Abstract
Purpose
Saliva is a good source of DNA for genomic research, and leukocytes are a predominant source of DNA in human saliva. Advanced human immunodeficiency virus (HIV)-type 1 infection disrupts tonsillar architecture and depletes tonsillar lymphocytes. We tested whether HIV-1 infection reduces extracted human DNA yield from saliva.
Methods
Approximately 2 mL of expectorated saliva was collected from HIV-infected adults during routine primary care clinic visits, and from healthy, HIV-negative controls. Human DNA was manually extracted, and was specifically quantified by assaying for the RNAse P gene.
Results
Seventy-five individuals were studied, including 25 HIV-infected adults with <200 CD4+ T cells/mm3 (i.e. acquired immunodeficiency syndrome), 25 with >200 CD4+ T cells/mm3, and 25 HIV-negative controls. Overall DNA yield was 64.7 μg [29.0 – 139.7 μg] (median [interquartile range]). Yields were comparable among HIV-infected individuals with lower CD4+ T cell counts (74.3 μg [39.4 – 151.4 μg]), higher CD4+ T cell counts (63.9 μg [29.2 – 172.1 μg]), and HIV-negative controls (61.4 μg [28.4 – 123.4 μg], P > 0.05).
Conclusion
Infection with HIV-1 does not reduce human DNA yield from saliva. Expectorated saliva should provide sufficient extracted native DNA for genomic studies in HIV-infected individuals.
Keywords: saliva, repository, DNA, HIV-1, AIDS
INTRODUCTION
An estimated 1 million individuals in the United States and 33 million worldwide are living with human immunodeficiency virus type 1 (HIV-1) 1, 2. Mortality from untreated HIV-1 infection approaches 100%. Fortunately, antiretroviral therapy (ART) markedly reduces acquired immunodeficiency syndrome (AIDS) mortality 3. Access to safe and effective ART is a cornerstone in the global struggle against AIDS 4, 5.
Variable responses to medications are influenced, at least in part, by human genetic polymorphisms 6-11. Because suboptimal response to ART can have devastating consequences, it is important to characterize the predictive value of human genetic variants for HIV treatment response. For persons affected by HIV to fully benefit from the human genomic revolution, DNA banks must be established that represent HIV-infected populations in resource-limited countries.
Expectorated saliva can provide ample native DNA for genomic analyses, and has some advantages over whole blood, particularly in resource-limited settings. As compared to whole blood, saliva collection is non-invasive, and with appropriate preservatives is stable for long-term storage at ambient temperature. This allows cost-efficient storage and shipping. In contrast, whole blood must either be extracted within a few days, or stored at −70°C until extracted. In an uncontrolled observation we found unexpectedly low extracted human DNA yields from saliva obtained from HIV-infected individuals in a resource-limited country.
In a previous study to define the cellular composition of saliva, flow cytometry of saliva from 258 (presumably HIV-negative) samples showed leukocytes to be more abundant than epithelial cells 12. During AIDS, viral replication in tonsils and other lymphoid tissues disrupts tonsillar architecture, causing follicular hypoplasia and depletion of lymphocytes 13. This may result in collagen deposition and scarring 14, and tonsillar abnormalities may persist despite antiretroviral therapy. This could explain reduced human DNA yields from saliva during HIV-1 infection.
The present study was designed to assess whether human DNA yields from expectorated saliva differ between HIV-positive and HIV-negative individuals, and to determine whether, among HIV-infected individuals, DNA yields are less in those with <200 CD4 T cells/mm3 in peripheral blood.
MATERIAL AND METHODS
Study Subjects and Design
We recruited HIV-positive volunteers during routine primary care visits at the Vanderbilt-affiliated Comprehensive Care Center. We also enrolled a control group of individuals not known to be HIV-positive healthy volunteer. Participants were at least 18 but no more than 65 years of age.
Expectorated saliva was collected using Oragene kits (DNA Genotek Inc.), according to the manufacturer’s guidelines. Participants did not eat or drink for 30 minutes before saliva collection. The HIV-positive donors were directly observed by study personnel as they expectorated approximately 2 mL of saliva. Specimens from HIV-positive donors were stratified based on whether the most recent previous peripheral blood CD4+ T cell count (with ~3 months) was >200 or <200 cells/mm3 (The latter cut-off meets the definition of AIDS15). Specimens were completely anonymized prior to DNA extraction. Two-way comparisons between groups were by Mann-Whitney U test, using a two-tailed test and Stata/IC version 10.0 (College Station, TX).
The study was approved by the Vanderbilt Institutional Review Board. Because specimens were anonymized written informed consent was not required.
DNA extraction and quantitation
Approximately 2 mL of saliva was collected into Oragene kits, and DNA stabilized mixing with Oragene DNA solution (2 mL) by promptly inverting for a few seconds following the manufacturer’s instructions 16, 17. The specimen was kept at room temperature until processed. From the ~4 mL final solution, DNA was manually extracted from 500 μL 18. Briefly, 500 μL of the each sample was incubated in a 1.5 mL microfuge tube at 50°C in a water bath for at least 1 hour. Then 20 μL of Oragene DNA Purifier was added and the sample mixed by vortexing. Samples were incubated on ice for 10 minutes, then microfuged at room temperature for 5 minutes at 15,000 × g. The supernatant was pipetted into a separate microfuge tube containing 5μl of glycogen and the pellet discarded. To 500 μL of supernatant, 500 μL of 100% ethanol at room-temperature was added and mixed by gentle inversion 10 times. The sample was incubated at room temperature for 10 minutes to precipitate DNA. The tube was microfuged at room temperature for 2 minutes at 15,000 × g. The supernatant was carefully removed and discarded. The pellet was soaked in 250 μL of 70% ethanol for 1 minute to remove impurities. To dissolve the DNA pellet, 100 μL of DNA buffer was added followed by vortexing for at least 5 seconds. The DNA was stored at −80°C. For selected samples the remaining ~3.5 mL sample was robotically extracted on an AutoPure LS instrument.
Saliva contains both human and non-human DNA. Human DNA was therefore specifically quantified by assaying the RNAse P gene by real-time PCR. Specimens were assayed using 384-well plates. To each well was added 4 μL of universal master mix (5.5 μL of No AmpErase® UNG, 0.55 μL of TaqMan® RNase P Control Reagents, and 2.75 μL of MilliQ water) and 1 μL of extracted DNA or 1 μL of TaqManR Control Genomic DNA (Qiagen) at (0, 0.5, 1, 5 and 10 ng). Standard curves were extrapolated to encompass DNA amounts in the saliva specimens, and assays were repeated on samples with threshold cycle (CT) values greater than or less than −1.0 or 1.0, respectively. The plate was sealed, then centrifuged for 3 minutes at 800 × g. Amount of DNA was quantified with an Applied Biosystems 7900HT Fast Real-Time PCR System. All specimens were assayed in duplicate.. The difference in RNAse P gene assay results between paired samples was 18.8%±15.5% (mean ± SD).
RESULTS
Seventy-five volunteers were studied, including 25 HIV-infected individuals with <200 CD4+ T cells/mm3 (meeting the case definition for AIDS15), 25 with >200 CD4+ T cells/mm3, and 25 HIV-negative controls. Overall DNA yield was 64.7 μg [29.0 – 139.7 μg] (median [interquartile range]). As shown in the Figure, DNA yields were comparable among HIV-infected individuals with CD4+ T cell counts <200 cells/mm3 (74.3 μg, [39.4 – 151.4 μg]), CD4+ T cell counts >200 cells/mm3 (63.9 μg [29.2 – 172.1 μg]), and HIV-negative controls (61.4 μg [28.4 – 123.4 μg], P > 0.05 for each 2-way comparison). Of 6 specimens with human DNA yields <1.25 μg from 500 μL (including 4 with no detectable human DNA), yield was not improved substantially by extracting the entire remaining specimen (data not shown).
Figure. Extracted human DNA yield from expectorated saliva.
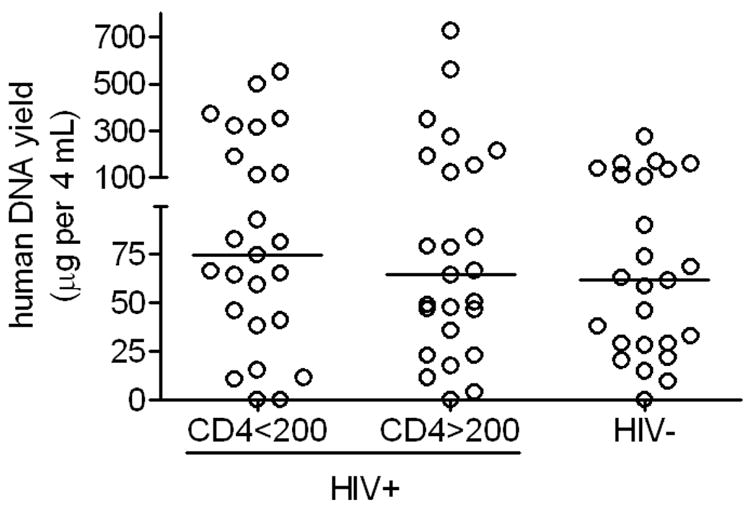
Each marker represents a different study participant. DNA was extracted from 500 μL of each ~4 mL specimen, which comprised ~2 mL of saliva plus 2 mL of Oragene DNA solution. Total human DNA yield was estimated by multiplying yield from 500 μL by a factor of 8.
DISCUSSION
The AIDS pandemic is having a devastating impact in many resource-limited countries. Better understanding of the importance of human genetic variation for HIV disease pathogenesis and treatment response may ultimately improve treatment strategies for HIV-1 and its complications. Progress in this area requires the establishment of robust repositories of human DNA. The present study establishes that, across a range of CD4 T cell counts, expectorated saliva provides comparable yields of human DNA from HIV-negative and HIV-positive individuals, including those with AIDS.
This study has practical implications. For genomic studies, the most frequently used sources of DNA are peripheral blood and oral specimens. We have shown that, among over 10,000 HIV-infected adults in the United States who have contributed whole blood specimens to the NIH-funded AIDS Clinical Trials Group (ACTG) Human DNA Repository, 15-mL whole blood samples yield a median of ~400 μg extracted DNA 19. However, whole blood is not a suitable source of DNA in many resource-limited settings due to the need for continuous cryopreservation at −70°C, and shipping on dry ice until extracted. Small quantities of DNA may be obtained from blood spot cards or buccal swabs, which may be stored and shipped at ambient temperature, and are sufficient for many assays. Unfortunately, native DNA yields from such specimens are insufficient to establish DNA repositories, necessitating whole genome amplification. Because native high-molecular-weight DNA is necessary for optimal performance of some genomic assays, whole genome amplified DNA is not a preferred repository specimen.
Although DNA yields from expectorated saliva are generally less than from whole blood, saliva has advantages. Because no equipment, freezers or refrigerators are used during specimen collection or storage, high quality specimens can be collected in diverse settings, including rural areas or locales where electricity supplies may be unreliable. In addition, because venipuncture is not required, specimens can be readily obtained away from the research clinic. With sufficient instructions samples may be collected remotely, without direct contact with research personnel.
The present study had limitations. Volunteers were enrolled in a large HIV primary care clinic in the United States. We cannot exclude the possibility that, in resource limited countries, malnutrition or other co-morbidities among HIV-infected individuals might reduce human DNA yields. In the present study, specimens were stored and processed under optimal conditions. It is possible that, in resource limited countries, exposure of specimens to extremes of temperature during storage and shipping might affect human DNA yields or integrity.
Acknowledgments
The authors are grateful to the persons who volunteered for this study. This study was supported in part by NIH grants AI068636 (DR, DWH), AI54999 (RB, DWH), AI077505 (DWH), AI069439 (RB, DWH), and RR024975 (DR, CS, DWH).
Footnotes
David W. Haas has received research grants from Bavarian Nordic, Boehringer-Ingelheim, Bristol-Myers Squibb, Gilead Sciences, Merck, Tanox, and Tibotec. He is on scientific advisory boards for Glaxo Smith Kline and Tibotec. Rebecca Basham, Danielle Richardson and Cara Sutcliffe have no conflicts.
References
- 1.Centers for Disease Control and Prevention. [Accessed February 12, 2009];HIV/AIDS Statistics and Surveillance. 2008 Available at: http://www.cdc.gov/hiv/topics/surveillance/index.htm.
- 2.Report on the Global AIDS Epidemic. [Accessed February 12, 2009];United Nations Programme on HIV/AIDS. 2007 Available at: http://www.unaids.org/en/
- 3.Palella FJJ, Delaney KM, Moorman AC, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. New Engl J Med. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 4.U.S.Department of State; 2005. [Accessed February 12, 2009]. The President’s Emergency Plan for AIDS Relief: Five-year strategy. Available at: http://www.state.gov/documents/organization/57420.pdf. [Google Scholar]
- 5.World Health Organization. [Accessed February 12, 2009];WHO and HIV/AIDS. Available at: http://www.who.int/hiv/en/
- 6.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 7.Sachidanandam R, Weissman D, Schmidt SC, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 8.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 9.Syvanen AC. Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat Rev Genet. 2001;2:930–942. doi: 10.1038/35103535. [DOI] [PubMed] [Google Scholar]
- 10.Phillips KA, Veenstra DL, Oren E, Lee JK, Sadee W. Potential role of pharmacogenomics in reducing adverse drug reactions: a systematic review. JAMA. 2001;286:2270–2279. doi: 10.1001/jama.286.18.2270. [DOI] [PubMed] [Google Scholar]
- 11.Haas DW. Pharmacogenomics and HIV therapeutics. J Infect Dis. 2005;191:1397–1400. doi: 10.1086/429303. [DOI] [PubMed] [Google Scholar]
- 12.Aps JK, Van den MK, Delanghe JR, Martens LC. Flow cytometry as a new method to quantify the cellular content of human saliva and its relation to gingivitis. Clin Chim Acta. 2002;321:35–41. doi: 10.1016/s0009-8981(02)00062-1. [DOI] [PubMed] [Google Scholar]
- 13.Haase AT, Henry K, Zupancic M, et al. Quantitative image analysis of HIV-1 infection in lymphoid tissue. Science. 1996;274:985–989. doi: 10.1126/science.274.5289.985. [DOI] [PubMed] [Google Scholar]
- 14.Schacker TW, Nguyen PL, Beilman GJ, et al. Collagen deposition in HIV-1 infected lymphatic tissues and T cell homeostasis. J Clin Invest. 2002;110:1133–1139. doi: 10.1172/JCI16413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. 1993 revised classification system for HIV infection and expanded surveillance case definition for AIDS among adolescents and adults. MMWR Recomm Rep. 1992;41:1–19. [PubMed] [Google Scholar]
- 16.OrageneR DNA Self-Collection Kit User Instructions (OG-250 Disc Format) [Accessed February 12, 2009];DNA Genotek. Available at: http://www.dnagenotek.com/techsupport_donor.htm.
- 17.Maximizing DNA yield with OrageneR DNA (PD-PR-010 Issue 1.6) [Accessed February 12, 2009];2004 Available at: http://www.dnagenotek.com/pdf_files/PDPR010_MaximizingDNAYield.pdf.
- 18.Laboratory Protocol for Manual Purification of DNA from 0.5 mL of OrageneR•DNA/saliva (PD-PR-006 Issue 3.2) [Accessed February 12, 2009];2006 Available at: http://www.dnagenotek.com/pdf_files/PD-PR-006%20Issue%203.4_Protocol%20Manual%20Purification%20of%20DNA.pdf.
- 19.Haas DW, Wilkinson GR, Kuritzkes DR, et al. A multi-investigator/institutional DNA bank for AIDS-related human genetic studies: AACTG protocol A5128. HIV Clin Trials. 2003;4(5):287–300. doi: 10.1310/MUQC-QXBC-8118-BPM5. [DOI] [PubMed] [Google Scholar]


