Abstract
Rationale
Exercise capacity is a physiological characteristic associated with protection from both cardiovascular and all-cause mortality. p53 regulates mitochondrial function and its deletion markedly diminishes exercise capacity, but the underlying genetic mechanism orchestrating this is unclear. Understanding the biology of how p53 improves exercise capacity may provide useful insights for improving both cardiovascular as well as general health.
Objective
To understand the genetic mechanism by which p53 regulates aerobic exercise capacity.
Methods and Results
Using a variety of physiological, metabolic and molecular techniques, we further characterized maximum exercise capacity and the effects of training, measured various non-mitochondrial and mitochondrial determinants of exercise capacity, and examined putative regulators of mitochondrial biogenesis. As p53 did not affect baseline cardiac function or inotropic reserve, we focused on the involvement of skeletal muscle and now report a wider role for p53 in modulating skeletal muscle mitochondrial function. p53 interacts with Mitochondrial Transcription Factor A (TFAM), a nuclear-encoded gene important for mitochondrial DNA (mtDNA) transcription and maintenance, and regulates mtDNA content. The increased mtDNA in p53+/+ compared to p53−/− mice was more marked in aerobic versus glycolytic skeletal muscle groups with no significant changes in cardiac tissue. These in vivo observations were further supported by in vitro studies showing over-expression of p53 in mouse myoblasts increases both TFAM and mtDNA levels while depletion of TFAM by shRNA decreases mtDNA content.
Conclusions
Our current findings indicate that p53 promotes aerobic metabolism and exercise capacity by utilizing different mitochondrial genes and mechanisms in a tissue-specific manner.
Keywords: aerobic, exercise, mitochondrial DNA, p53, TFAM
Across populations aerobic exercise capacity inversely correlates with cardiovascular disease and all-cause mortality.1–3 Our report of a marked reduction in the maximal exercise capacity of p53 homozygous knockout (p53−/−) mice and subsequent confirmation by others provided physiologic evidence for p53 as an important mediator of aerobic metabolism.4,5 We previously showed that p53 promotes mitochondrial respiration in human and murine cells by regulating the transcription of Synthesis of Cytochrome c Oxidase 2 (SCO2), a gene essential for oxidative phosphorylation.4,6 A concurrent report demonstrating that p53 directly suppresses glycolysis through TIGAR, a p53-dependent regulator of glycolysis and apoptosis, suggested that p53 can coordinate aerobic and glycolytic metabolism.7
Multiple factors contribute to aerobic exercise capacity, but one major determinant is the mitochondrial content of skeletal muscle as demonstrated by a genetic selection experiment.8 A recent study showed decreased mitochondrial density in the skeletal muscle of p53 deficient mice,5 but the genetic mechanism orchestrating this change has remained unclear. A number of other studies have also associated p53 with exercise response and mitochondrial function. For example, p53 levels are increased after acute exercise, and the transition from glycolysis to oxidative metabolism during development is dependent on p53.9,10 p53 can transactivate ribonucleotide reductase p53R2 (RRM2B) which is important for maintaining mtDNA in skeletal muscle.11 Additionally, two recent studies have shown associations between p53 and mitochondrial DNA (mtDNA) content using cell models.12,13
Mitochondrial Transcription Factor A (TFAM) is another essential gene for mtDNA replication and transcription. p53 has been shown to physically interact with TFAM protein for mtDNA maintenance,14 however, the levels of p53 in mitochondria are normally very low. Here, we report that p53 can regulate TFAM transcription by interacting with its p53 binding site in myoblasts, determines mtDNA content in skeletal muscle, and confers higher maximum exercise capacity, either at baseline or after exercise training. Taken together, these data suggest that p53 affects mitochondrial function through more than one pathway and strengthens its role as a general promoter of aerobic capacity, an important determinant of cardiovascular function and health.
Materials and Methods
Animals and Cell Lines
All mice were maintained and handled in accordance with NHLBI Animal Care and Use Committee. p53−/− mice were obtained from Jackson Laboratories (C57BL/6J strain), and male mice were tested at 8–12 weeks of ages unless otherwise specified. C2C12 cells were obtained from ATCC, primary myoblasts were isolated from hind limb muscles of 2-week old mice, and mouse embryo fibroblasts (MEFs) were isolated from 13.5 to 14.5-d embryos as previously described.15,16 p53−/− MEFs were transiently transfected with 0.4 μg wild-type or mutant (mt135) p53 plasmid in 6-well plates (Clontech) using Effectene (Qiagen) and TFAM mRNA was quantified by RT-PCR after 24 h.
Mouse Exercise Characterization, Metabolic and Mitochondrial Studies
Details for mouse exercise testing, training, other phenotypic characterization, metabolic and mitochondrial studies are presented in detail in supplemental materials.
Antibodies and Western Blotting
Antibodies were from the following sources: rabbit control IgG serum (SC-2027), rabbit polyclonal anti-p53 (against full-length protein, FL-393, SC-6243), goat polyclonal anti-TFAM (A-17) (Santa Cruz Biotech); rabbit polyclonal anti-PGC-1α (101707, Cayman); rabbit polyclonal anti-p53R2 (Abcam); rabbit polyclonal anti-SCO2 as described4; rabbit polyclonal anti-TFAM (a generous gift from Dr. Eric A. Shoubridge, McGill University), and mouse monoclonal anti-tubulin (Clone B-5-1-2, Sigma) antibodies. Proteins samples were homogenized in ice cold RIPA lysis buffer with protease inhibitor cocktail (Roche), resolved by Tris-glycine SDS PAGE, and transferred to Immobilon-P membrane (Millipore) for standard ECL western blotting.
Identification of p53 Response Elements and Luciferase Transactivation Assay
The mouse TFAM genomic sequence was obtained from UCSC Genome Browser (http://genome.ucsc.edu/). Putative p53 responsive elements (p53REs) were identified using VectorNTI Advance 10 (Invitrogen). Only those matching more than 80% of the core consensus sequence (5′-RRRCWWGYYY-3′) with 0 to 13 bases between the two core binding motifs were evaluated further by reporter assay (R, purine; Y, pyrimidine; W, A or T).
40-bp long oligonucleotides containing the putative TFAM p53RE sequence (or mutated core binding sequence RRRAWWAYYY) were synthesized, annealed and cloned them into the pTA-luciferase vector (Clontech). Transactivation was measured 36 h after Lipofectamine 2000 (Invitrogen)-mediated cotransfection with pGL4.74 containing TK promoter and Renilla luciferase as a transfection efficiency control in C2C12 myoblasts. For p53 knockdown experiments, ON-TARGET Plus mouse p53-specific and non-specific siRNAs (Dharmacon RNAi Technologies) were mixed with the above reporter constructs and co-transfected.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was carried out using ChIP-IT Express (Active Motif) according to the manufacturer’s protocol. C2C12 myoblasts were treated with 100 μg/ml 5-fluorouracil for 48 h, fixed in 1% formaldehyde containing complete medium for 10 min at room temperature, and sonicated to obtain the nuclear lysates. Rabbit control IgG serum or polyclonal anti-p53 antibody was used at 10 μg/ml concentration to immunoprecipitate the fixed chromatin for PCR amplification. Primer sequences for APOE (nonspecific genomic control), TFAM and p21 p53REs are provided in online supplemental material.
Real-time PCR mRNA and mtDNA Quantification
Details for mRNA and mtDNA quantification and primer sequences are provided in the online supplemental material.
cDNA Transfection or Lentiviral Transduction of C2C12 Cells
Mouse myoblast C2C12 cells were transfected with empty vector or pLEX containing mutated (MUT) or wild-type (WT) p53 using FuGENE 6 (Roche Diagnostics) according to the manufacture’s protocol and allowed to recover for 24 h prior to changing the medium for myotube differentiation. Plasmids containing the sequences for non-specific shRNA (SHC002, Sigma-Aldrich) and TFAM shRNA (TRCN0000086066, Open Biosystems) (sequences in supplemental material) were used to prepare lentivirus according to manufacturer’s protocol (Sigma-Aldrich). Cells were transduced with lentivirus (MOI ~1) for 24 h followed by puromycin selection. For differentiating into myotubes prior to mtDNA and protein analysis, cells were incubated for 3 d in DMEM containing 2% horse serum.
Statistical Analysis
Data are presented as mean ± SEM. All P-values were calculated using two-tailed distribution Student’s t test and considered to be significant if P < 0.05. One-way analysis of variance (ANOVA) was performed for comparisons among the different genotype groups followed by Tukey’s post hoc test using Instat v3.06 software (Graph pad).
Results
p53 Determines Maximum Exercise Capacity via Modulation of Skeletal Muscle
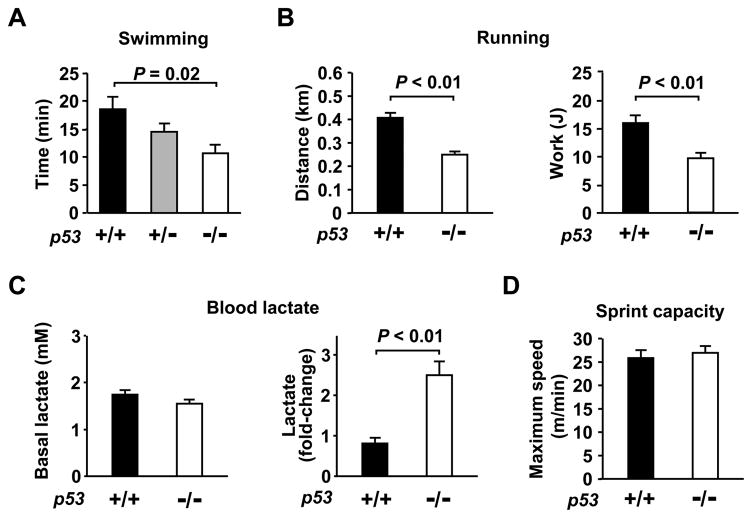
To quantify the effect of p53 on aerobic exercise capacity, we examined its gene dosage effect on swimming endurance using three different genotypes of mice: wild-type (p53+/+), heterozygous (p53+/−) and homozygous (p53−/−) knockout animals. We observed a direct relationship between p53 copy number and swimming duration; relative to p53+/+ mice, the swim times were reduced by 22% ± 8% (P = 0.14) and 42% ± 8% (P < 0.05) in p53+/− and p53−/− mice, respectively (one-way ANOVA, P < 0.05, Figure 1A). Although the p53+/− mice were useful for demonstrating a gene dosage effect for the swimming test, in subsequent experiments we focused our studies on p53+/+ and p53−/− animals that gave more consistent and significant changes and to avoid the more variable phenotype associated with the haploinsufficient state.
Figure 1.
Aerobic exercise capacity is p53 dependent. A, Effect of p53 gene dose on maximum swimming duration (min). p53 genotype: +/+ (black); +/− (gray); −/− (white). One-way ANOVA, P < 0.05. B, Maximum treadmill running capacity in p53+/+ (dark) and p53−/− (light) mice expressed as distance (km) and work (J) (n = 6–8 each). C, Measurements of resting blood lactate levels (mM) under resting conditions (left panel) and after sub-maximum exercise (fold-change of resting state) (right panel), n = 9 each, mean ± SEM. D, Similar sprint capacity as measured by maximum treadmill speed tolerated by p53+/+ (dark) and p53−/− (light) mice, n = 9 each.
To confirm the observed change in swimming endurance, we employed a treadmill running protocol as a different exercise modality and observed a similar reduction in distance running and work capacity of p53−/− mice compared to p53+/+ mice (Figure 1B). After sub-maximum exercise p53−/− mice had up to a three-fold higher rise in blood lactate compared to p53+/+ mice, providing biochemical evidence that these isogenic mice have decreased aerobic exercise capacity (Figure 1C). Despite the decrease in endurance exercise capacity of p53−/− mice, one prediction would be that these mice have preserved sprint capability as this form of exercise is less dependent on mitochondrial function. Indeed, p53−/− mice were equally capable of high intensity sprinting as their wild-type counterparts (Figure 1D). Another question that arises is whether there are changes in the glycogen stores of p53−/− skeletal muscle in association with altered endurance capacity as glycolytic muscles contain higher glycogen levels. Although we observed higher glycogen content in the more glycolytic tibialis anterior (TA) muscle group, we did not observe any significant differences by p53 genotype in either aerobic or glycolytic muscle groups (Online Figure I). Thus, it appeared that the decreased aerobic capacity of p53−/− mice manifests itself only when challenged by maximum endurance testing.
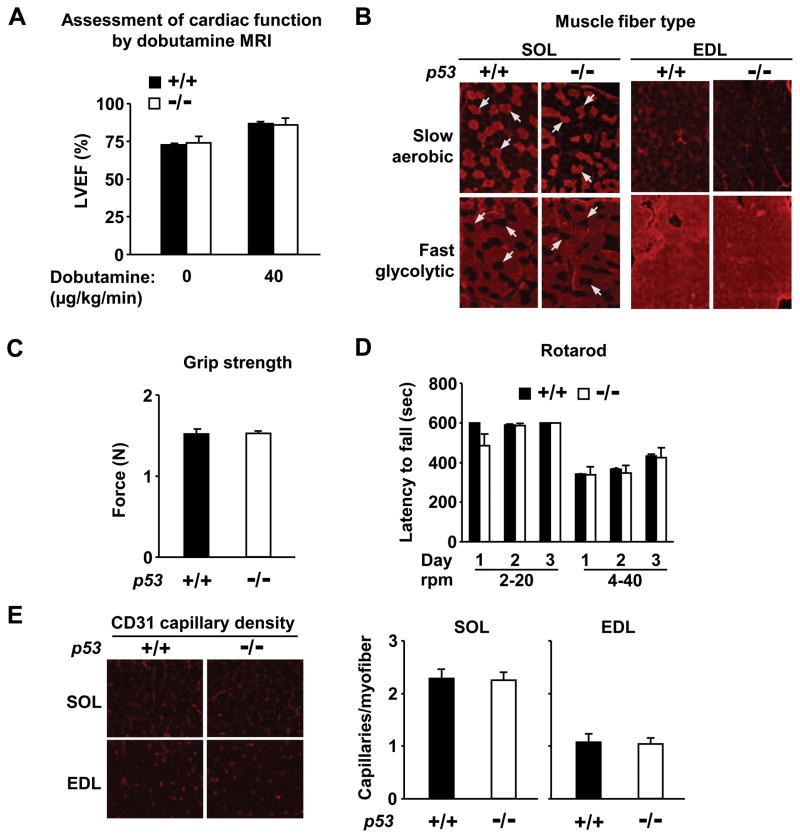
Because p53−/− mice appear overtly normal, with similar body composition and mass to their p53+/+ counterparts (Online Figure II), we closely examined some of the major determinants of exercise capacity. As cardiac output is an important determinant of exercise capacity, we examined both baseline left ventricular function and inotropic reserve using dobutamine MRI (Figure 2A). No significant differences were detected between p53+/+ and p53−/− mice in cardiac function nor in the gross functioning of their neuromuscular and vascular systems as assessed by motor coordination and muscle strength, fiber type composition and capillary density of both aerobic and glycolytic skeletal muscle groups (Figure 2A to 2E). Other factors such as hematocrit and hemoglobin concentration were also similar between p53+/+ and p53−/− mice; 48±2 % (14.9±0.4 g/dL) and 49±3 % (15.6±0.7 g/dL), respectively. The absence of significant alterations in these important physiological parameters in p53−/− mice was striking given the profound reduction in exercise capacity, and underscored the potential contribution of p53-dependent regulation of mitochondrial respiration to the observed exercise phenotype.
Figure 2.
Evaluation of cardiovascular and neuromuscular determinants of exercise performance. A, Left ventricular ejection fraction (LVEF) assessed by MRI at rest and peak dobutamine infusion to mimic exercise-induced cardiac response, n = 3 each. P < 0.05 for 0 versus 40 μg/kg/min infusion for both p53+/+ and p53−/− mice. B, Fiber types of soleus (SOL, aerobic muscle) and extensor digitorum longus (EDL, glycolytic muscle) were visualized using anti-MHC I (slow fiber, aerobic) or anti-MHC II (fast fiber, glycolytic) antibody. Arrows highlight the complementary nature of the slow and fast muscle fibers that is similar in serial sections of both p53+/+ and p53−/− soleus muscle. C, Forelimb grip strength (N, Newton) was assessed using a digital grip strength meter, n = 7–9 each. D, Motor coordination was assessed by latency to fall on a rotarod at varying rotational speeds over a 3-day test period in p53+/+ (dark) and p53−/− (light) mice, n = 4–5 each. E, Capillary density was visualized by anti-CD31 (PECAM-1) antibody immunofluorescence of soleus (SOL) and extensor digitorum logus (EDL) muscles. p53+/+ (dark) and p53−/− (light). Mean ± SEM.
Training Accentuates the Lower Exercise Capacity of p53−/− Mice
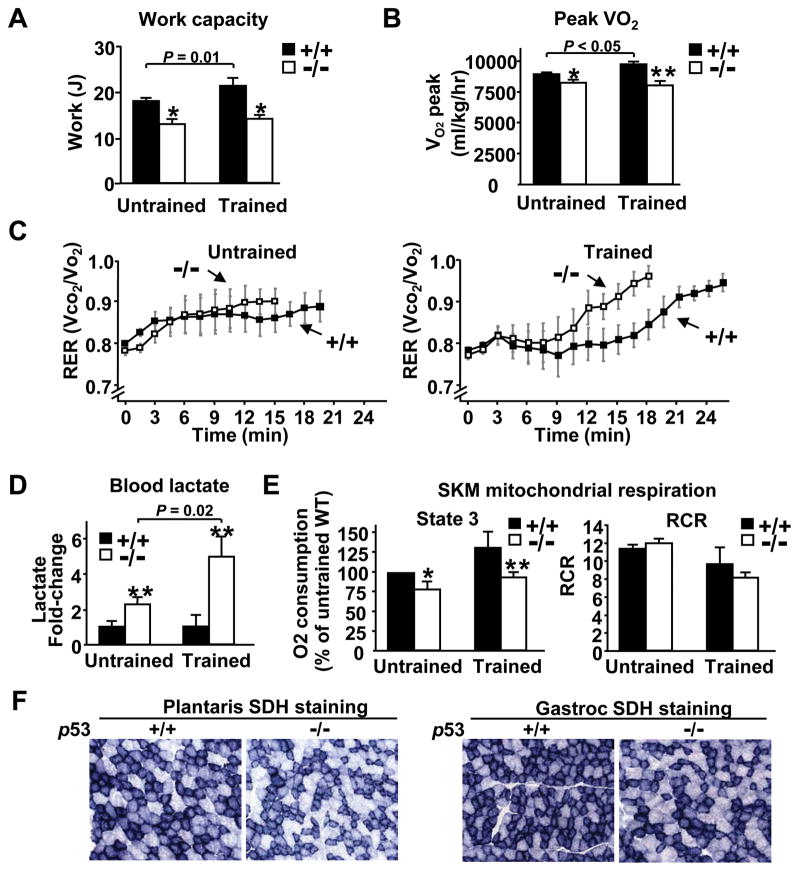
As skeletal muscle is a major determinant of aerobic exercise capacity8 and exercise training increases mitochondrial biogenesis,17 we reasoned that p53 may influence its adaptive responses to exercise training. To test this hypothesis, the maximum exercise capacity (calculated as work) was evaluated in p53+/+ and p53−/− mice after a 5 week period of treadmill training protocol. The maximum work capacity of p53+/+ mice was significantly increased after training as expected, but the work capacity of p53−/− was relatively unresponsive to training, accentuating the difference observed between p53+/+ and p53−/− mice (Figure 3A). Direct measurement of aerobic capacity by oxygen consumption (Vo2) and respiratory exchange ratio (RER, CO2 production/O2 consumption expressed as Vco2/Vo2) at maximum exercise revealed differences between p53+/+ and p53−/− mice that paralleled the work capacity differences of the respective untrained and trained groups (Figure 3B and 3C). The relatively greater increase in blood lactate levels of p53−/− mice compared to p53+/+ mice in the trained versus untrained group further complemented the differences observed in RER (Figure 3D). Collectively, these findings reemphasized that p53 is required to complete the adaptive changes in aerobic metabolism that are necessary to increase exercise capacity in response to training. It is also notable that the relative difference in peak Vo2 between trained p53+/+ and p53−/− mice and its impact on exercise capacity were comparable to a model of exercise enhancement by the over-expression of mitochondrial biogenesis regulator PGC-1α.18
Figure 3.
p53 augments the increase in maximum exercise capacity and oxygen consumption with training. A, Maximum work capacity of untrained and trained mice by treadmill running. Untrained p53+/+ and p53−/− mice, n = 11 and 9, respectively; trained p53+/+ and p53−/− mice, n = 5 each. B, Peak Vo2 was measured during maximum treadmill exercise protocol, n = 3–4. C, RER (CO2 production/O2 consumption) was measured during peak Vo2 exercise testing, n = 3–4. D, Blood lactate levels were measured after sub-maximum treadmill exercise in untrained and trained mice and are shown as fold-change relative to p53+/+ mice within each group, n = 5 each. E, Relative oxygen consumption of mitochondria isolated from skeletal muscle of untrained and trained mice in state 3 respiration, normalized to the weight of skeletal muscle tissue (nmol O2/min/g tissue). Data shown are relative to untrained p53+/+ mice (left panel). RCR was estimated from the ratio of state 3 to state 2 respiration (right panel). *P < 0.05 or **P < 0.01 between corresponding p53+/+ and p53−/− mice, n = 3–4. Mean ± SEM. F, Succinate dehydrogenase (SDH) enzymatic activity staining in representative frozen sections of plantaris and gastrocnemius muscles. The p53+/+ and p53−/− muscle sections were placed on the same glass slide, thus, all pairs underwent similar treatment. Magnification, 20×.
To understand the biology underlying the increase in aerobic capacity following training, we examined skeletal muscle mitochondrial oxygen consumption in untrained and trained states. When normalized to skeletal muscle mass to account for differences in tissue mitochondrial volume density, we observed increases in mitochondrial state 3 respiration in the setting of similar respiratory control ratios (RCR) that correlated well with peak work capacity and Vo2 changes (Figure 3A, 3B and 3E). The higher oxygen consumption in p53+/+ mice was consistent with the recently reported increase in mitochondrial volume density in the mixed fiber gastrocnemius muscle measured by electron microscopy,5 which we also confirmed in the more aerobic soleus muscle group (Online Figure III). In addition to increased mitochondrial density, succinate dehydrogenase staining of plantaris, another aerobic muscle group, and gastrocnemius supported relatively increased mitochondrial oxidative capacity in p53+/+ compared to p53−/− skeletal muscles (Figure 3F).
p53 Interacts with TFAM Gene
We had previously reported that decreased oxygen consumption in p53−/− liver mitochondria was primarily mediated by SCO2.4 However, unlike in liver, mitochondria prepared from p53−/− skeletal muscle did not show a significant decrease in SCO2 mRNA or protein levels (Online Figure IVA). These findings suggested that in skeletal muscle p53 affects mitochondrial function by regulating the expression of proteins other than SCO2. Although p53-dependent ribonucleotide reductase (RRM2B, p53R2) was an obvious candidate gene given its previous association with skeletal muscle mtDNA homeostasis,11 we did not observe significant differences in p53R2 levels or in the expression of two additional p53 targets ferredoxin reductase (FDXR) and Tp53-induced glycolysis and apoptosis regulator (TIGAR) previously associated with mitochondrial and metabolic functions (Online Figure IVB to IVD).7,19 We also failed to detect a significant effect of p53 genotype on the abundance of mitochondrial biogenesis regulators PPARγ coactivator-1α (PGC-1α), PGC-1β, NRF1 and NRF220 and representative members of both nuclear- and mitochondrial-encoded respiratory chain components at baseline or after exercise (Online Figure V). These negative results led us to speculate the involvement of other factors in skeletal muscle.
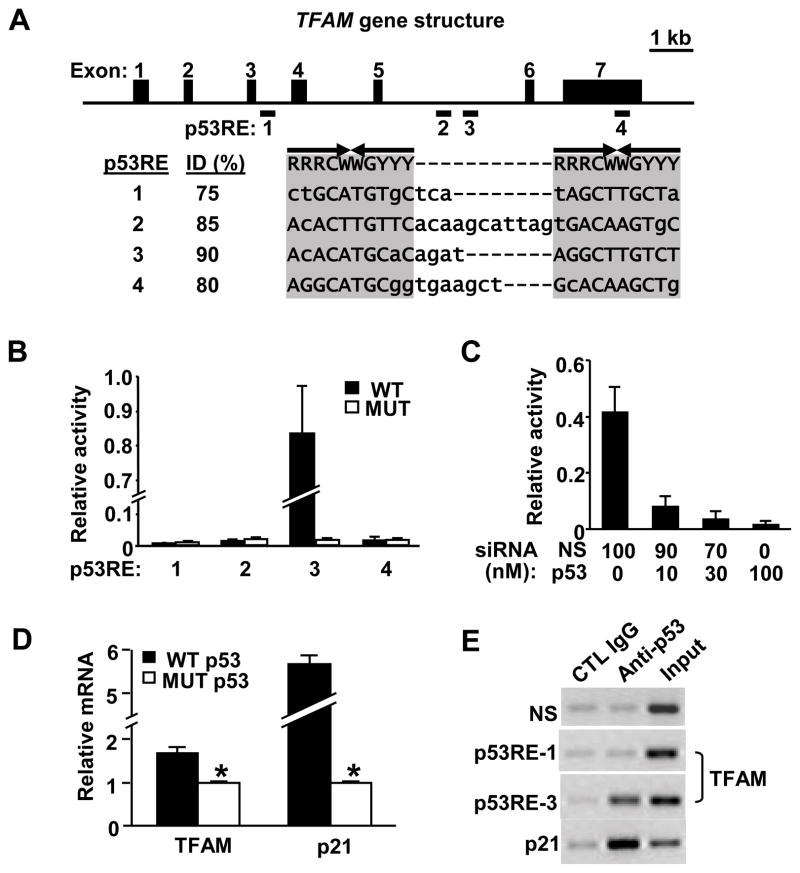
Mitochondrial biogenesis is also mediated by mitochondrial transcription factors (TFAM, TFBs).21 The central importance of TFAM to the transcription and maintenance of mtDNA, which is correlated to aerobic exercise capacity in both mouse and man, made it an attractive candidate for mediating some of the effects of p53.18,22 Indeed, in silico analysis of the murine TFAM genomic locus revealed four putative p53 response element (p53RE) consensus sequences (Figure 4A). These putative p53 response elements in TFAM were tested by creating luciferase reporter constructs with either wild-type (WT) or mutated (MUT) p53 binding sequences and transfecting them into C2C12 murine myoblasts, which harbor wild-type p53 alleles. Only p53RE-3 located in intron 5, exhibiting the closest homology (90% identity) to the p53 binding consensus sequence, displayed significant luciferase activity (Figure 4B). The introduction of point mutations that disrupted the p53 binding sequence eliminated the luciferase signal, demonstrating sequence specificity (Figure 4B). Furthermore, the dependence of p53RE-3 activity on the presence of endogenous wild-type p53 protein in C2C12 cells was confirmed by dose-dependent siRNA-mediated knockdown of p53 abundance (Figure 4C).
Figure 4.
p53 interacts with TFAM gene. A, Genomic structure of mouse TFAM with seven exons (black boxes). The putative p53 responsive elements (p53REs) relative to bp +1 position of the ATG start site are: 1, +2475 to +2497; 2, +6538 to +6568; 3, +7087 to +7110; and 4, +11631 to 11657. Consensus matching p53RE bases are shown in capital letters (ID, % identity). R, purine; Y, pyrimidine; W, A or T base. B, Transcriptional activities of wild-type (WT) or mutated (MUT) TFAM p53REs were determined by luciferase reporters after transfection into C2C12 myoblasts. C, Knockdown of endogenous p53 decreases p53RE luciferase activity. The p53RE-3 luciferase construct was co-transfected with indicated p53-targeted and non-specific (NS) siRNA (100 nM total). D, TFAM mRNA response to transient transfection of exogenous wild-type or mutant p53 into p53−/− MEF cells. E, Chromatin immunoprecipitation (ChIP) assay to demonstrate in vivo interaction between p53 protein and TFAM p53RE-3 in C2C12 myoblasts. Nonspecific (NS) genomic and TFAM p53RE-1 primers were used as negative controls; the p53RE of p21 gene was used as a positive control. Rabbit non-immune serum (Control IgG) and polyclonal antibody against full-length p53 (Anti-p53) were used for immunoprecipitation; INPUT, crude chromatin. *P < 0.05. Mean ± SEM.
We next examined whether the TFAM gene could be transactivated in cells by transiently transfecting wild-type or mutant p53 into p53−/− MEFs. The expression of wild-type p53 significantly increased TFAM transcript levels relative to mutant p53 (Figure 4D). A corresponding increase in the transcript level of p21, the prototypical target of p53, was also observed. To establish that p53 interacts with the putative p53 binding sequences in TFAM, we performed chromatin immunoprecipitation (ChIP) in C2C12 murine myoblasts. In contrast to nonspecific control IgG, anti-p53 antibody pulled down the TFAM p53RE-3 genomic DNA fragment detected by PCR amplification, with the p21 gene promoter serving as positive control (Figure 4E). A nonspecific genomic region in the APOE gene and TFAM p53RE-1 (unresponsive to p53 by luciferase assay) served as negative controls. In summary, these data supported the notion that p53 is capable of interacting with and regulating the expression of the TFAM gene, thereby contributing to mitochondrial biogenesis.
p53 Regulates TFAM Expression and mtDNA Content in Skeletal Muscle
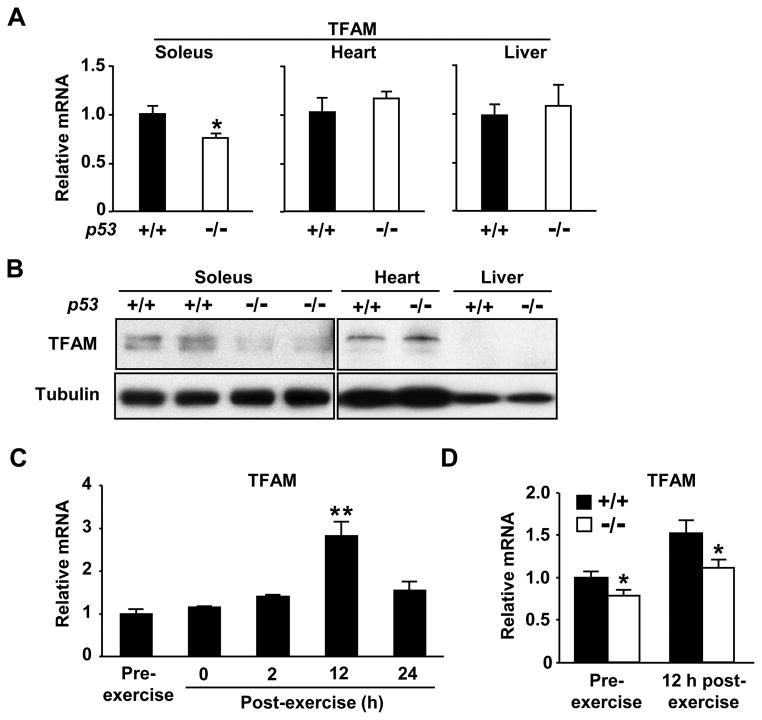
Under basal (i.e. no exercise) state, the absence of p53 reduced both TFAM mRNA and protein levels in soleus skeletal muscle but not in heart or liver, supporting our in vitro interaction studies (Figure 5A and 5B). TFAM protein in liver was not detected at this level of sensitivity consistent with its low mtDNA content (Figure 6A). Based on the requirement of p53 for the increase in aerobic capacity with training, we examined TFAM mRNA expression after a single session of exercise in untrained mice. TFAM expression peaked around 12 h after exercise stimulation (Figure 5C), at which point the relative increase in TFAM mRNA was comparable in p53+/+ and p53−/− mice (Figure 5D). While these data suggest that factors other than p53 such as p38 MAPK and PGC-1α23 are important in initiating the exercise-induced change in TFAM expression, p53 may enhance its expression.
Figure 5.
p53 modulates TFAM expression in skeletal muscle. A, p53 affects the basal expression of TFAM mRNA in soleus muscle but not in heart or liver, n = 5–9 each. B, p53 affects the level of TFAM protein in soleus muscle but not in heart or liver. (Note: TFAM protein in liver is undetectable at this level of sensitivity consistent with its low mtDNA content shown in Figure 6A; TFAM resolves as a single or double band in mouse skeletal muscle or heart tissue, respectively) C, Acute exercise (14 m/min for 1 h) increases TFAM mRNA expression in soleus muscle of p53+/+ mice. **P < 0.01 versus pre-exercise level; n = 3–5. D, Relative TFAM mRNA expression in soleus 12 h after acute exercise. *P < 0.05 between corresponding p53+/+ (dark) and p53−/− (light) mice; n = 6. Mean ± SEM.
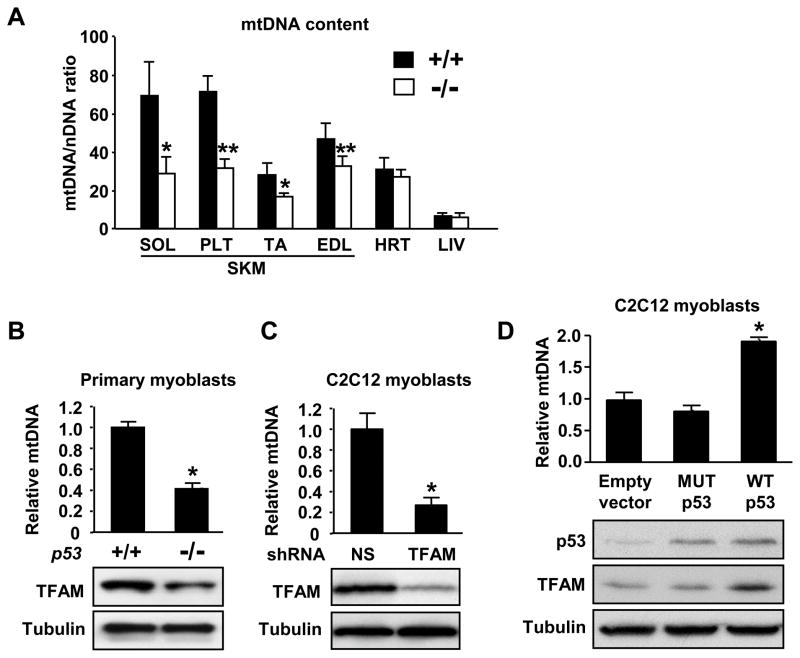
Figure 6.
p53 increases mtDNA copy number and TFAM level. A, Relative mtDNA content expressed as a function of total genomic DNA (nDNA) in various skeletal muscle (SKM) groups, heart (HRT) and liver (LIV) of p53+/+ (dark) and p53−/− (light) mice. Aerobic muscle groups are represented by soleus (SOL) and plantaris (PLT); glycolytic muscle groups are represented by tibialis anterior (TA) and extensor digitorum longus (EDL). n = 5 each. B, Relative mtDNA content and corresponding TFAM protein levels of primary mouse myoblasts. C, Stable knock down of TFAM by shRNA concomitantly decreases TFAM protein and mtDNA levels in C2C12 cells. D, Conversely, transient over-expression of wild-type (WT) but not mutant (MUT) p53 in C2C12 cells increases TFAM protein and relative mtDNA content in parallel. *P < 0.05; **P < 0.01; n = 3. Mean ± SEM.
We measured the tissue content of mtDNA relative to nuclear DNA and observed significantly lower levels in various skeletal muscle groups of p53−/− mice (Figure 6A). A notable pattern emerged: the relative difference in mtDNA content associated with p53 copy number was greater for aerobic muscles (soleus, plantaris) versus glycolytic muscles (tibialis anterior, extensor digitorum longus) (P < 0.01 between soleus and EDL), suggesting the adaptive changes for aerobic metabolism in muscle may require p53. In contrast, heart and liver mtDNA content was not significantly affected further demonstating that p53 may modulate various aspects of mitochondrial function in a tissue-specific manner (Figure 6A). To control for the potential confounding effects associated with cellular heterogeneity in tissues, we analyzed mtDNA content and TFAM protein levels in myoblasts derived from p53+/+ and p53−/− mice and observed similarly decreased levels of both in the p53−/− state (Figure 6B). The relationship between p53, TFAM and mtDNA was further explored by the stable shRNA-mediated depletion of TFAM in C2C12 myoblasts which mimicked the reduced mtDNA content of p53−/−myoblasts, while the transient over-expression of wild-type, but not mutant, p53 increased TFAM protein and mtDNA levels in parallel (Figure 6C and 6D). These findings were consistent with well established mouse genetic studies showing that deletion of the TFAM gene results in mitochondrial DNA depletion and dysfunction, while over-expression of TFAM leads to increased mtDNA content.24,25
Discussion
Here, we have established that p53 determines maximum aerobic exercise capacity and regulates mtDNA content in skeletal muscle. Given the tight concordance between TFAM and mtDNA,24,25 p53 interaction with the TFAM regulatory sequences may contribute to the observed differences in mtDNA content of skeletal muscle. The effect of p53 on TFAM and mtDNA appears to be tissue-specific and distinct from what we have previously described for p53 regulation of SCO2 in cytochrome c oxidase assembly in liver.4
The mechanisms underlying the tissue-specific alterations in mtDNA content are likely to be complex due to the interaction of p53 dependent and independent components expressed in tissues relevant to exercise physiology. p53 has many transcriptional target genes and indirectly influences the expression of many others,26 thus, our proposed regulation of the mitochondria by p53 in mouse skeletal muscle is unlikely to be the sole mechanism. Rather than a promoter, the p53 binding site in TFAM likely serves as a regulatory element such as an enhancer given its location in an intron 8 kb downstream of the transcription start site.26,27 It should be noted that a recent publication did not detect significant differences in TFAM mRNA levels in p53 knocked down cells 12 which is not consistent with the lower levels that we observed in p53−/− adult mouse skeletal muscle. However, the knock down of p53 in cultured fibroblasts promotes cell proliferation which likely impacts mitochondrial biogenesis including TFAM expression by known regulators such as NRF1 and NRF2.20 Thus, this discrepancy highlights the importance of considering the cellular context of gene expression in comparing in vitro to in vivo observations.
An additional layer of complexity is introduced by the translocation of p53 to the mitochondria and its potential interaction with TFAM or other components important for mtDNA maintenance.28–32 However, the greater abundance of TFAM protein compared to the relatively negligible pool of mitochondrial p53 under normal state raises questions about their stoichiometric relationship.33 Our observation that p53 can enhance TFAM transcription provides a genetic insight into skeletal muscle mitochondria biogenesis, an important determinant of exercise capacity. Although we have proposed a transcriptional mechanism for TFAM regulation by p53, there may be additional mechanisms by which p53 and TFAM interact.14
A recent study has demonstrated the proteomic diversity of tissue mitochondria which may reflect the need for mitochondria to fulfill functions that are unique to different tissues.34 While liver mitochondria mainly serve biosynthetic or general metabolic functions, skeletal muscle mitochondria are suited for oxidative phosphorylation to supply energy for contractile work. Thus, mitochondria are likely to have different regulatory requirements depending on their function. We propose that the targeting of SCO2 and TFAM by p53 exemplify two distinct pathways by which it may influence mitochondrial biogenesis in a tissue-specific manner. p53 regulation of TFAM expression may contribute by augmenting the overall program of mitochondrial biogenesis initiated by p53-independent mediators of exercise training such as p38 MAPK and PGC-1α.23
There is increasing evidence that link mitochondrial function with other well-known regulators of cell growth. These include mitochondrial regulation by ataxia-telangiectasia mutated kinase (ATM) via ribonucleotide reductase R1 subunit,35 retinoblastoma (Rb) via mitochondrial biogenesis coactivator PGC-1α,36 and PTEN via mitochondrial protein PTEN-induced kinase 1 (PINK1).37 Studies showing the role of mitochondrial uncoupling proteins in both cardiovascular diseases and cancer further indicate the importance of mitochondrial function in these processes.38,39 It is conceivable that the optimization of mitochondrial function that reduces reactive oxygen species (ROS) generation, DNA damage, or bioenergetic stress ultimately result in improvement of exercise capacity. Such mitochondrial adaptations in a wide range of chronic conditions such as atherosclerosis and congestive heart failure would appear important. Thus, understanding the molecular basis for maintaining or expanding aerobic capacity may thereby provide further insights into a host of debilitating cardiovascular diseases as well as potential therapeutic strategies against them.
Supplementary Material
Acknowledgments
We wish to thank S. Matoba, C. Birdsall and O. Gavrilova for advice and assistance and the NHLBI Electron Microscopy Core Facility for technical support. We are grateful to M. Sack, F. Bunz and T. Finkel for critical review and advice.
Sources of Funding
This research was supported by the Division of Intramural Research, National Heart, Lung, and Blood Institute, NIH, and a postdoctoral fellowship to T.M. from the Japan Society for the Promotion of Science
Non-standard Abbreviations and Acronyms
- mtDNA
mitochondrial DNA
Footnotes
Disclosures
None.
References
- 1.Sandvik L, Erikssen J, Thaulow E, Erikssen G, Mundal R, Rodahl K. Physical fitness as a predictor of mortality among healthy, middle-aged Norwegian men. N Engl J Med. 1993;328:533–7. doi: 10.1056/NEJM199302253280803. [DOI] [PubMed] [Google Scholar]
- 2.Erikssen G, Liestol K, Bjornholt J, Thaulow E, Sandvik L, Erikssen J. Changes in physical fitness and changes in mortality. Lancet. 1998;352:759–62. doi: 10.1016/S0140-6736(98)02268-5. [DOI] [PubMed] [Google Scholar]
- 3.Kokkinos P, Myers J, Kokkinos JP, Pittaras A, Narayan P, Manolis A, Karasik P, Greenberg M, Papademetriou V, Singh S. Exercise capacity and mortality in black and white men. Circulation. 2008;117:614–22. doi: 10.1161/CIRCULATIONAHA.107.734764. [DOI] [PubMed] [Google Scholar]
- 4.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–3. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 5.Saleem A, Adhihetty PJ, Hood DA. Role of p53 in mitochondrial biogenesis and apoptosis in skeletal muscle. Physiol Genomics. 2009;37:58–66. doi: 10.1152/physiolgenomics.90346.2008. [DOI] [PubMed] [Google Scholar]
- 6.Papadopoulou LC, Sue CM, Davidson MM, Tanji K, Nishino I, Sadlock JE, Krishna S, Walker W, Selby J, Glerum DM, Coster RV, Lyon G, Scalais E, Lebel R, Kaplan P, Shanske S, De Vivo DC, Bonilla E, Hirano M, DiMauro S, Schon EA. Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat Genet. 1999;23:333–7. doi: 10.1038/15513. [DOI] [PubMed] [Google Scholar]
- 7.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–20. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 8.Wisloff U, Najjar SM, Ellingsen O, Haram PM, Swoap S, Al-Share Q, Fernstrom M, Rezaei K, Lee SJ, Koch LG, Britton SL. Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science. 2005;307:418–20. doi: 10.1126/science.1108177. [DOI] [PubMed] [Google Scholar]
- 9.Ibrahim MM, Razmara M, Nguyen D, Donahue RJ, Wubah JA, Knudsen TB. Altered expression of mitochondrial 16S ribosomal RNA in p53-deficient mouse embryos revealed by differential display. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1998;1403:254–264. doi: 10.1016/s0167-4889(98)00066-4. [DOI] [PubMed] [Google Scholar]
- 10.Chen YW, Nader GA, Baar KR, Fedele MJ, Hoffman EP, Esser KA. Response of rat muscle to acute resistance exercise defined by transcriptional and translational profiling. J Physiol. 2002;545:27–41. doi: 10.1113/jphysiol.2002.021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, Chretien D, de Lonlay P, Paquis-Flucklinger V, Arakawa H, Nakamura Y, Munnich A, Rotig A. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet. 2007;39:776–80. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 12.Lebedeva MA, Eaton JS, Shadel GS. Loss of p53 causes mitochondrial DNA depletion and altered mitochondrial reactive oxygen species homeostasis. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbabio.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulawiec M, Ayyasamy V, Singh KK. p53 regulates mtDNA copy number and mitocheckpoint pathway. J Carcinog. 2009;8:8. doi: 10.4103/1477-3163.50893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Itoh H, Kang D, Kohno K. P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 2003;63:3729–34. [PubMed] [Google Scholar]
- 15.Rando TA, Blau HM. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J Cell Biol. 1994;125:1275–87. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol. 2003;5:741–7. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holloszy JO, Coyle EF. Adaptations of skeletal muscle to endurance exercise and their metabolic consequences. J Appl Physiol. 1984;56:831–8. doi: 10.1152/jappl.1984.56.4.831. [DOI] [PubMed] [Google Scholar]
- 18.Calvo JA, Daniels TG, Wang X, Paul A, Lin J, Spiegelman BM, Stevenson SC, Rangwala SM. Muscle-specific expression of PPARgamma coactivator-1alpha improves exercise performance and increases peak oxygen uptake. J Appl Physiol. 2008;104:1304–12. doi: 10.1152/japplphysiol.01231.2007. [DOI] [PubMed] [Google Scholar]
- 19.Hwang PM, Bunz F, Yu J, Rago C, Chan TA, Murphy MP, Kelso GF, Smith RA, Kinzler KW, Vogelstein B. Ferredoxin reductase affects p53-dependent, 5-fluorouracil-induced apoptosis in colorectal cancer cells. Nat Med. 2001;7:1111–7. doi: 10.1038/nm1001-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–68. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 21.Shadel GS. Expression and maintenance of mitochondrial DNA: new insights into human disease pathology. Am J Pathol. 2008;172:1445–56. doi: 10.2353/ajpath.2008.071163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Hiatt WR, Barstow TJ, Brass EP. Relationships between muscle mitochondrial DNA content, mitochondrial enzyme activity and oxidative capacity in man: alterations with disease. Eur J Appl Physiol Occup Physiol. 1999;80:22–7. doi: 10.1007/s004210050553. [DOI] [PubMed] [Google Scholar]
- 23.Akimoto T, Pohnert SC, Li P, Zhang M, Gumbs C, Rosenberg PB, Williams RS, Yan Z. Exercise stimulates Pgc-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. J Biol Chem. 2005;280:19587–93. doi: 10.1074/jbc.M408862200. [DOI] [PubMed] [Google Scholar]
- 24.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–6. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 25.Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson NG. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–44. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 26.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–12. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 27.Laptenko O, Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13:951–61. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- 28.Achanta G, Sasaki R, Feng L, Carew JS, Lu W, Pelicano H, Keating MJ, Huang P. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. Embo J. 2005;24:3482–92. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen D, Yu Z, Zhu Z, Lopez CD. The p53 pathway promotes efficient mitochondrial DNA base excision repair in colorectal cancer cells. Cancer Res. 2006;66:3485–94. doi: 10.1158/0008-5472.CAN-05-4103. [DOI] [PubMed] [Google Scholar]
- 30.Boopathi E, Srinivasan S, Fang JK, Avadhani NG. Bimodal protein targeting through activation of cryptic mitochondrial targeting signals by an inducible cytosolic endoprotease. Mol Cell. 2008;32:32–42. doi: 10.1016/j.molcel.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong TS, Rajagopalan S, Townsley FM, Freund SM, Petrovich M, Loakes D, Fersht AR. Physical and functional interactions between human mitochondrial single-stranded DNA-binding protein and tumour suppressor p53. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bakhanashvili M, Grinberg S, Bonda E, Simon AJ, Moshitch-Moshkovitz S, Rahav G. p53 in mitochondria enhances the accuracy of DNA synthesis. Cell Death Differ. 2008;15:1865–74. doi: 10.1038/cdd.2008.122. [DOI] [PubMed] [Google Scholar]
- 33.Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. Embo J. 2007;26:923–34. doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson DT, Harris RA, French S, Blair PV, You J, Bemis KG, Wang M, Balaban RS. Tissue heterogeneity of the mammalian mitochondrial proteome. Am J Physiol Cell Physiol. 2007;292:C689–97. doi: 10.1152/ajpcell.00108.2006. [DOI] [PubMed] [Google Scholar]
- 35.Eaton JS, Lin ZP, Sartorelli AC, Bonawitz ND, Shadel GS. Ataxia-telangiectasia mutated kinase regulates ribonucleotide reductase and mitochondrial homeostasis. J Clin Invest. 2007;117:2723–34. doi: 10.1172/JCI31604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sankaran VG, Orkin SH, Walkley CR. Rb intrinsically promotes erythropoiesis by coupling cell cycle exit with mitochondrial biogenesis. Genes Dev. 2008;22:463–75. doi: 10.1101/gad.1627208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gautier CA, Kitada T, Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci U S A. 2008;105:11364–9. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernal-Mizrachi C, Gates AC, Weng S, Imamura T, Knutsen RH, DeSantis P, Coleman T, Townsend RR, Muglia LJ, Semenkovich CF. Vascular respiratory uncoupling increases blood pressure and atherosclerosis. Nature. 2005;435:502–6. doi: 10.1038/nature03527. [DOI] [PubMed] [Google Scholar]
- 39.Gates AC, Bernal-Mizrachi C, Chinault SL, Feng C, Schneider JG, Coleman T, Malone JP, Townsend RR, Chakravarthy MV, Semenkovich CF. Respiratory uncoupling in skeletal muscle delays death and diminishes age-related disease. Cell Metab. 2007;6:497–505. doi: 10.1016/j.cmet.2007.10.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.