L22 to RP11-163D23 (Fig. 2). PA47 showed a deletion of BAC KB-764F10 at 6q26 (Fig. 3). This deletion was confirmed by further analysis on a chromosome 6 tiling path array (Fig. 3 inset) and was shown to involve a 230-kb region, including BACs KB-764F10, RP11-266I4, RP11-307K1, RP11-494F24, and RP11-27G12. A further 10 cases (PA2, PA3, PA5, PA9, PA10, PA24, PA29, PA41, PA42, and PA48), chosen to represent a cross-section of ages, genders, and ploidy, were also screened with the chromosome 6 tiling path array. No further changes below the level of detection of the 0.97 Mb array were detected.
FIGURE 2.

Part of a whole genome microarray comparative genomic hybridization plot from sample PA20 showing DNA copy number ratio across chromosome 3, including a region of gain on 3p.
FIGURE 3.
Part of a whole genome microarray comparative genomic hybridization plot from sample PA47 showing DNA copy number ratio across chromosome 6. The inset shows the region from a chromosome 6 tiling path array corresponding to the region of loss at 6q26 seen on the 0.97 Mb resolution plot.
Microarray Comparative Genomic Hybridization of Cases in Patients Over 15 Years of Age (n = 12)
The median age of adult cases was 25 years (range, 16–33 years). Five of these cases (42%) had an apparently normal complement of DNA across the genome. The remaining 7 cases (58%) all showed multiple chromosomal gains involving between 4 and 9 chromosomes. In contrast to the childhood group, no case showed gain affecting only one chromosome. The most commonly involved chromosome was chromosome 5, which was gained in all cases showing any chromosomal gain, followed by chromosomes 6 and 7, which were each gained in 6 cases. In addition, PA35 showed a pericentromeric region of gain encompassing a maximum of 9.3 Mb in 3q11.2 including BACs RP11-88I7 and RP11-12A13 (Fig. 4).
FIGURE 4.

DNA copy number ratio across chromosome 3 for PA35 indicating a pericentromeric region of gain.
Statistical Comparison of Pilocytic Astrocytoma From Those Under and Over 15 Years Old
The occurrence of chromosomal gain was not evenly distributed by age. An overview of changes in adult and childhood cases, grouped to show patterns of chromosomal gain, is presented in Figure 5. In the adult group, 58% of cases (7 of 12) showed gain of at least one chromosome, whereas in the childhood group, only 22% of cases (7 of 32) showed whole chromosomal gain. This difference is statistically significant (p = 0.03, 2-tailed Fisher exact test). In the subset of cases that showed whole chromosomal gain, the number of chromosomes affected also differed between age groups. A median of 5 chromosomes was gained in the adult cases and a median of one in childhood cases with gain. This difference is also significant (p = 0.02, 2-tailed Mann-Whitney U test). Of further note is the fact that no chromosomal gains were seen in any case from a patient of less than 10 years of age.
FIGURE 5.
Summary of changes found in 44 primary tumor samples grouped to show similar patterns of chromosomal gain. Cases with no chromosomal gain are in descending age order. The age of each case is shown with the case number (n/a, not available). Adult cases are shown on the top and childhood cases underneath with chromosome number indicated below both. Each filled box represents a chromosomal gain. *, Small region of gain; ×, small region of loss.
Verification of Copy Number Change by Fluorescent In Situ Hybridization and Microsatellite Analysis
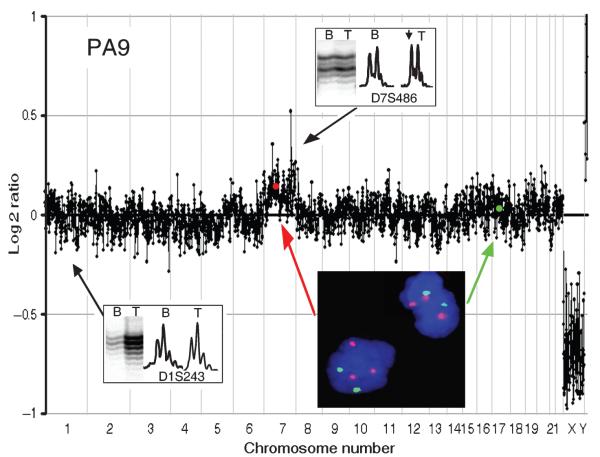
Because the level of copy number change seen on aCGH data is dependent on the tumor cell content of the sample (all tumor tissue will contain normal cells such as blood vessel components, inflammatory cells, macrophages, and so on; see next section), and some of the changes seen were subtle alterations in the tumor:normal DNA ratio, independent verification of copy number by other methods was considered essential. A subset of cases for which paraffin-embedded sections were available was analyzed by interphase FISH using centromeric probes. As an example, the whole genome plot for PA9 showed a subtle increase in log2 ratio, which was interpreted as indicating gain of chromosome 7 (Fig. 6). FISH analysis of a section from PA9 clearly showed trisomy of chromosome 7 (Fig. 6 inset).
FIGURE 6.
A whole genome plot for PA9 showing a subtle gain of chromosome 7 confirmed by the gain of intensity of the tumor allele seen on the inset. A fluorescent in situ hybridization image of a section from PA9 is also shown. Chr7 centromeric probes are shown in red and Chr17 centromeric probes in green with a blue DAPI counterstain. Positions of centromeres of chromosomes 7 and 17 on the microarray comparative genomic hybridization plot are shown with a red and green dot, respectively. After immunohistochemical analysis with CD68k and CD45, this case had a revised estimate of tumor cell content of only 66%.
In addition, a subset of cases showing subtle copy number changes was assessed by microsatellite analysis. Insets on Figures 1 and 6 show that allelic imbalance is observed for microsatellites in regions assessed as gain from the aCGH plot, and balance is seen when aCGH is judged to show normal copy number.
In all other cases analyzed with microsatellites and/or interphase FISH, these techniques confirmed the interpretation of the aCGH results. It is a feature of the sensitivity of microarray CGH and the quality of material for this tumor series that it was possible to determine these very subtle but independently verifiable changes in copy number.
Immunohistochemical Analysis of the Microglial/Macrophage Content of Tumors
To assess the contribution of microglia/macrophages to the normal cell component of tumor samples, antibodies against a macrophage marker (CD68k) and a more general immune cell marker (CD45) were used in an immunohistochemical analysis on a subset of tumors. All tumors showed some positively stained cells with a range for CD68K immunoreactive cells of 3% to 16% (mean, 10%) and for CD45 of 4% to 22% (mean, 15%). These results were used to calculate a revised figure for tumor cell content in these cases, and the size of any changes in log2 ratio seen on their aCGH plots correlated with the revised figure (Table 2).
TABLE 2.
Results of Immunohistochemical Analysis
| Case | CD68K Positive (%) | CD45 Positive (%) | Revised Tumor Content (%)* | Mean of Log2 Ratio of Change† |
|---|---|---|---|---|
| PA9 | 16 | 22 | 66 | 0.11 |
| PA12 | 3 | 9 | 79 | 0.24 |
| PA14 | 13 | 21 | 73 | N/A |
| PA57 | 9 | 18 | 67.5 | N/A |
| PA67 | 10 | 4 | 83 | 0.24 |
Results of immunohistochemical staining of a subset of cases with antibodies against a microglial marker (CD68k) and an immune cell marker (CD45). Greater than 200 cells were counted in each case.
This figure is calculated by subtracting the mean of the 2 staining figures from the tumor content estimate in Table 1.
When one or more chromosomal gains were seen on the aCGH plots of these cases, the mean log2 ratio of the change(s) is given.
N/A, data not available; aCGH, microarray comparative genomic hybridization.
DISCUSSION
The first striking feature of this study is the large number of tumors showing normal genetic copy number profiles across the genome on this 0.97 Mb resolution array. This indicates that in these tumors, alterations other than large regions of gain or loss of genetic material are responsible for tumorigenesis and that this applies to the majority of pilocytic astrocytomas. Possibilities include smaller changes than could be detected by this array, balanced translocations (which are not detectable by aCGH), and epigenetic changes such as gene silencing by promoter methylation. Hypermethylation of several tumor-suppressor genes has been reported in PAs, including ras association domain family 1a (RASSFIA), retinoblastoma gene (RB1), cyclin-dependent kinase 4 inhibitor A ( p16INK4A), and tumor protein p73 (31, 32).
The recurrent, nonrandom pattern of chromosomal copy number aberration identified in this study, albeit in only one-third of the cases, is also remarkable. The only previous study to suggest recurrent whole chromosome changes in this tumor type used FISH with a limited number of pericentromeric probes and found chromosomal gains, most commonly of 7 and 8, in 6 of 18 cases (23). Chromosome 5, which we have found together with chromosome 7 to be the 2 chromosomes most frequently affected in PA, was not investigated.
Trisomy 5 has been reported as an indicator of both good and poor prognosis in various lymphoreticular cancers (33-35). In sporadic digestive tract neuroendocrine tumors, gain of chromosome 5 is thought to be a late event associated with tumor progression and is often found in association with trisomy 7 (36). Trisomy 7 is found in many tumor types. Notably, it is one of the most frequently altered chromosomes in grade II–IV diffuse astrocytomas (37, 38). However, trisomy 7 is also found in a number of peritumoral and nonneoplastic tissues and has been documented in short-term cultures of normal brain (39, 40). Furthermore, it has been reported that gain of chromosome 7 in neoplastic and nonneoplastic tissues may be linked to age and not disease (41).
Aneuploidy, as seen in one-third of PAs in this study, is a common feature of solid tumor cells. The debate regarding the significance of aneuploidy as a driving force behind tumorigenesis as opposed to merely a bystander effect is still ongoing (42, 43). Also, aneuploidy is often thought of in connection with a breakdown of the checkpoints controlling chromosomal segregation leading to random chromosomal gains and losses at each cell division. Yet, once generated, individual chromosomal changes must be maintained through many generations if they are to show clonal expansion within the tumor.
Once generated, copy number changes may cause altered expression of genes on the affected chromosome(s), which will upset the balance of signals needed to maintain normal cell behavior. The embryonic lethality of aneuploidy of the majority of autosomes supports this hypothesis. This alteration in expression as opposed to chromosomal copy number changes per se may drive tumorigenesis. An increased incidence of cancer in sufferers of mosaic variegated aneuploidy, a rare recessive disorder characterized by constitutional alterations in DNA ploidy, lends weight to the suggestion that the effects of chromosomal imbalances may indeed play a causal role (44). However, the possibility remains that other genetic changes leading directly to tumorigenesis may coincidentally cause aneuploidy that then provides some growth advantage without being responsible for initiating the tumor.
The extent of aneuploidy seen in PAs differs significantly in its age distribution, being more common and affecting more chromosomes in older patients. One explanation for this might be that the PAs in the adults developed in childhood and remained undiagnosed such that the trisomies have been acquired with time. In this case, the aneuploidy might be a consequence not directly involved in the initial development of the tumor. This may also indicate a limited window of time during development in which there is the potential for initiation of a PA. The decreasing incidence of PAs with increasing age may support this hypothesis. Alternatively, the differing chemical and structural environments of childhood and adult brains may require the development of different genotypes and thus phenotypes to produce a histologically similar tumor. In this case, chromosomal copy number change may truly define a distinct genetic subgroup of this tumor type. Genetic subgroups of astrocytomas and other brain tumors have previously been reported (45-47), and it has been shown that these groups can correlate with prognosis (48, 49), possibly in an age-dependent manner (50). Interestingly, another grade I glioma, myxopapillary ependymoma, is also characterized by chromosomal gains but shows a different pattern in terms of the chromosomes involved (51).
Small regions of gain and loss were also seen in some tumors in our series, but they were not a feature of most cases. The region of gain on chromosome 3 in PA20 contains 49 gene entries in the Ensembl database (www.ensembl.org), which are listed in Supplemental Table 2. Gain of several of these genes would not intuitively suggest a growth advantage. For example, the von Hippel-Lindau tumor-suppressor gene, VHL, which is deleted in several tumor types (52), FANCD2 (involved in DNA repair), and CIDEC (a cell death effector protein). However, the region also includes the RAF1 (C-raf) protooncogene, which is implicated in several cellular processes important in oncogenesis (53).
The region of gain on chromosome 3 in PA35 is pericentromeric. The 3q clone closest to the centromere shows gain, whereas the most centromeric clone on 3p shows normal copy number. However, the resolution of our array is not as high around the centromeres as it is in euchromatic areas. The maximal gained region contains 11 confirmed gene entries in Ensembl (Supplemental Table 3), including the EPHA3 gene, which encodes the ephrin type A receptor 3 and which may play a role in tumor angiogenesis and progression (54).
The deleted region of chromosome 6 in PA47 spans roughly 230 kb on 6q26. This region is within the fragile site FRA6E and includes parts of introns 1 and 2 and all of exon 2 of the PARK2 gene. This gene is an E3 ubiquitin ligase implicated in autosomal-recessive juvenile Parkinsonism and contains 12 exons spaced over nearly 1.5 Mb. It has also been shown to be altered in several cancer types, including breast, ovarian, hepatocellular, and nonsmall cell lung cancer (55-57). Homozygous deletions affecting exon 2 specifically have been reported in 2 lung adenocarcinoma cell lines, Calu-3 and H-1573 (55). The FRA6E region has also recently been shown to be altered in higher-grade astrocytomas (29, 58). Other potential regions of gain or loss spanning only one clone on the 0.97 Mb array are awaiting confirmation at higher resolution.
Of the 44 cases examined in this study, 40 were also subject to a previous metaphase CGH analysis. Except for one case, all previous changes were identified in the current study, and the aCGH method also detected many more changes than previously reported. The one discrepant case showed genomewide normal copy number in the present study, whereas the previous analysis appeared to indicate a number of regions of gain. Repeat array hybridizations for the case gave identical results.
The degree of change in log2 ratio seen in an aCGH plot is dependent on the normal cell content of the tumor sample. In effect, there is a dilution of tumor cell DNA by normal DNA from cell types such as macrophages. Cells of the vasculature are accounted for in the tumor content figures given in Table 1, but other normal cell types that are difficult to identify histologically without immunohistochemistry are not. Thus, as the normal cell content of the tumor samples may be higher than originally estimated, we carried out immunohistochemistry for CD45 and CD68 on a subset of the cases to identify macrophages and inflammatory cells. Activated microglia have long been known to infiltrate gliomas with one study reporting that up to 52% of cells in areas described grossly as pilocytic astrocytoma may in fact be macrophages (59, 60). A revised figure for tumor cell content was calculated for this subset (Table 2), and it is interesting to note that the size of changes in the log2 ratio on the aCGH plot correlated with this revised figure. This provides an explanation for the subtlety of the copy number changes seen in several cases. An additional explanation for the subtlety of changes may be that there are subpopulations of tumor cells present with and without a given abnormality. A combination of these 2 effects may be the true cause.
Expression array analysis of childhood PAs has indicated 2 subgroups in this tumor type, which may differ in their clinical and biologic behavior in terms of likelihood of recurrence (61). It would be of great interest to see if the subgroups proposed here, based on genomic status, also show common differences in their gene expression profiles. Characterization of genetic changes in a series of PAs with very long-term follow up would also be of value to determine whether whole chromosomal gains correlate with recurrent behavior or, in the rare cases when it does occur, malignant transformation of PAs. In addition, further studies to investigate the possibility of very small copy number changes, balanced translocations, and epigenetic alterations will also be necessary to improve our understanding of the tumorigenic processes leading to the formation of pilocytic astrocytomas.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. M. G. McCabe for his technical support and advice; the Mapping Core, Map Finishing, and Microarray Facility groups of the Wellcome Trust Sanger Institute, Hinxton, UK, for initial clone supply and verification; the Centre for Microarray resources in the Department of Pathology, University of Cambridge, for printing of the arrays; and David A. Carter for excellent technical assistance.
This work was supported by grants from Cancer Research UK, the Jacqueline Seroussi Memorial Foundation for Cancer Research, the Samantha Dickson Research Trust, CAMPOD, and the Ludwig Institute for Cancer Research.
Footnotes
Supplementary tables are available online at http:www.jneuropath.com.
REFERENCES
- 1.Office for National Statistics The Health of Children and Young People. Mar 31, 2004. [Accessed June 2, 2005]. Available at: www.statistics.gov.uk/children/
- 2.Kleihues P, Cavenee WK. Pathology and Genetics of Tumours of the Nervous System: World Health Organization Classification of Tumours. IARC Press; Lyon, France: 2000. [Google Scholar]
- 3.Central Brain Tumour Registry of the United States . Statistical Report: Primary Brain Tumours in the United States, 1997–2001. CBTRUS; Chicago: 2005. [Google Scholar]
- 4.Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. 2005;64:479–89. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- 5.Claus D, Sieber E, Engelhardt A, et al. Ascending central nervous spreading of a spinal astrocytoma. J Neurooncol. 1995;25:245–50. doi: 10.1007/BF01053158. [DOI] [PubMed] [Google Scholar]
- 6.Kleinman GM, Schoene WC, Walshe TM, III, et al. Malignant transformation in benign cerebellar astrocytoma. Case report. J Neurosurg. 1978;49:111–18. doi: 10.3171/jns.1978.49.1.0111. [DOI] [PubMed] [Google Scholar]
- 7.Tomlinson FH, Scheithauer BW, Hayostek CJ, et al. The significance of atypia and histologic malignancy in pilocytic astrocytoma of the cerebellum: A clinicopathologic and flow cytometric study. J Child Neurol. 1994;9:301–10. doi: 10.1177/088307389400900317. [DOI] [PubMed] [Google Scholar]
- 8.Krieger MD, Gonzalez-Gomez I, Levy ML, et al. Recurrence patterns and anaplastic change in a long-term study of pilocytic astrocytomas. Pediatr Neurosurg. 1997;27:1–11. doi: 10.1159/000121218. [DOI] [PubMed] [Google Scholar]
- 9.Dirven CM, Mooij JJ, Molenaar WM. Cerebellar pilocytic astrocytoma: A treatment protocol based upon analysis of 73 cases and a review of the literature. Childs Nerv Syst. 1997;13:17–23. doi: 10.1007/s003810050033. [DOI] [PubMed] [Google Scholar]
- 10.Giannini C, Scheithauer BW. Classification and grading of low-grade astrocytic tumors in children. Brain Pathol. 1997;7:785–98. doi: 10.1111/j.1750-3639.1997.tb01064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aldape K, Simmons ML, Davis RL, et al. Discrepancies in diagnoses of neuroepithelial neoplasms: The San Francisco Bay Area Adult Glioma Study. Cancer. 2000;88:2342–49. [PubMed] [Google Scholar]
- 12.Agamanolis DP, Malone JM. Chromosomal abnormalities in 47 pediatric brain tumors. Cancer Genet Cytogenet. 1995;81:125–34. doi: 10.1016/0165-4608(94)00123-s. [DOI] [PubMed] [Google Scholar]
- 13.Bhattacharjee MB, Armstrong DD, Vogel H, et al. Cytogenetic analysis of 120 primary pediatric brain tumors and literature review. Cancer Genet Cytogenet. 1997;97:39–53. doi: 10.1016/s0165-4608(96)00330-5. [DOI] [PubMed] [Google Scholar]
- 14.Bigner SH, McLendon RE, Fuchs H, et al. Chromosomal characteristics of childhood brain tumors. Cancer Genet Cytogenet. 1997;97:125–34. doi: 10.1016/s0165-4608(96)00404-9. [DOI] [PubMed] [Google Scholar]
- 15.Blaeker H, Rasheed BK, McLendon RE, et al. Microsatellite analysis of childhood brain tumors. Genes Chromosomes Cancer. 1996;15:54–63. doi: 10.1002/(SICI)1098-2264(199601)15:1<54::AID-GCC8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 16.Debiec-Rychter M, Alwasiak J, Liberski PP, et al. Accumulation of chromosomal changes in human glioma progression. A cytogenetic study of 50 cases. Cancer Genet Cytogenet. 1995;85:61–67. doi: 10.1016/0165-4608(95)00129-8. [DOI] [PubMed] [Google Scholar]
- 17.Jenkins RB, Kimmel DW, Moertel CA, et al. A cytogenetic study of 53 human gliomas. Cancer Genet Cytogenet. 1989;39:253–79. doi: 10.1016/0165-4608(89)90192-1. [DOI] [PubMed] [Google Scholar]
- 18.Ransom DT, Ritland SR, Kimmel DW, et al. Cytogenetic and loss of heterozygosity studies in ependymomas, pilocytic astrocytomas, and oligodendrogliomas. Genes Chromosomes Cancer. 1992;5:348–56. doi: 10.1002/gcc.2870050411. [DOI] [PubMed] [Google Scholar]
- 19.Roberts P, Chumas PD, Picton S, et al. A review of the cytogenetics of 58 pediatric brain tumors. Cancer Genet Cytogenet. 2001;131:1–12. doi: 10.1016/s0165-4608(01)00483-6. [DOI] [PubMed] [Google Scholar]
- 20.Sanoudou D, Tingby O, Ferguson-Smith MA, et al. Analysis of pilocytic astrocytoma by comparative genomic hybridization. Br J Cancer. 2000;82:1218–22. doi: 10.1054/bjoc.1999.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shlomit R, Ayala AG, Michal D, et al. Gains and losses of DNA sequences in childhood brain tumors analyzed by comparative genomic hybridization. Cancer Genet Cytogenet. 2000;121:67–72. doi: 10.1016/s0165-4608(00)00218-1. [DOI] [PubMed] [Google Scholar]
- 22.Szymas J, Wolf G, Peterson S, et al. Comparative genomic hybridisation indicating two distinct subgroups of pilocytic astrocytomas. Neurosurg Focus. 2000;8 Clinical Pearl 2. [Google Scholar]
- 23.White FV, Anthony DC, Yunis EJ, et al. Nonrandom chromosomal gains in pilocytic astrocytomas of childhood. Hum Pathol. 1995;26:979–86. doi: 10.1016/0046-8177(95)90087-x. [DOI] [PubMed] [Google Scholar]
- 24.Zattara-Cannoni H, Gambarelli D, Lena G, et al. Are juvenile pilocytic astrocytomas benign tumors? A cytogenetic study in 24 cases. Cancer Genet Cytogenet. 1998;104:157–60. doi: 10.1016/s0165-4608(97)00455-x. [DOI] [PubMed] [Google Scholar]
- 25.Ichimura K, Schmidt EE, Goike HM, et al. Human glioblastomas with no alterations of the CDKN2A (p16INK4A, MTS1) and CDK4 genes have frequent mutations of the retinoblastoma gene. Oncogene. 1996;13:1065–72. [PubMed] [Google Scholar]
- 26.Fiegler H, Carr P, Douglas EJ, et al. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer. 2003;36:361–74. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- 27.McCabe M, Ichimura K, Liu L, et al. High resolution array-based comparative genomic hybridisation of medulloblastomas and supratentorial primitive neuroectodermal tumours. J Neuropathol Exp Neurol. doi: 10.1097/00005072-200606000-00003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seng TJ, Ichimura K, Liu L, et al. Complex chromosome 22 rearrangements in astrocytic tumors identified using microsatellite and chromosome 22 tile path array analysis. Genes Chromosomes Cancer. 2005;43:181–93. doi: 10.1002/gcc.20181. [DOI] [PubMed] [Google Scholar]
- 29.Ichimura K, Mungall AJ, Fiegler H, et al. Small regions of overlapping deletions on 6q26 in human astrocytic tumours identified using chromosome 6 tile path array-CGH. Oncogene. 2006;25:1261–71. doi: 10.1038/sj.onc.1209156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyakawa A, Ichimura K, Schmidt EE, et al. Multiple deleted regions on the long arm of chromosome 6 in astrocytic tumours. Br J Cancer. 2000;82:543–49. doi: 10.1054/bjoc.1999.0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez-Gomez P, Bello MJ, Lomas J, et al. Epigenetic changes in pilocytic astrocytomas and medulloblastomas. Int J Mol Med. 2003;11:655–60. [PubMed] [Google Scholar]
- 32.Yu J, Zhang H, Gu J, et al. Methylation profiles of thirty four promoter-CpG islands and concordant methylation behaviours of sixteen genes that may contribute to carcinogenesis of astrocytoma. BMC Cancer. 2004;4:65. doi: 10.1186/1471-2407-4-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris RL, Harrison CJ, Martineau M, et al. Is trisomy 5 a distinct cytogenetic subgroup in acute lymphoblastic leukemia? Cancer Genet Cytogenet. 2004;148:159–62. doi: 10.1016/s0165-4608(03)00272-3. [DOI] [PubMed] [Google Scholar]
- 34.Schlegelberger B, Zwingers T, Hohenadel K, et al. Significance of cytogenetic findings for the clinical outcome in patients with T-cell lymphoma of angioimmunoblastic lymphadenopathy type. J Clin Oncol. 1996;14:593–99. doi: 10.1200/JCO.1996.14.2.593. [DOI] [PubMed] [Google Scholar]
- 35.Schlegelberger B, Zwingers T, Harder L, et al. Clinicopathogenetic significance of chromosomal abnormalities in patients with blastic peripheral B-cell lymphoma. Kiel-Wien-Lymphoma Study Group. Blood. 1999;94:3114–20. [PubMed] [Google Scholar]
- 36.Terris B, Meddeb M, Marchio A, et al. Comparative genomic hybridization analysis of sporadic neuroendocrine tumors of the digestive system. Genes Chromosomes Cancer. 1998;22:50–56. doi: 10.1002/(sici)1098-2264(199805)22:1<50::aid-gcc7>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 37.Liu L, Ichimura K, Pettersson EH, et al. Chromosome 7 rearrangements in glioblastomas; loci adjacent to EGFR are independently amplified. J Neuropathol Exp Neurol. 1998;57:1138–45. doi: 10.1097/00005072-199812000-00005. [DOI] [PubMed] [Google Scholar]
- 38.Ichimura K, Ohgaki H, Kleihues P, et al. Molecular pathogenesis of astrocytic tumours. J Neurooncol. 2004;70:137–60. doi: 10.1007/s11060-004-2747-2. [DOI] [PubMed] [Google Scholar]
- 39.Johansson B, Heim S, Mandahl N, et al. Trisomy 7 in nonneoplastic cells. Genes Chromosomes Cancer. 1993;6:199–205. doi: 10.1002/gcc.2870060402. [DOI] [PubMed] [Google Scholar]
- 40.Heim S, Mandahl N, Jin Y, et al. Trisomy 7 and sex chromosome loss in human brain tissue. Cytogenet Cell Genet. 1989;52:136–38. doi: 10.1159/000132863. [DOI] [PubMed] [Google Scholar]
- 41.Broberg K, Toksvig-Larsen S, Lindstrand A, et al. Trisomy 7 accumulates with age in solid tumors and non-neoplastic synovia. Genes Chromosomes Cancer. 2001;30:310–15. [PubMed] [Google Scholar]
- 42.Matzke MA, Mette MF, Kanno T, et al. Does the intrinsic instability of aneuploid genomes have a causal role in cancer? Trends Genet. 2003;19:253–56. doi: 10.1016/s0168-9525(03)00057-x. [DOI] [PubMed] [Google Scholar]
- 43.Marx J. Debate surges over the origins of genomic defects in cancer. Science. 2002;297:544–46. doi: 10.1126/science.297.5581.544. [DOI] [PubMed] [Google Scholar]
- 44.Hanks S, Rahman N. Aneuploidy-cancer predisposition syndromes: A new link between the mitotic spindle checkpoint and cancer. Cell Cycle. 2005;4:225–27. [PubMed] [Google Scholar]
- 45.Hirose Y, Aldape KD, Chang S, et al. Grade II astrocytomas are subgrouped by chromosome aberrations. Cancer Genet Cytogenet. 2003;142:1–7. doi: 10.1016/s0165-4608(02)00791-4. [DOI] [PubMed] [Google Scholar]
- 46.Misra A, Pellarin M, Nigro J, et al. Array comparative genomic hybridization identifies genetic subgroups in grade 4 human astrocytoma. Clin Cancer Res. 2005;11:2907–18. doi: 10.1158/1078-0432.CCR-04-0708. [DOI] [PubMed] [Google Scholar]
- 47.Nigro JM, Misra A, Zhang L, et al. Integrated array-comparative genomic hybridization and expression array profiles identify clinically relevant molecular subtypes of glioblastoma. Cancer Res. 2005;65:1678–86. doi: 10.1158/0008-5472.CAN-04-2921. [DOI] [PubMed] [Google Scholar]
- 48.Shiraishi T, Tabuchi K. Genetic alterations of human brain tumors as molecular prognostic factors. Neuropathology. 2003;23:95–108. doi: 10.1046/j.1440-1789.2003.00477.x. [DOI] [PubMed] [Google Scholar]
- 49.Smith JS, Tachibana I, Passe SM, et al. PTEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst. 2001;93:1246–56. doi: 10.1093/jnci/93.16.1246. [DOI] [PubMed] [Google Scholar]
- 50.Batchelor TT, Betensky RA, Esposito JM, et al. Age-dependent prognostic effects of genetic alterations in glioblastoma. Clin Cancer Res. 2004;10:228–33. doi: 10.1158/1078-0432.ccr-0841-3. [DOI] [PubMed] [Google Scholar]
- 51.Mahler-Araujo MB, Sanoudou D, Tingby O, et al. Structural genomic abnormalities of chromosomes 9 and 18 in myxopapillary ependymomas. J Neuropathol Exp Neurol. 2003;62:927–35. doi: 10.1093/jnen/62.9.927. [DOI] [PubMed] [Google Scholar]
- 52.Kondo K, Kaelin WG., Jr. The von Hippel-Lindau tumor suppressor gene. Exp Cell Res. 2001;264:117–25. doi: 10.1006/excr.2000.5139. [DOI] [PubMed] [Google Scholar]
- 53.Beeram M, Patnaik A, Rowinsky EK. Raf: A strategic target for therapeutic development against cancer. J Clin Oncol. 2005;23:6771–90. doi: 10.1200/JCO.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 54.Brantley DM, Cheng N, Thompson EJ, et al. Soluble Eph A receptors inhibit tumor angiogenesis and progression in vivo. Oncogene. 2002;21:7011–26. doi: 10.1038/sj.onc.1205679. [DOI] [PubMed] [Google Scholar]
- 55.Cesari R, Martin ES, Calin GA, et al. Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25-q27. Proc Natl Acad Sci U S A. 2003;100:5956–61. doi: 10.1073/pnas.0931262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Picchio MC, Martin ES, Cesari R, et al. Alterations of the tumor suppressor gene Parkin in non-small cell lung cancer. Clin Cancer Res. 2004;10:2720–24. doi: 10.1158/1078-0432.ccr-03-0086. [DOI] [PubMed] [Google Scholar]
- 57.Wang F, Denison S, Lai JP, et al. Parkin gene alterations in hepatocellular carcinoma. Genes Chromosomes Cancer. 2004;40:85–96. doi: 10.1002/gcc.20020. [DOI] [PubMed] [Google Scholar]
- 58.Mulholland PJ, Fiegler H, Mazzanti C, et al. Genomic profiling identifies discrete deletions associated with translocations in glioblastoma multiforme. Cell Cycle. 2006;5:783–91. doi: 10.4161/cc.5.7.2631. [DOI] [PubMed] [Google Scholar]
- 59.Penfield W. Microglia and the process of phagocytosis in gliomas. Am J Pathol. 1925;1:77–97. [PMC free article] [PubMed] [Google Scholar]
- 60.Sasaki A, Yamaguchi H, Horikoshi Y, et al. Expression of glucose transporter 5 by microglia in human gliomas. Neuropathol Appl Neurobiol. 2004;30:447–55. doi: 10.1111/j.1365-2990.2004.00556.x. [DOI] [PubMed] [Google Scholar]
- 61.Wong KK, Chang YM, Tsang YT, et al. Expression analysis of juvenile pilocytic astrocytomas by oligonucleotide microarray reveals two potential subgroups. Cancer Res. 2005;65:76–84. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.