Summary
Genetic factors, specifically the VKORC1 and GGCX genes, have been shown to contribute to the interindividual variability in response to the vitamin K-antagonist, warfarin, which influences the dose required to achieve the desired anticoagulation response. These differences in warfarin sensitivity may be explained by differences in vitamin K status. Men and women (n = 416, 60–80 y), primarily of European descent, were genotyped for common polymorphisms in VKORC1 and GGCX. Cross-sectional associations exist between polymorphisms and biochemical markers of vitamin K [plasma phylloquinone, percent undercarboxylated osteocalcin (%ucOC)]. VKORC1 rs8050894 GG homozygotes had significantly higher cross-sectional measures of plasma phylloquinone than carriers of the CG or CC genotypes (plasma phylloquinone geometric means: GG 0.874±0.092 versus CG/CC 0.598± 0.044; p = 0.020), whereas carriers of VKORC1 rs7294 AA or AG had significantly lower plasma phylloquinone concentrations compared to GG homozygotes (plasma phyllo-quinone geometric means: 0.579±0.045 versus 0.762±0.057; p = 0.035). Cross-sectional analyses also revealed that heterozygous carriers of GGCX rs10187424 and rs7568458 had significantly lower %ucOC relative to either homozygous group. Polymorphisms in genes encoding enzymes involved in vitamin K metabolism may modulate plasma concentrations of phylloquionone and percent carboxylation of osteocalcin.
Keywords: vitamin K epoxide reductase, gamma glutamyl carboxylase, vitamin K, warfarin, polymorphism
Warfarin is the most widely prescribed anticoagulant for thromboembolic therapy in North America and Europe (1). Warfarin functions as a vitamin K antagonist and thus inhibits the activity of the vitamin K-dependent coagulation proteins (2). It is used for treatment of many health conditions, including atrial fibrillation, deep vein thrombosis, and recurrent stroke (3). Large interindividual differences exist in warfarin dose requirements, which present challenges in its management (4, 5). Warfarin dose requirements are influenced by ethnicity, age, gender, body mass index (BMI), and concurrent use of other medications (4, 5). In addition, several studies have revealed genetic components that influence the dose of warfarin necessary for a therapeutic response. Genetic variation in CYP2C9 (6–9), epoxide hydrolase (10), calumernin (8, 11), apolipoprotein E (12, 13), vitamin K epoxide reductase (VKORC1) (8, 9, 14), and γ-glutamyl carboxylase (GGCX) (11, 14, 15) have all been shown to be associated with warfarin sensitivity.
Of those genetic factors influencing warfarin sensitivity, VKORC1 and GGCX are critical enzymes responsible for the action of vitamin K as a cofactor in the γ-carboxylation of vitamin K-dependent proteins, including those involved in coagulation (16). The γ-carboxylation of vitamin K-dependent proteins is necessary for the proper functioning of these proteins (2, 17). The γ-carboxylation of these proteins is catalyzed by GGCX with reduced vitamin K (hydroquinone) acting as a cofactor. In this reaction, hydroquinone is oxidized to vitamin K 2,3 epoxide. Reduction of vitamin K 2,3 epoxide back to the active hydroquinone is catalyzed by VKORC1 (16).
It has been hypothesized that the associations between VKORC1 polymorphisms and warfarin sensitivity are due to differences in the availability of vitamin K for γ-carboxylation reactions (8, 15, 18, 19). Similarly, associations of warfarin sensitivity with GGCX genetic variation has been attributed to alterations in GGCX activity that subsequently influence functionality of vitamin K-dependent proteins (5, 15). However, direct evidence supporting putative mechanisms of the effects of genetic variation in VKORC1 and GGCX on warfarin sensitivity is lacking. Some evidence of altered VKORC1 mRNA has been reported (18); however effects of differential VKORC1 gene expression or protein concentrations on plasma phylloquinone (vitamin K1) concentrations are not known. Similarly, the association of genetic variation in GGCX and the degree of carboxylation of a vitamin K-dependent protein has not been assessed.
In this study, we investigated the cross-sectional associations of genetic variation in VKORC1 and GGCX with two measures of vitamin K status, plasma phylloquinone and serum percent undercarboxylated osteocalcin (%ucOC) prior to randomization to a vitamin K supplementation study. Secondary analyses were conducted on baseline measures of proteins induced by vitamin K absence–factor II, PIVKA-II, the degree of undercarboxylation of prothrombin. Serum %ucOC is a measure of vitamin K carboxylation in extra-hepatic proteins and is a sensitive measure of vitamin K nutritional status (20), whereas PIVKA-II is a measure of hepatic vitamin K status. We put forth two main hypotheses: first, that genetic variation in VKORC1 will influence vitamin K recycling, hence availability of phylloquinone in circulation; and second that genetic variation in GGCX will influence the γ-carboxylation of vitamin K-dependent proteins, as measured by differences in %ucOC and PIVKA-II.
EXPERIMENTAL
Study participants
Free-living men and postmenopausal women, 60–80 y of age, were enrolled in a 3-y, double-blind, randomized, placebo-controlled trial to assess the impact of vitamin K supplementation on age-related bone loss and vascular calcification (21). Subjects in this study were primarily of European descent (n = 387; 93%). In addition, 13 (3.1%) Black, 10 (2.4%) Asian, 4 (1.0%) Hispanic, and 2 (0.5%) American Indian subjects were included in the study. Exclusion criteria included women <5 y postmenopausal or on estrogen replacement; a terminal illness; renal or liver disease; a kidney stone in the past 5 y; current oral anticoagulant use and a history of heart disease or osteoporosis. Of the 452 participants enrolled in this study, 36 subjects were excluded due to inability to obtain DNA and/or written consent for genotyping. The Tufts Medical Center Institutional Review Board approved the protocol.
At the time of the baseline visit prior to randomization, information regarding current medication use, medical history and smoking status was collected. Height and weight were measured while the subjects stood. Body mass index (BMI) was calculated as the weight in kilograms divided by the square of the height in meters. Smoking status was defined as current smokers or non-smokers (Y/N). Current smokers were defined as subjects who reported smoking cigarettes on a regular basis during the previous year. Habitual intakes of phylloquinone were assessed by the Willett semi-quantitative food frequency questionnaire (FFQ), which has been validated for a relative estimate of vitamin K status in population-based studies (22). Information about vitamin and mineral supplement use was documented on medical forms.
Biochemical measurements
Fasting blood samples (>10 h) were collected between the hours of 7:00 and 9:30 am. Plasma and serum samples were stored at −70°C, and were analyzed upon first thaw. Plasma phylloquinone concentrations were determined by reversed-phase HPLC using post-column, solid phase chemical reduction of phylloquinone to its hydroquinone, followed by fluorometric detection (23). The lower limit of detection for phylloquinone with this assay is 0.05 nmol/L. This assay has a 5.3% coefficient of variation for 10 replicates of control plasma. The normal range for plasma phylloquinone concentrations in our laboratory is 0.4–2.6 nmol/L (24).
Osteocalcin was measured in serum using a radioimmunoassay (20). OC uses human OC for standard and tracer and a polyclonal antibody directed to intact human OC. The antibody recognizes intact OC and the large N-terminal-mid molecule fragment. Undercarboxylated OC (ucOC) is a marker of extrahepatic vitamin K status and is determined in this assay as plasma OC that does not bind in vitro to hydroxyapatite. Binding in vitro to hydroxyapatite varies with the amount of total OC in the sample, so ucOC was expressed as the percentage of total OC (%ucOC) to minimize this discrepancy. High %ucOC is indicative of poor vitamin K status. The normal range for %ucOC in our laboratory is 0–50% (unpublished data based on analysis of 2,000 free-living adults). This assay has an 11.8% coefficient of variation for 5 replicates of control plasma.
PIVKA-II (proteins induced by vitamin K absence–factor II), a functional measure of the biological activity of vitamin K in a hepatic vitamin K–dependent protein, was analyzed in citrated plasma with an enzyme-linked immunosorbent assay from American Bioproducts Company (Parsippany, NJ). This assay has a 7.5% coefficient of variation for 10 replicates of control plasma.
Triglyceride-rich lipoproteins are thought to be the primary transporters of phylloquinone (25, 26). Plasma lipid profiles included enzymatic measurement of total cholesterol and triglyceride concentrations (27) using a COBAS Mira (Roche Instruments, Belleville, NJ).
Genotyping
There were 416 study participants who provided written, informed consent for genotyping. Genomic DNA was isolated from blood and purified for PCR analysis using the QIAamp DNA Mini Kit (Qiagen Inc., Chatsworth, CA). Genotyping was carried out using the ABI prism 7900 using single nucleotide polymorphism (SNP) discrimination assays designed by Applied Biosystems (ABI) (Applied Biosystems, Foster City, CA). Taqman probe-based 5′ nuclease assay chemistry unites PCR amplification and signal generation into a single step (28). Accuracy of genotype was tested by introducing 5% dummy duplicates and blank controls.
We selected publicly available SNPs in the vitamin K epoxide reductase (VKORC1) and γ-glutamyl carboxylase (GGCX) genes from the National Center for Biotechnology Information (NCBI) website. SNPs that had previously been published in the literature or had a minor allele frequency greater than 20% were considered for genotyping. SNPs within putative transcription factor binding sites were also considered.
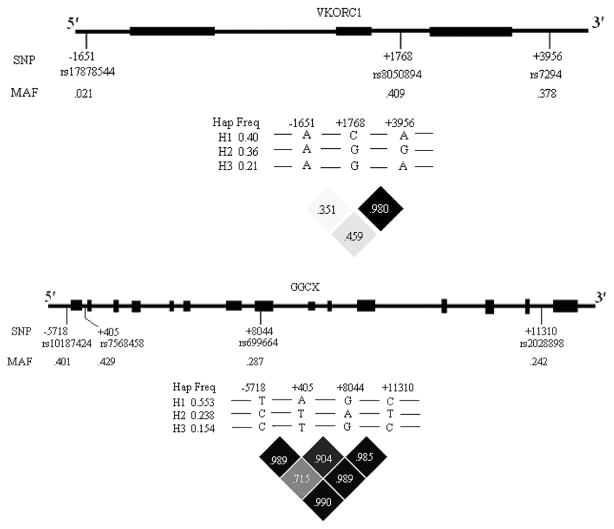
VKORC1 is located on chromosome 16p11.2 and spans about 5 kbp (29) (Fig. 1). The VKORC1 SNPs genotyped for this study include dbSNP rs17878544 (−1651A>G) located upstream −1651 relative to the mRNA start site (GenBank accession number AY587020); two intron 1 SNPs dbSNP rs2884737 (723T>G) and dbSNP rs17708472 (924G>A); dbSNP rs8050894 (1768G>C) located in intron 2; and dbSNP rs7294 (3956G>A) located in the 3′-UTR.
Fig. 1.
Scheme of the genes encoding VKORC1 and GGCX. Black boxes indicate exons; solid black lines indicate introns; single nucleotide polymorphisms (dbSNP) and minor allele frequency are shown. LD and common haplotypes across each gene are also shown. LD values represent Lewontin’s D′.
GGCX is located on chromosome 2p12 and spans approximately 13 kbp (30). (Fig. 1) The GGCX SNPs genotyped for this study include dbSNP rs10187424 (−5718C>T) located upstream −5718 relative to the mRNA start site; dbSNP rs7568458 (405A>T) located in intron 1; dbSNP rs699664 (8044A>G), a non-synonymous (Arg>Gly) polymorphism located in exon 8 and dbSNP rs2028898 (11310C>T) located in intron 14. PCR conditions and primers and probes used for each assay are listed in the supplementary table available online.
Statistical analysis
The genotype frequencies for all markers were evaluated for Hardy-Weinberg Equilibrium (HWE) using the χ2 test. Two SNPs in intron 1 of the VKORC1 gene (rs2884737 and rs17708472) were not in HWE and were therefore excluded from further analysis. All other SNPs were in HWE (p>0.05). Linkage disequilibrium (LD) between each pair of SNPs within each gene was evaluated using Lewontin’s D′ (31). Haplotypes were inferred by the EM-algorithm using the Helixtree software package (Golden Helix, Inc., Boseman, MT).
Analyses were first conducted separately for different ethnic groups. As magnitude and direction of associations with genotypes was similar in the ethnic groups (data not shown), we combined subjects into a single analysis and controlled for ethnicity. To evaluate the relationships between biochemical markers of vitamin K status and polymorphisms in the VKORC1 and GGCX genes, we used analysis of covariance (ANCOVA), while adjusting for age, gender, plasma triglycerides, dietary phylloquinone, BMI, smoking status, and ethnicity, which are known determinants of vitamin K status (32). Plasma phylloquinone and serum %ucOC were the outcome variables of interest and each VKORC1 and GGCX marker was used as a primary independent variable. The SNPs associated with biochemical measures of vitamin K status (p<0.10) in the fully adjusted models were further analyzed assuming dominant, recessive, or additive modes of inheritance. Interaction terms were also explored. Secondary analyses using multivariate regression were conducted with baseline measures of PIVKA-II.
To address the skewed distribution of plasma phylloquinone concentrations, logarithmic transformation was applied prior to analysis, and all results are reported as geometric means. We also performed haplotype analyses evaluating the association of plasma phylloquinone and serum %ucOC with the combination of the VKORC1 and GGCX markers in LD using Helixtree.
Statistical analysis was performed in SPSS for Windows (v. 14, Chicago, IL). Tests with p-value ≤0.05 were considered statistically significant.
RESULTS
Characteristics of the 245 women and 171 men in the present study are presented in Table 1. The mean age of the subjects was 68±5.5 y. Although subjects were excluded from this study for major health problems, such as liver, renal, or heart disease, a mean BMI of 28 kg/m2 suggests that a large number were overweight or obese. Gender-specific differences were detected in total, HDL and LDL cholesterol. Plasma phylloquinone concentrations were not significantly different between men and women; however, women had a significantly higher %ucOC than men (Table 1). There were no statistically significant interactions between SNPs and gender for models with %ucOC as the dependent variable, and the genotype effects on %ucOC showed similar trends for men and women. Therefore, gender specific analyses were not conducted for this variable.
Table 1.
Sample characteristics of subjects at baseline.
| Men (n = 171) | Women (n = 245) | p | |
|---|---|---|---|
| Age, y | 68.9±0.4 | 67.6±0.3 | 0.022 |
| Plasma phylloquinone, nmol/L | 1.2±0.1 | 1.1±0.1 | 0.403 |
| Serum ucOC, % | 37±1.2 | 41±1.1 | 0.009 |
| Dietary phylloquinone, μg | 151±6 | 198±8 | <0.001 |
| BMI, kg/m2 | 28.0±0.3 | 27.9±0.3 | 0.843 |
| Total cholesterol, mg/dL | 188.9±2.5 | 215.1±2.4 | <0.001 |
| HDL cholesterol, mg/dL | 50.6±0.9 | 61.2±0.9 | <0.001 |
| LDL cholesterol, mg/dL | 114.8±2.2 | 131.6±1.9 | <0.001 |
| Triglycerides, mg/dL | 118.8±5.7 | 112.8±4.1 | 0.381 |
| PIVKA-II, ng/mL | 2.43±0.06 | 2.44±0.06 | 0.883 |
| Current smoking, % | 4.9 | 7.0 | 0.617 |
Values are presented as mean±SE unless otherwise noted.
Continuous traits were compared using the t-test; smoking status was compared using the χ2 test.
Determinants of vitamin K status, including age, gender, plasma triglyceride, BMI and smoking status, were not significantly different in participants genotyped for this study as compared to those for whom there were no genotype data available (n = 36; data not shown). However, mean self-reported dietary phylloquinone intake for participants not analyzed was 141 μg/d compared to 179 μg/d reported by participants in the study (p = 0.046). The %ucOC was also higher in the participants not analyzed compared to the participants included in the study (47.5% versus 40.1%, respectively; p = 0.01). Baseline outcomes of participants with genotype data are presented in Table 2.
Table 2.
Baseline measures of plasma phylloquinone and serum %ucOC by VKORC1 and GGCX genotype.
| Gene SNP | Allele | n (%) | Plasma phylloquinone1,2, nmol/L | p | Serum ucOC1, % | p |
|---|---|---|---|---|---|---|
| VKORC1 | AA | 398 (96) | 0.64 (0.56–0.73) | 40±1 | ||
| rs17878544 | AG | 18 (4) | 0.61 (0.33–1.13) | 0.876 | 40±4 | 0.966 |
| VKORC1 | CC | 154 (37) | 0.59 (0.48–0.72) | 42±1 | ||
| rs8050894 | CG | 183 (44) | 0.60 (0.50–0.72) | 41±1 | ||
| GG | 79 (19) | 0.87 (0.65–1.15) | 0.0203 | 37±2 | 0.0433 | |
| VKORC1 | GG | 160 (38) | 0.76 (0.62–0.92) | 40±1 | ||
| rs7294 | AG | 197 (47) | 0.57 (0.48–0.69) | 40±1 | ||
| AA | 59 (14) | 0.57 (0.41–0.80) | 0.0354 | 42±2 | 0.792 | |
| GGCX | TT | 150 (36) | 0.56 (0.46–0.69) | 42±1 | ||
| rs10187424 | CT | 198 (48) | 0.67 (0.56–0.81) | 38±1 | ||
| CC | 68 (16) | 0.73 (0.54–0.99) | 0.268 | 43±2 | 0.018 | |
| GGCX | AA | 137 (33) | 0.54 (0.44–0.67) | 42±1 | ||
| rs7568458 | AT | 201 (48) | 0.69 (0.57–0.82) | 38±1 | ||
| TT | 78 (19) | 0.71 (0.54–0.94) | 0.192 | 43±2 | 0.012 | |
| GGCX | GG | 212 (51) | 0.60 (0.50–0.71) | 41±1 | ||
| rs699664 | GA | 169 (40) | 0.64 (0.53–0.71) | 40±1 | ||
| AA | 35 (8) | 0.95 (0.62–1.45) | 0.134 | 39±2 | 0.659 | |
| GGCX | CC | 235 (56) | 0.59 (0.50–0.69) | 40±1 | ||
| rs2028898 | CT | 160 (38) | 0.70 (0.57–0.85) | 40±1 | ||
| TT | 21 (5) | 0.89 (0.51–1.53) | 0.198 | 42±4 | 0.860 |
Adjusted for age, gender, BMI, smoking, plasma triglycerides, dietary phylloquinone, ethnicity.
Presented as geometric mean (95% CI).
Recessive model for G allele.
Dominant model for A allele.
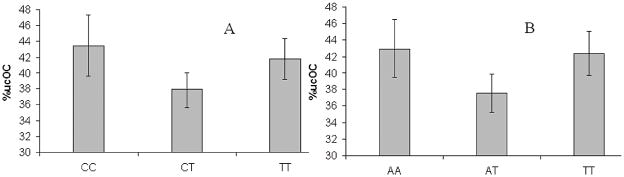
VKORC1 SNPs rs8050894 and rs7294, separated by about 2 kb and in LD with each other (D′ = 0.98), showed significant associations with baseline plasma phylloquinone concentrations. The minor allele of rs8050894 (G) was associated with higher concentrations of plasma phylloquinone whereas the minor allele of rs7294 (A) was associated with lower concentrations of phylloquinone. Subjects homozygous for rs8050894 GG had significantly higher plasma phylloquinone concentrations compared to those with the CC or GC genotype (recessive model for the G allele) (geometric means: 0.874±0.092 versus 0.598±0.044; p = 0.020) (Fig. 2). On the other hand, carriers of rs7294 A (dominant model for the A allele) had significantly lower plasma phylloquinone concentrations (geometric means: 0.579±0.045 versus 0.762±0.057; p = 0.035) (Fig. 2). In addition, recessive modeling of VKORC1 rs8050894 showed a significant association with %ucOC. Homozygous carriers of the G allele had significantly lower %ucOC compared to those with the CC or GC genotype (36.8±1.9 versus 41.1±0.9; p = 0.043) (Fig. 2). These associations remained significant after adjusting for covariates that may influence vitamin K status, including triglycerides, smoking status, gender, age, ethnicity, BMI, and dietary phylloquinone. Significant association between VKORC1 rs8058094 and both phenotypes of interest is consistent with the fact that %ucOC is modestly, but significantly, inversely related to plasma phylloquinone (Spearman correlation −0.193, p<0.001).
Fig. 2.
Histograms of mean plasma phylloquinone concentrations and %ucOC for VKORC1 SNPs at baseline. Histograms A and B show the recessive model for the G allele of VKORC1 rs8050894 for plasma phylloquinone (p = 0.02) and %ucOC (p = 0.043), respectively. Histogram C shows dominant modeling for the A allele of VKORC1 rs7294 and plasma phylloquinone (p = 0.035). Plasma phylloquinone is reported as the geometric mean.
Statistically significant fully adjusted associations were found with GGCX rs10187424 and rs7568458 and %ucOC (p = 0.018 and 0.012, respectively). These SNPs, separated by about 6 kb, are in linkage equilibrium (D′ = 0.98). Heterozygous carriers of these SNPs had significantly lower %ucOC relative to either homozygous group (Fig. 3). No significant differences within GGCX genotype group were found for plasma phylloquinone.
Fig. 3.
Histograms showing mean %ucOC values for GGCX SNPs at baseline. GGCX SNPs rs10187424 (p = 0.018) (Histogram A) and rs7568458 (p = 0.012) (Histogram B).
No significant associations were found in secondary analyses of VKORC1 and GGCX polymorphisms and PIVKA-II (Table 3).
Table 3.
Baseline measures of plasma PIVKA-II by VKORC1 and GGCX genotype.
| Gene SNP | Allele | n (%) | PIVKA-II1, ng/mL | p |
|---|---|---|---|---|
| VKORC1 rs17878544 | AA | 398 (96) | 2.19 (1.85–2.55) | |
| AG | 18 (4) | 1.89 (1.36–2.43) | 0.233 | |
| VKORC1 rs8050894 | CC | 152 (37) | 2.07 (1.70–2.45) | |
| CG | 180 (44) | 2.26 (1.90–2.62) | ||
| GG | 77 (19) | 2.07 (1.70–2.43) | 0.105 | |
| VKORC1 rs7294 | GG | 156 (38) | 2.21 (1.84–2.50) | |
| AG | 197 (47) | 2.15 (1.79–2.50) | ||
| AA | 58 (14) | 1.99 (1.59–2.39) | 0.311 | |
| GGCX rs10187424 | TT | 150 (36) | 2.13 (1.76–2.49) | |
| CT | 198 (48) | 2.17 (1.81–2.53) | ||
| CC | 68 (16) | 2.10 (1.71–2.49) | 0.793 | |
| GGCX rs7568458 | AA | 137 (33) | 2.10 (1.75–2.47) | |
| AT | 201 (48) | 2.18 (1.83–2.54) | ||
| TT | 78 (19) | 2.11 (1.71–2.49) | 0.666 | |
| GGCX rs699664 | GG | 212 (51) | 2.07 (1.72–2.43) | |
| GA | 169 (40) | 2.17 (1.80–2.54) | ||
| AA | 35 (8) | 2.28 (1.86–2.71) | 0.329 | |
| GGCX rs2028898 | CC | 235 (56) | 2.10 (1.75–2.45) | |
| CT | 160 (38) | 2.17 (1.80–2.54) | ||
| TT | 21 (5) | 2.26 (1.78–2.74) | 0.583 |
Adjusted for age, gender, BMI, smoking, plasma triglycerides, dietary phylloquinone, ethnicity, and presented as mean (95% CI).
To further explore the associations between VKORC1 and GGCX SNPs and baseline and 3-y measures of vitamin K status, we performed haplotype analyses. In haplotype analyses of VKORC1, we identified three common haplotypes (frequency>5%): Hap1-rs17878544 A-rs8050894 C-rs7294 A (frequency 40%); Hap2-rs17878544 A-rs8050894 G-rs7294 G (frequency 36%), and Hap3-rs17878544 A-rs8050894 G-rs7294 A (frequency 21%) (Fig. 1). In haplotype analysis of GGCX, we identified three common haplotypes (frequency>5%): Hap1-rs10187424 T-rs7568458 A-rs699664 G-rs2028898 C (frequency 55%); Hap2-rs10187424 C-rs7568458 T-rs699664 A-rs2028898 T (frequency 24%); Hap3-rs10187424 C-rs7568458 T-rs699664 G-rs2028898 C (frequency 15%) (Fig. 1). However, VKORC1 and GGCX haplotype analyses did not increase the variance explained by the models relative to analyses of single SNPs.
DISCUSSION
In the present study, genetic variation in VKORC1 was associated with variation in plasma phylloquinone concentrations. Furthermore, genetic variations in VKORC1 and GGCX were associated with variation in the carboxylation of osteocalcin, a vitamin K-dependent protein in bone.
The availability of the reduced form of vitamin K for the γ-carboxylation of vitamin K-dependent clotting proteins is vital for the proper functioning of the blood coagulation cascade. Warfarin acts by interfering with the vitamin K cycle, specifically by decreasing the activity of VKORC1 (33). Of the VKORC1 polymorphisms examined in this study, homozygous carriers of the minor allele of rs8050894 (G) had significantly higher concentrations of plasma phylloquinone than carriers of the C allele. Consistent with this observation, VKORC1 rs8050894 GG homozygotes had a marginally, although significantly, lower %ucOC (i.e. greater carboxylation) than carriers of the C allele. This suggests adequate availability of reduced substrate for carboxylation in extrahepatic tissues. Similarly, homozygous carriers of the major allele of rs7294 (G) were significantly associated with higher concentrations of plasma phylloquinone, but not %ucOC, compared to carriers of the A allele. The minor allele of VKORC1 rs8050894 (G) and the major allele of VKORC1 rs7294 (G) are linked and have been reported to be part of haplotype sequences that are associated with decreased warfarin dose requirements (18, 34). It was an unexpected finding that a haplotype sequence associated with higher plasma phylloquinone concentrations and lower %ucOC would be associated with lower warfarin dose requirements. However, it has been shown that VKORC1 is less active in extrahepatic tissues compared to the liver where the coagulation proteins are synthesized (35), and the influence of polymorphisms in this enzyme may also be tissue specific. We were unable to detect a genotype effect on baseline measures of PIVKA-II, but there is a narrow range of PIVKA-II measured among individuals with normal coagulation (36). Therefore, it is not unexpected that we were unable to detect a genotype effect on PIVKA-II in this population, whereas an effect of VKORC1 genetic variation on carboxylation of hepatic proteins may be more pronounced in an individual undergoing anticoagulant therapy.
A role for GGCX genetic variation in the interindividual variability in warfarin dose requirement has been suggested based on data from several studies. A recent study in Japanese patients indicated an association with the number of microsatellite repeats (CAA)n in intron 6 of the GGCX gene and mean warfarin daily dose requirements (15). Others have found small, but significant associations between GGCX SNPs and warfarin dose requirements (11, 14). The associations of these polymorphisms with warfarin dose requirements are thought to be due to alterations in enzyme activity. Decreased GGCX activity would lead to less fully carboxylated, and thus less functional, vitamin K-dependent proteins. It is presumed that the functionality of the vitamin K-dependent coagulation proteins would influence warfarin dose requirements in order to achieve stable oral anticoagulation. We examined the association of GGCX SNPs with %ucOC, a measure of vitamin K-dependent carboxylation in extra-hepatic proteins, and PIVKA-II, a measure of carboxylation of prothrombin. GGCX rs10187424 and rs7568458 showed significant associations with baseline measures of %ucOC. These results revealed lower mean %ucOC in the heterozygous group for both SNPs than in either homozygous group. This phenomenon, known as heterosis, has been widely discussed in the literature (for review, see Comings and MacMurray (37)). There are a number of possible explanations for this result, including an intermediate or wider range of expression levels occurring in heterozygote individuals. The GGCX SNPs we examined were not significantly associated with plasma phylloquinone concentrations, and they remained highly significantly associated with %ucOC when plasma phylloquinone was added to the statistical model. This suggests that the association of these GGCX SNPs and %ucOC is not due to availability of vitamin K as a cofactor, but may indicate a direct influence on the γ-carboxylation of osteocalcin. These findings are consistent with a recent report that a functional SNP in the GGCX (Arg325Gln) was associated with lower forearm BMD among women older than 75 y compared to the more common variant (325-Gln), which the authors attribute to the lower carboxylase activity associated with Arg325Gln (38). In contrast, we were unable to detect an association between GGCX SNPs and PIVKA-II, most likely due to the narrow range in serum PIVKA-II concentrations within participants in this study. The assay for %ucOC is sensitive to both dietary phylloquinone restriction and supplementation, whereas the PIVKA-II assay only responds to dietary restriction (36). The participants in this study had self-reported dietary phylloquinone intakes that were higher than the average intakes for older adults (32), which would have limited our ability to detect differences in PIVKA-II among the individual SNPs.
The mixed ethnicity of the study participants is a limitation of this study. Our subject population was predominately of European descent, although other ethnic groups were also represented in accordance with the racial and ethnic composition of older adults in eastern Massachusetts at the time of participant recruitment (U.S. Census Bureau. Release date 09-15-99) (38). Recent studies suggest that the majority of genes studied in more than one ethnic group, and found to be significant in at least one group, have similar effects across ethnic groups (39, 40). In our analysis we pooled data across ethnic groups to improve statistical power as compared to analyses restricted to one ethnicity. Further study will be needed to replicate our findings in different populations.
Previous studies show that the large inter-individual variation in warfarin dose requirement has a strong genetic component. In particular, genetic variation in VKORC1 and GGCX, enzymes involved in the vitamin K cycle, have been shown to influence warfarin sensitivity. However, the biologic mechanisms by which variants in the VKORC1 and GGCX genes are associated with inter-individual variation in warfarin dose requirement are not known. As hypothesized, we found that VKORC1 polymorphisms are associated with plasma vitamin K concentrations in a cohort of primarily European descent. We also showed that GGCX polymorphisms influence the γ-carboxylation of osteocalcin, a surrogate measure of extra-hepatic vitamin K-dependent proteins. Further studies are necessary to fully elucidate the mechanisms behind these associations.
Supplementary Material
Acknowledgments
Based upon work supported by the U.S. Department of Agriculture, Agricultural Research Service under Cooperative Agreement No. 58-1950-7-707, National Institute of Aging (AG14759, HL69272) and NIDDK (T32DK62032). Any opinions, findings, conclusions or recommendations expressed in this publication are those of the authors, and do not necessarily reflect the view of the U.S. Department of Agriculture. The authors would like to thank Caren Gundberg of Yale University for her interpretation of these data.
References
- 1.Smith P, Arnesen H, Holme I. The effect of warfarin on mortality and reinfarction after myocardial infarction. N Engl J Med. 1990;323:147–152. doi: 10.1056/NEJM199007193230302. [DOI] [PubMed] [Google Scholar]
- 2.Berkner KL. The vitamin K-dependent carboxylase. J Nutr. 2000;130:1877–1880. doi: 10.1093/jn/130.8.1877. [DOI] [PubMed] [Google Scholar]
- 3.Hirsh J, Dalen JE, Anderson DR, Poller L, Bussey H, Ansell J, Deykin D, Brandt JT. Oral anticoagulants: mechanism of action, clinical effectiveness, and optimal therapeutic range. Chest. 1998;114:445S–469S. doi: 10.1378/chest.114.5_supplement.445s. [DOI] [PubMed] [Google Scholar]
- 4.Loebstein R, Yonath H, Peleg D, Almog S, Rotenberg M, Lubetsky A, Roitelman J, Harats D, Halkin H, Ezra D. Interindividual variability in sensitivity to warfarin—Nature or nurture? Clin Pharmacol Ther. 2001;70:159–164. doi: 10.1067/mcp.2001.117444. [DOI] [PubMed] [Google Scholar]
- 5.Wadelius M, Sorlin K, Wallerman O, Karlsson J, Yue QY, Magnusson PK, Wadelius C, Melhus H. Warfarin sensitivity related to CYP2C9, CYP3A5, ABCB1 (MDR1) and other factors. Pharmacogenomics J. 2004;4:40–48. doi: 10.1038/sj.tpj.6500220. [DOI] [PubMed] [Google Scholar]
- 6.Hillman MA, Wilke RA, Yale SH, Vidaillet HJ, Caldwell MD, Glurich I, Berg RL, Schmelzer J, Burmester JK. A prospective, randomized pilot trial of model-based warfarin dose initiation using CYP2C9 genotype and clinical data. Clin Med Res. 2005;3:137–145. doi: 10.3121/cmr.3.3.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sconce EA, Khan TI, Wynne HA, Avery P, Monkhouse L, King BP, Wood P, Kesteven P, Daly AK, Kamali F. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005;106:2329–2333. doi: 10.1182/blood-2005-03-1108. [DOI] [PubMed] [Google Scholar]
- 8.Vecsler M, Loebstein R, Almog S, Kurnik D, Goldman B, Halkin H, Gak E. Combined genetic profiles of components and regulators of the vitamin K-dependent gamma-carboxylation system affect individual sensitivity to warfarin. Thromb Haemost. 2006;95:205–211. doi: 10.1160/TH05-06-0446. [DOI] [PubMed] [Google Scholar]
- 9.Michaud V, Vanier MC, Brouillette D, Roy D, Verret L, Noel N, Taillon I, O’Hara G, Gossard D, Champagne M, Goodman K, Renaud Y, Brown A, Phillips M, Ajami AM, Turgeon J. Combination of phenotype assessments and CYP2C9-VKORC1 polymorphisms in the determination of warfarin dose requirements in heavily medicated patients. Clin Pharmacol Ther. 2008;83:740–748. doi: 10.1038/sj.clpt.6100434. [DOI] [PubMed] [Google Scholar]
- 10.Loebstein R, Vecsler M, Kurnik D, Austerweil N, Gak E, Halkin H, Almog S. Common genetic variants of microsomal epoxide hydrolase affect warfarin dose requirements beyond the effect of cytochrome P450 2C9. Clin Pharmacol Ther. 2005;77:365–372. doi: 10.1016/j.clpt.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Kimura R, Miyashita K, Kokubo Y, Akaiwa Y, Otsubo R, Nagatsuka K, Otsuki T, Okayama A, Minematsu K, Naritomi H, Honda S, Tomoike H, Miyata T. Genotypes of vitamin K epoxide reductase, gamma-glutamyl carboxylase, and cytochrome P450 2C9 as determinants of daily warfarin dose in Japanese patients. Thromb Res. 2006;120:181–186. doi: 10.1016/j.thromres.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 12.Kohnke H, Sorlin K, Granath G, Wadelius M. Warfarin dose related to apolipoprotein E (APOE) genotype. Eur J Clin Pharmacol. 2005;61:381–388. doi: 10.1007/s00228-005-0936-3. [DOI] [PubMed] [Google Scholar]
- 13.Sconce EA, Daly AK, Khan TI, Wynne HA, Kamali F. APOE genotype makes a small contribution to warfarin dose requirements. Pharmacogenet Genomics. 2006;16:609–611. doi: 10.1097/01.fpc.0000220567.98089.b5. [DOI] [PubMed] [Google Scholar]
- 14.Wadelius M, Chen LY, Downes K, Ghori J, Hunt S, Eriksson N, Wallerman O, Melhus H, Wadelius C, Bentley D, Deloukas P. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 2005;5:262–270. doi: 10.1038/sj.tpj.6500313. [DOI] [PubMed] [Google Scholar]
- 15.Shikata E, Ieiri I, Ishiguro S, Aono H, Inoue K, Koide T, Ohgi S, Otsubo K. Association of pharmacokinetic (CYP2C9) and pharmacodynamic (factors II, VII, IX, and X; proteins S and C; and gamma-glutamyl carboxylase) gene variants with warfarin sensitivity. Blood. 2004;103:2630–2635. doi: 10.1182/blood-2003-09-3043. [DOI] [PubMed] [Google Scholar]
- 16.Ferland G. The vitamin K-dependent proteins: an update. Nutr Rev. 1998;56:223–230. doi: 10.1111/j.1753-4887.1998.tb01753.x. [DOI] [PubMed] [Google Scholar]
- 17.Presnell SR, Stafford DW. The vitamin K-dependent carboxylase. Thromb Haemost. 2002;87:937–946. [PubMed] [Google Scholar]
- 18.Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, Blough DK, Thummel KE, Veenstra DL, Rettie AE. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 19.Geisen C, Watzka M, Sittinger K, Steffens M, Daugela L, Seifried E, Muller CR, Wienker TF, Oldenburg J. VKORC1 haplotypes and their impact on the inter-individual and inter-ethnical variability of oral anticoagulation. Thromb Haemost. 2005;94:773–779. doi: 10.1160/TH05-04-0290. [DOI] [PubMed] [Google Scholar]
- 20.Gundberg CM, Nieman SD, Abrams S, Rosen H. Vitamin K status and bone health: an analysis of methods for determination of undercarboxylated osteocalcin. J Clin Endocrinol Metab. 1998;83:3258–3266. doi: 10.1210/jcem.83.9.5126. [DOI] [PubMed] [Google Scholar]
- 21.Booth SL, Dallal G, Shea MK, Gundberg CG, Peterson J, Dawson-Hughes B. Effect of vitamin K supplementation on bone loss in elderly men and women. J Clin Endocrinol Metab. 2008;93:1217–1223. doi: 10.1210/jc.2007-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKeown NM, Jacques PF, Gundberg CM, Peterson JW, Tucker KL, Kiel DP, Wilson PWF, Booth SL. Dietary and non-dietary determinants of vitamin K biochemical measures in men and women. J Nutr. 2002;132:1329–1334. doi: 10.1093/jn/132.6.1329. [DOI] [PubMed] [Google Scholar]
- 23.Davidson KW, Sadowski JA. Determination of vitamin K compounds in plasma or serum by high-performance liquid chromatography using postcolumn chemical reduction and fluorimetric detection. Methods Enzymol. 1997;282:408–421. doi: 10.1016/s0076-6879(97)82124-6. [DOI] [PubMed] [Google Scholar]
- 24.Sadowski JA, Hood SJ, Dallal GE, Garry PJ. Phylloquinone in plasma from elderly and young adults: factors influencing its concentration. Am J Clin Nutr. 1989;50:100–108. doi: 10.1093/ajcn/50.1.100. [DOI] [PubMed] [Google Scholar]
- 25.Erkkila AT, Lichtenstein AH, Dolnikowski GG, Grusak MA, Jalbert SM, Aquino KA, Peterson JW, Booth SL. Plasma transport of vitamin K in men using deuterium-labeled collard greens. Metabolism. 2004;53:215–221. doi: 10.1016/j.metabol.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 26.Kohlmeier M, Salomon A, Saupe J, Shearer MJ. Transport of vitamin K to bone in humans. J Nutr. 1996;126:1192S–1196S. doi: 10.1093/jn/126.suppl_4.1192S. [DOI] [PubMed] [Google Scholar]
- 27.McNamara JR, Schaefer EJ. Automated enzymatic standardized lipid analyses for plasma and lipoprotein fractions. Clin Chim Acta. 1987;166:1–8. doi: 10.1016/0009-8981(87)90188-4. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Flood SJ, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 29.Li T, Chang CY, Jin DY, Lin PJ, Khvorova A, Stafford DW. Identification of the gene for vitamin K epoxide reductase. Nature. 2004;427:541–544. doi: 10.1038/nature02254. [DOI] [PubMed] [Google Scholar]
- 30.Kuo WL, Stafford DW, Cruces J, Gray J, Solera J. Chromosomal localization of the gamma-glutamyl carboxylase gene at 2p12. Genomics. 1995;25:746–748. doi: 10.1016/0888-7543(95)80024-g. [DOI] [PubMed] [Google Scholar]
- 31.Lewontin RC. The interaction of selection and linkage. Optimum models. Genetics. 1964;50:757–782. doi: 10.1093/genetics/50.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shea MK, Benjamin EJ, Dupuis J, Massaro JM, Jacques PF, D’Agostino RB, Sr, Ordovas JM, O’Donnell CJ, Dawson-Hughes B, Vasan RS, Booth SL. Genetic and non-genetic correlates of vitamins K and D. Eur J Clin Nutr. 2007 doi: 10.1038/sj.ejcn.1602959. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin DY, Tie JK, Stafford DW. The conversion of vitamin K epoxide to vitamin K quinone and vitamin K quinone to vitamin K hydroquinone uses the same active site cysteines. Biochemistry. 2007;46:7279–7283. doi: 10.1021/bi700527j. [DOI] [PubMed] [Google Scholar]
- 34.D’Andrea G, D’Ambrosio RL, Di Perna P, Chetta M, Santacroce R, Brancaccio V, Grandone E, Margaglione M. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose-anticoagulant effect of warfarin. Blood. 2005;105:645–649. doi: 10.1182/blood-2004-06-2111. [DOI] [PubMed] [Google Scholar]
- 35.de Boer-van den Berg MA, Thijssen HH, Vermeer C. The in vivo effects of acenocoumarol, phenprocoumon and warfarin on vitamin K epoxide reductase and vitamin K-dependent carboxylase in various tissues of the rat. Biochim Biophys Acta. 1986;884:150–157. doi: 10.1016/0304-4165(86)90238-2. [DOI] [PubMed] [Google Scholar]
- 36.Sokoll LJ, Sadowski JA. Comparison of biochemical indexes for assessing vitamin K nutritional status in a healthy adult population. Am J Clin Nutr. 1996;63:566–573. doi: 10.1093/ajcn/63.4.566. [DOI] [PubMed] [Google Scholar]
- 37.Comings DE, MacMurray JP. Molecular heterosis: a review. Mol Genet Metab. 2000;71:19–31. doi: 10.1006/mgme.2000.3015. [DOI] [PubMed] [Google Scholar]
- 38.Population Estimates for Counties by Age Group: July 1, 1999.
- 39.Ioannidis JP, Ntzani EE, Trikalinos TA. ‘Racial’ differences in genetic effects for complex diseases. Nat Genet. 2004;36:1312–1318. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 40.Goldstein DB, Hirschhorn JN. In genetic control of disease, does ‘race’ matter? Nat Genet. 2004;36:1243–1244. doi: 10.1038/ng1204-1243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.