Abstract
Near-infrared spectroscopy (NIRS) is a noninvasive neuroimaging tool for studying evoked hemodynamic changes within the brain. By this technique, changes in the optical absorption of light are recorded over time and are used to estimate the functionally evoked changes in cerebral oxyhemoglobin and deoxyhemoglobin concentrations that result from local cerebral vascular and oxygen metabolic effects during brain activity. Over the past three decades this technology has continued to grow, and today NIRS studies have found many niche applications in the fields of psychology, physiology, and cerebral pathology. The growing popularity of this technique is in part associated with a lower cost and increased portability of NIRS equipment when compared with other imaging modalities, such as functional magnetic resonance imaging and positron emission tomography. With this increasing number of applications, new techniques for the processing, analysis, and interpretation of NIRS data are continually being developed. We review some of the time-series and functional analysis techniques that are currently used in NIRS studies, we describe the practical implementation of various signal processing techniques for removing physiological, instrumental, and motion-artifact noise from optical data, and we discuss the unique aspects of NIRS analysis in comparison with other brain imaging modalities. These methods are described within the context of the MATLAB-based graphical user interface program, HomER, which we have developed and distributed to facilitate the processing of optical functional brain data.
1. Introduction
Over the past three decades, near infrared spectroscopy (NIRS) has been developing as a method for the noninvasive investigation of the changes in cerebral hemodynamic levels associated with brain activity (reviewed in [1–5]). This optically based imaging method offers several additional features that compliment other existing techniques such as functional magnetic resonance imaging (fMRI). In particular, NIRS offers a higher temporal resolution and additional spectroscopic information about both oxyhemoglobin and deoxyhemoglobin changes in comparison with fMRI. However, with the development of this technology, there are also several unique obstacles and technology-specific limitations that must be addressed. While many of the methods that have been developed for the analysis and interpretation of similar neuroimaging experiments, such as fMRI, have been borrowed and applied to optical experiments, additional caveats for dealing with the specific issues of optical technology must also be considered. In particular, motion artifacts, the presence of superficial physiological signals, limited depth sensitivity of optical measurements, and possible cross talk between oxyhemoglobin and deoxyhemoglobin concentrations have created unique challenges for the development of NIRS analysis methods.
In this paper, we review the current state of time-series analysis methods used in functional NIRS to overcome these challenges. We will describe the practical implementation of some of these signal processing and analysis techniques by presenting these methods in the context of HomER, a MATLAB-based graphical user interface program for processing functional brain data from NIRS that we have developed and distributed via the Internet [6].
A. What is NIRS?
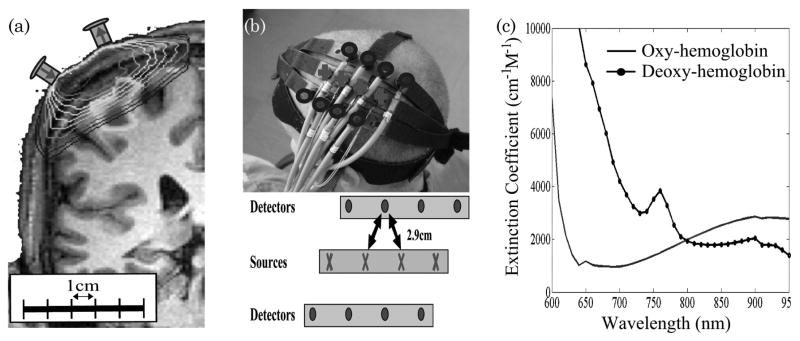
The NIRS technique [7–9] uses low level lights (typically between 5 and 10 mW) within the wavelength region of 650–850 nm light to measure optical absorption changes over time. This is done by noninvasively placing optodes on the surface of the sample (the head); the optodes send and receive light, recording the changes in returning light that has traversed through the sample (i.e., the head). Because of the low optical absorption in biological tissue at these wavelengths, often referred to as the “near-infrared window,” light can penetrate up to several centimeters of tissue as shown in Fig. 1. This optical window allows light to penetrate deep enough to sample the outer 1.5–2 cm of the head through skin and skull and reach the outer approximately 5–10 mm of brain tissue [12–14]. Thus, the NIRS technique can be sensitive to hemoglobin changes in the outermost cortex, allowing optical measurements to be used for imaging brain function. For NIRS brain imaging experiments, a spatially distributed array of laser sources are placed on the surface of the subject’s head and are used to transmit light into the brain. Since the tissue of the adult human head is highly scattering, this light does not travel straight through the tissue as it would for γ- or x-ray beams of light. Instead, near-infrared photons repeatedly change directions as they scatter throughout the tissue. Only a very small fraction of this light (approximately one out of every 109 photons) reaches the surface of the head at specific detector positions, usually 3–4 cm away from the entry source position, where it can be recorded. Thus, the light entering at a source position and exiting the head at a detector position samples a diffuse volume between these positions. Over time, changes in the amount of detected light exiting the surface occur due to changes in optical absorption of the underlying tissue along this path between a source and detector pair; these changes alter the probability of a photon’s reaching the detector before being absorbed. In brain activation studies, these changes are related to the functionally evoked changes in oxyhemoglobin and deoxyhemoglobin concentrations in the brain, which can be estimated from optical measurements of these absorption changes at multiple wavelengths (colors) by virtue of the unique spectroscopic signatures of the two forms of hemoglobin [15]. Figure 1 shows the depth of optical penetration into the adult human head and primary motor cortex region based on simulations of photon propagation [16] through a segmented anatomical model as described in [17].
Fig. 1.
Typical setup for a NIRS experiment. (a) Sensitivity of a NIRS measurement determined by the propagation of light emanated from a source position and recorded by a detector placed several centimeters away. (b) Sample NIRS probe used to measure the primary motor cortex [10]. (c) Absorption spectrum (extinction coefficients) for oxyhemoglobin and deoxyhemoglobin over the range of wavelengths typically used for optical imaging [11].
In 1977, Jöbsis [7] first demonstrated optical measurements of evoked cerebral changes, and, over the past several decades since this publication, NIRS has been applied to study a variety of cerebral regions including the visual [18–20], auditory [21–23], and somatosensory cortices [24,25]. Other areas of investigation have included the motor [26–29], prefrontal or cognitive [30–33], and language systems [34]. In addition to its use in functional brain experiments, NIRS is beginning to be applied to study several clinical problems, in part motivated by the portability and bedside applicability of NIRS instruments. Clinically oriented NIRS applications have addressed the prevention and treatment of seizures [35–40], have addressed Alzheimer’s disease [41–43], and have provided a potential tool to monitor neonate brain status [44] and stroke rehabilitation [45–52]. NIRS has also been applied to psychiatric health concerns such as depression [30,53–57] and schizophrenia [57–62].
The recent growth of this field has been in part the result of the increased availability of instruments. There are currently a number of commercially available optical systems: from Hamamatsu, the NIRO 500 and the Somentics INVOS 3100 systems; NIRx Medical technologies, the Dynot system; Hitachi Medical Corporation, the ETG Optical Topography System system; ISS Inc., the Imagent system; and Techen Inc., the NIRS2 and CW5 systems, to cite a few. This growth may also be due in part to an increased recognition of several advantages of optical imaging that enhance other existing neuroimaging modalities, such as fMRI. In particular, NIRS experiments are less costly to perform than conventional fMRI. Generally, optical instruments are also smaller, more portable, less expensive to purchase, and require little to no scheduled maintenance costs (unlike MRI or positron emission tomography scanners). Additionally, NIRS experimental procedures are conducted in a more experimentally controllable environment than the noisy and confining scanners used in MRI and other neuroimaging methods. Finally, NIRS can tolerate subject motion to a larger extent than fMRI, provided that the NIRS head probe remains fixed to the head of the subject. This has allowed the NIRS technique to more easily study subject populations that have been traditionally difficult to study with fMRI, including infants [23,63,64], children [31,65], and the elderly [43]. This has also allowed NIRS to be used for studies requiring subject motion, for example, to study the effects of exercise [66,67] or posture [68] on cerebral signals.
As the NIRS technology continues to advance, there is an increasing need for the development of standardized processing and analysis tools. In this paper, we review several methods of analysis and signal processing that have been introduced in recent years for dealing with specific issues of NIRS experiments. In the context of the discussion of these methods, we describe the practical implementation of these methods within HomER, a program for functional NIRS data analysis and visualization. HomER, which is an acronym for hemodynamic optically measured evoked response, is a suite of open-source MATLAB (MathWorks, Natick, Massachusetts) programs interfaced together by a front-end graphical user interface. For basic analysis, the HomER program allows the user to calculate hemodynamic changes in oxyhemoglobin and deoxyhemoglobin concentrations from raw light intensity time series acquired at different wavelengths. The program contains an assortment of signal processing tools for more advanced analysis, including bandpass filters to remove instrumental or physiological noise contributions, subject motion correction filters [64], and principal component-based filters to remove systemic physiology [69,70]. HomER also contains various tools for block averaging and linear regression of stimulus epochs for conventional analysis of functional data, as blocked, event-related, and multiple condition experimental designs. Both single subject and group-averaged responses can be estimated with relative statistical analysis. Last, HomER incorporates the tools for basic optical image reconstruction, allowing for both backprojection and tomographic reconstructions of functional data. These latter features allow the users to visualize static images and movies of functional activity.
B. Theory of Optical Imaging
Although they were already introduced in the previous section, here we will define the fundamentals of optical imaging and, in particular, the theory describing the relationships between measured signals at the instrument and underlying physiological quantities of interest, namely, hemoglobin changes. In this paper, we will focus on continuous wave (CW) optical measurements, as these are most often used in brain imaging experiments because of their achievable higher temporal resolution and the lower cost of photon detectors, which allows a greater number of spatially resolved measurements. CW NIRS imaging uses a stable light source, which sends a continuous beam of light into the tissue. This light is monitored as it passes through the sample (e.g., tissue), and the intensity of the detected light is used to determine the amount of optical absorption by the sample. Other forms of NIRS imaging, such as time-domain [71] and frequency-domain [72] methods, use more complex sources of light to measure the phase of the returning light or the temporal distribution of photons after they have migrated through the sample. In a typical CW NIRS brain functional study, a grid of light sources and photon detectors is positioned on the head of a subject and used to noninvasively monitor optical absorption changes in the tissue volume between each source and detector pair, as illustrated in Fig 1. By using diffusion approximations and finite-element or Monte-Carlo-based computational methods [12,13,16,73–76], we can estimate the sensitivity profile for each source–detector pair on the basis of the optical properties of the tissue as shown in Fig 1(a). From the surface of the head, light reaches the outer 5–8 mm of brain tissue after passing through the scalp (0.5–1 cm), skull (0.5–1 cm), and cerebral spinal fluid (0–2 mm) layers. The thickness of these layers, especially skull and cerebral spinal fluid, can vary depending on age, gender, or other intersubject factors. Since, the thickness and composition of these tissue layers influences how light propagates through the head, regional or inter-subject differences may affect the sensitivity of a NIRS measurement to brain activity and may create potential biases in the spatial localization of brain activity [77]. This is one of the drawbacks of noninvasive optical imaging. In recent years, several groups [3,78–81] have introduced methods to account for this by incorporating anatomical volumetric data from MRI or computerized tomography to define the spatial boundaries of the skull and scalp and thus improve knowledge of where the light has traveled within the head. Several Monte Carlo and finite-element codes have been described to use such anatomical information to generate solutions of the optical forward problem and to determine how a change in absorption within a tissue layer projects into a change in detected light intensity [12,13,16,73–76]. For more information about the optical forward problem, the recent review by Gibson et al.[4] offers an introduction to this problem.
During brain activation, increases in blood flow and volume and oxygen metabolism compete to raise and lower the blood oxygen saturation, respectively (see Buxton et al. [82] for a review of the use of vascular biophysical models of these relationships). As the levels of oxyhemoglobin and deoxyhemoglobin change, light is differentially absorbed compared with the rest state (baseline). The change in light absorption, referred to as delta optical density, ΔOD, can be calculated from the normalized changes in the light incident on a detector from a source position. Formally, ΔOD is defined as
| (1a) |
| (1b) |
where Φ̂ (intensity) describes the amount of light that was emitted from a source position (i) that becomes incident on a given detector position (j). Δμabs is the change in the absorption coefficient with respect to the baseline level. These changes are specific to the wavelength of light examined, which is indexed by λ. The spatial integrals in Eqs. (1) are functional path integrals over the volume sampled by the ensemble of possible photon trajectories through the tissue, which in turn is defined by the stochastic diffusion of light in tissue. In general, the detected light intensity is proportional to the amplitude of the voltage signal reported by the photon detectors, assuming that the instability and possible nonlinearity of the photon detectors of the NIRS instrument used can be neglected. By normalizing the intensity measurements to the incident light at the start of the experiment (in practice, usually the mean of the signal over the course of the study is used), one can neglect detector efficiency, detector analog amplification (gain), and the instrument’s absolute incident laser power, since these appear as scaling factors that multiply both the numerator and denominator in Eq. (1b). This is an important attribute of functional imaging by CW NIRS, as this normalization removes the need to calibrate the instrument. However, this normalization also restricts CW NIRS to the determination of evoked changes rather then absolute levels of hemoglobin.
For most optical functional brain measurements, absorption changes due to hemoglobin are assumed to be small and, thus, are thought to not perturb the path of light through the tissue. For a typical vascular change due to brain activation (around 1–5 μM in oxyhemoglobin), the expected change in optical density is 0.1%–1.0% cm−1. In this limit, absorption changes can be expressed as a linear combination of the changes in oxyhemoglobin and deoxyhemoglobin (HbO2 and HbR, respectively) by replacing the path integral in Eq. (1a) with the multiplication by an effective mean path-length term that represents the average path of light traveled through the illuminated region [15,83]. The resulting linear relationship between changes in optical density and changes in the underlying concentration of absorbing species in the sample is referred to as the Beer–Lambert law:
| (2) |
where the variable ε is the wavelength-dependent extinction coefficient for each hemoglobin species. In Eq. (2), Lij is the mean path length traveled by the light from the source i to the detector j. However, since light propagation through biological tissue follows a diffuse rather than a straight path, the Beer–Lambert law must be adjusted for the additional effective distance traveled by light as it scatters through the tissue. Also, since the tissue region where the absorption is presumed to be changing (e.g., the brain) is only a small fraction of the volume that the diffuse light samples, partial volume corrections must also be applied to rescale the magnitude of absorption changes. The combination of path length and partial volume scaling can be incorporated into a term denoted the differential path-length factor (DPF) [15] within the modified Beer–Lambert law (MBLL). Advantages and weakness of this simplified method have been extensively discussed in several papers [15,84,85]. The net absorption change at each wavelength due to changes in hemoglobin concentration is given by the expression
| (3) |
where the term DPF (differential path-length factor) has been added to account for differences between the linear distance between the source and detector pair (Lij) and the true effective mean path length that light travels as is scatters through the tissue. This term has wavelength dependence, since the effective mean path length depends on the scattering properties of the illuminated tissue. To estimate the change in optical density related to brain activity, one must also account for partial volume scaling due to the fact that hemoglobin changes are not uniformly distributed along the illuminated tissue but are limited to a restricted cortical volume. This partial volume effect results in an underestimation of the effective hemoglobin changes in the cortex. The effects of scattering and partial volume can be accounted for by multiplying the linear source–detector separation by additional differential and partial path-length factors [3,84–86]. This scaling factors adjust for the effective path length of light through the region of interest by approximating that all absorption changes occurs within a subset of the total sampled volume (i.e., brain activity is confined to the cortical layers). Knowledge of these factors, which depend on the detailed anatomy of the individual subject’s head, limit the quantitative accuracy of the NIRS method and can introduce systematic biases in the estimate of the evoked signal that came from within the brain.
In most NIRS brain activation studies, multiple wavelength measurements of optical absorption are often converted to changes in oxyhemoglobin and deoxyhemoglobin by solving the series of linear equations posed by the MBLL. Hemoglobin changes can be estimated by a minimum weighted least-squares solution to Eq. (3) from data at two or more wavelengths. This is given by the equation
| (4a) |
where
| (4b) |
The matrix E contains the extinction coefficients for oxyhemoglobin and deoxyhemoglobin at each wavelength measured. In Eqs. (4), the matrix R is defined as the a priori estimate of the covariance of the measurement error. This equation arises from a weighted least-squares cost function, and the effect of normalization by this measurement error term is to precondition the measurements by weighting according to their expected measurement noise levels. For example, the larger the error in a particular single measurement, the less weight it should have in determining the solution to the optical reconstruction of hemoglobin [81]. In particular, the signal-to-noise ratio and measurement errors are likely to vary between measured wavelengths. When the number of wavelengths measured exceeds the number of unknowns, as is the case for three or more wavelengths, there may no longer be a unique solution of the MBLL that simultaneously satisfies all measurements. As shown by Yalavarthy et al. [87] in the context of optical mammography, a more accurate estimate of the two hemoglobin species can be achieved by weighting the multiwavelength measurements according to the errors in each measurement performed by using this approach. This approach is believed to be particularly advantageous in cases where the signal-to-noise ratio differs greatly between measurement pairs, which is often the case with real-world data sets [81].
2. Limitations to the Quantification of Hemoglobin by using Near-Infrared Spectroscopy
The quantification of absolute (e.g., micromolar) hemoglobin changes is dependent on knowledge of the differential path-length and partial volume factors (reviewed in [88,89]). Although the propagation of light through tissue can either be modeled by using light transport theory [13,14,16] or be measured experimentally [83,90], most NIRS experiments do not have access to this information, as this generally requires an additional MRI or computed axial tomography scan to provide anatomical information (e.g., [3,78,91]). Several groups have tabulated values of the differential and partial path-length factors [17,83,86,90]. Nevertheless, uncertainty in these factors will ultimately limit the quantitative ability of NIRS to measure absolute hemoglobin concentration changes because the path traveled by light through cortical tissue varies with the wavelength-dependent scattering properties of the head and head layers [3]. This can give rise to cross-talk errors in the separation of oxyhemoglobin and deoxyhemoglobin estimates [3,86,92] and may be minimized by an optimal choice of wavelength pairs as discussed in [3,86,93].
3. Time-Series Methods in Optical Imaging
Whereas a strength of functional MRI is its spatial resolution (or more accurately, the ability to directly solve the MRI image reconstruction problem as compared with the ill-posed nature of spatial reconstruction in diffuse optical methods), strengths of optical methods are certainly temporal resolution and the ability to measure both hemoglobin species more directly. Sophisticated time-series analysis of functional optical studies is thus very important for interpreting such studies. Although this topic is the focus of the remainder of this report, we will begin by acknowledging that much work is still needed in this field. In particular, the analysis of optical measurements poses several unique challenges.
Because fMRI and optical experiments usually have similar designs and hypotheses, many analysis approaches suited for fMRI have often been applied with little modification to optical data [94]. Indeed, much of the analysis of optical imaging has benefited from similar advances in fMRI. However, because of the different biophysics associated with the optical and fMRI techniques, there are a number of specific issues, limitations, and methods specific to the optical technology. Unlike fMRI analysis, which often draws statistical information from the spatial proximity of measurements (e.g., voxels in the volume images) and temporal compression methods from the assumption of canonical temporal shapes of the evoked response (e.g., the Γ function response [95]), analysis of optical data has generally focused on more traditional time-series methods, including bandpass filtering, temporal smoothing, and linear deconvolution, to preserve temporal information about the evoked functional hemodynamic response while trying to remove specific artifacts in the measurements, such as cardiac pulsation signals. In the following sections, we discuss the analysis of the temporal information in optical measurements and emphasize the differences in comparison with fMRI data analysis.
4. Sources of Noise in Near-Infrared Spectroscopy
There are three distinct sources of noise that characterize NIRS measurements: instrumental noise, experimental errors, and systemic physiological artifacts. These three types of error have different properties and, thus, fit into different parts of a typical optical analysis stream. For example, instrument noise and most experimental errors, such as motion artifacts, are not related to true underlying absorption changes and should be removed prior to conversion of signals into oxyhemoglobin and deoxyhemoglobin via the Beer–Lambert equation [Eq. (3)]. This will avoid propagation of these errors across wavelengths, which would cause additional cross talk in the separation of oxyhemoglobin and deoxyhemoglobin. In contrast, physiological noise arises from biological hemoglobin oscillations. Since these sources of error affect the multiple optical wavelengths in a manner consistent with the Beer–Lambert equation, there is expected to be a partial correlation between wavelengths due to their dependence on a common underlying hemoglobin change. Thus, biological signals are better removed after the MBLL conversion of the raw signals to hemoglobin units, where one can take advantage of the differential contribution to these signals from the arterial and venous vascular compartments.
A. Instrumental Noise
Electrical noise from the computer or other hardware in the instrument or from the surrounding space can create noise in the measurements. In general, such electrical noise is often assumed to have either a uniform frequency spectrum or one that can be measured using an ex vivo phantom. Since NIRS instruments often sample at much higher rates than typical hemodynamic changes, most of the signal power at the higher sampled frequencies is from instrumental noise, which can allow an estimate of the measurement noise properties of the data by assuming a known instrument noise profile (e.g., uniform or white instrument noise). Much of the high frequency of this instrument noise can be separated from physiological signals by low-pass filtering the data. In general, experimental designs can be optimized for the particular NIRS system by adjusting the laser intensities of each color of light and detector gain amplification (if available); external light and electrical interference can reduce instrumental noise to nearly negligible compared with the other sources of noise, particularly physiological noise.
B. Experimental Errors
There are several types of artifact, which we call “experimental errors,” that may also contribute to the errors in the analysis of optical studies. For example, experimental errors may arise as the result of subject motion or noncompliance with the stimulus paradigm. In comparison with functional MRI, in which motion refers to the movement of the subject relative to the MRI scanner, motion artifacts in optical imaging arise from the potential of the optical fibers to move on the head, creating a large jump in the detected light signal. Note that if the optical probe is well secured to the head, the subject may freely move during the experiment, and several groups have used this feature in studies involving movement or subject posture that would be difficult to perform in fMRI, for example, allowing NIRS to be used to study brain function during mobility challenges [96]. As a practical example, we have found that anchoring the optical fibers to an additional place on the subject’s body, for example, by securing these to a backpack or harness rather than allowing the fibers to dangle directly from the subject’s head, can be used to reduce motion artifacts by refocusing any tension in the fiber optics away from the point of contact between the scalp and the fiber. Tightly wrapping the optodes on the subject’s head by using bandages or other wraps to prevent motion artifacts is not recommended, since this can make the head gear less comfortable to wear and thus may promote anxiety and movement by the participant.
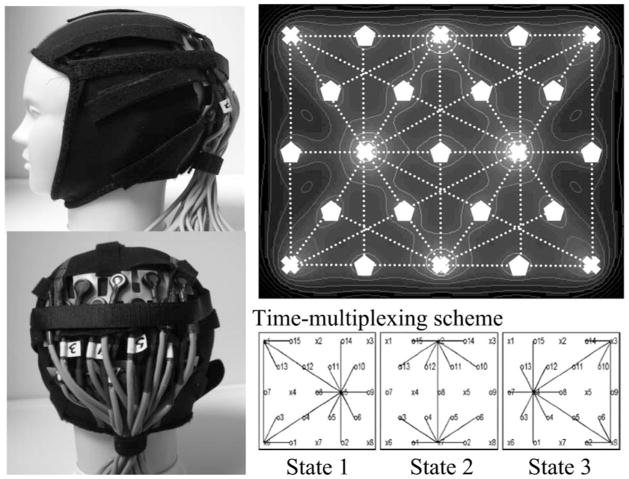
Over the past years, we have worked on several optical probe designs in an attempt to balance the ability to quickly position the probe on the head, to maintain stability and avoid movement of the probe on the head, to provide increased comfort for the subject (which will reduce subject restlessness and movement), and to allow flexibility to locally adjust the optode positions and displace hair from beneath the optodes to make good contact with the skin. In Fig. 2, we demonstrate recent optical head probes. For this probe, our fiber optics are embedded in a plastic strip, which is affixed with Velcro to a neoprene hood. The Velcro can be loosened to allow access to move the hair beneath the optical fibers. This probe was used in studies of visual activation and used a tomographic (overlapping) measurement geometry to obtain better spatial resolution [98].
Fig. 2.
Tomographic optical probe: sample arrangement for a tomographic-style probe [97] used for a study of visual activation. To acquire optical signals from multiple source–detector distances, a time-multiplexing scheme is used in which our system switches between three sets of laser on–off states and detector gains (lower right). In our current system, a complete cycle can be imaged at up to 3 Hz. Overlapping (tomographic) measurements provide more uniform spatial sensitivity and coverage of the underlying brain. The theoretical sensitivity profile for this probe is shown in the upper right-hand panel with contour lines at 5 dB intervals based on a semi-infinite homogeneous (μa = 0.1 cm−1 and μs′ = 10 cm−1) slab geometry.
In general, we recommend that the probe design match the experiment. For example, if the subject is going to move around as part of the experiment (e.g., [96]), the probe needs to be more rigid and secure to the head, which implies that more time may be needed to place the probe and adjust the hair and positioning to obtain adequate signals. In contrast, in other studies quickness of setup may be a more important design parameter, for example, for imaging children who will be less patient to sit through a lengthy setup procedure to place the cap on the head. In addition, particular care must be taken to avoid motions caused by a cued task as part of the experimental procedure, for example, a specific head motion, facial expression, or speech, which can involve facial muscles beneath the straps holding the probe to the head. A chin strap can be used to hold the probe on the head more securely (as shown in Fig. 2); however, in our experience, this may cause additional problems if the task involves speech.
In addition, experimental errors can also be introduced by the design of the functional task. Although in many ways experimental design for optical studies is similar to that used in fMRI (also a hemodynanic based imaging method), there are several additional considerations regarding unique attributes of optical signals. In particular, if filtering and other signal processing methods will be used in the analysis of the optical signals, it is important to anticipate these during the design of the experiment. For example, improper design of the timing of an experimental paradigm can introduce errors associated with numerical instabilities or underdetermination of the equations associated with functional analysis and deconvolution techniques [99–102]. In general, this requires that the timing of stimulus presentation is designed such that either events are spaced widely apart in time, as in slow event-related and most blocked design experiments, or the timing is optimally jittered to reduce collinearity between columns of the experimental design matrix. This procedure is reviewed in [99]. While longer-duration stimulation gives better statistical power because the response builds to a higher level, this type of design is suboptimal for addressing hypotheses based on the timing of the transients of the evoked hemodynamic response especially since temporal resolution is a potential advantage of optical methods. If a rapid event-related design is used, it is also important to consider any linearity assumptions of the evoked hemodynamic response that must go into regression analysis, such as the assumption that repeated evoked responses will add linearly. For event-related stimulus designs, the hemodynamic responses may be approximated by this linear summation, provided that the interstimulus interval is longer than around 4 s (e.g., [103]). Last, the reproducibility of results can be affected by a difference in the positioning of the NIRS probe on the subject’s head, which can introduce partial-volume and path-length differences in single-subject longitudinal studies [104]. Vascular and anatomical differences between individuals affect the comparisons of results across multiple subjects [105,10]. These sources of error often translate to loss of data and need to be dealt with before data acquisition. Operator expertise, careful design of probes, and optimized experimental paradigms can reduce many of these experimental sources of error. Some, like motion artifacts, can also be dealt after data acquisition by signal processing techniques, as will be discussed below.
C. Physiological Fluctuations
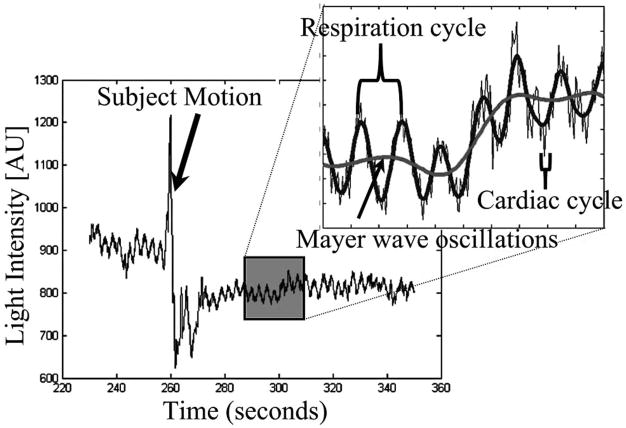
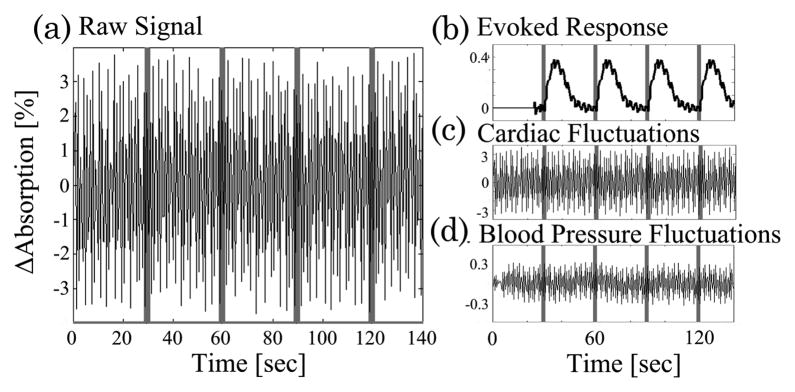
Since diffuse optical light spreads as the light penetrates deeper into the tissue, NIRS measurements are much more sensitive to absorption changes in the superficial layers compared with at the ~2 cm depth needed for functional measurements. Thus, a significant source of noise in optical studies is due to physiological signals in these superficial layers. These signals include systemic physiological hemodynamic fluctuations such as cardiac pulsation, respiratory signals, and blood pressure changes, which are present in the scalp and underlying cerebral tissue [69,106,107]. Figure 3 demonstrates such cardiac, respiratory, and blood pressure (Mayer wave) oscillations seen in a representative NIRS measurement. Although often these physiological fluctuations are considered to be noise because they interfere with the estimate of evoked functional responses, the sensitivity of NIRS to these physiological fluctuations in other applications is not necessarily a negative attribute. Recently, several groups have begun to use NIRS to explore normal and abnormal vascular physiology, such as vasomotion or autonomic regulation [69,108–110] in healthy subjects and stroke patients. We will defer discussion of these signals until later in this paper when we further discuss specific filtering methods.
Fig. 3.
Physiological fluctuations in optical signals: physiology fluctuations are generally the dominant source of noise in NIRS measurements because of the superficial sensitivity of the technique. This figure illustrates cardiac, respiratory, and blood pressure (Mayer wave) oscillations recorded during a resting period for the subject. The data also demonstrate a motion artifact, where the probes shifted during recording and generated a large perturbation of the signal intensity.
5. Reduction of Noise in Optical Signals
A. Methods for Motion Correction
As previously mentioned, there are two points in the analysis of optical data where noise can be removed, namely, before and after the conversion to physiological hemoglobin concentrations via the MBLL. We believe that this distinction is important, since many of the analysis methods that will be discussed in the following sections are designed to take advantage of a particular attribute of the noise source. For example, if motion artifacts are not removed prior to the signal normalization to obtain optical density [Eqs. (1)], then the magnitude of the optical density changes will be biased and cross talk may be introduced in the estimates of oxyhemoglobin and deoxyhemoglobin via the Beer–Lambert equation. While the best opportunities to deal with such motion artifacts occur during the data acquisition and reflect subject compliance, experimental design, and the experimenter’s expertise in positioning and securing the NIRS probe, motion artifacts will undoubtedly still appear in certain studies. Motion is particularly common in dealing with infant or animal subjects, where subject compliance is more difficult to control. Probe design, as discussed in the previous section, plays an important role in reducing but often not completely eliminating these errors. In general, motion artifacts are represented by very large jumps in the amplitudes of optical signals as shown in Fig. 3. Often these are several orders of magnitude larger then the measurement variance expected from physiological sources of error within the head, and this feature can be used to help detect regions affected by motion artifacts. Aside from qualitative visual inspection, which is often sufficient to identify large motion errors, it is possible to detect motion by using more defined criteria such as the analysis of studentized residuals. Studentized signals are a method of outlier detection based on the number of standard deviations between the residual error of a single measurement and the mean signal; for example, the studentized residual is defined by (y − ŷ)/σ, where y is the measurement, ŷ is the model of the data (which can be generated by filtering or taking the mean of the data or can be the model generated by linear regression to find the evoked hemodynamic signals), and σ is the unbiased estimate of root-mean-squared error of the measurements. This method can be used to identify extreme outliers by objective criteria, say, rejecting points that are greater then 3 standard deviations from the mean. Outlier detection can be preformed on either direct signals or on the temporal derivative of the signal, providing a means to specifically reject large, rapid jumps in the signal that are due to motion. Once identified, the temporal regions affected by motion artifacts can be excluded from further analysis by either removing entire stimulus blocks within the region, by removing columns from the linear regression model used in the functional analysis, or by excluding the entire data file or subject. Note, however, that since subject movement can physically move the NIRS probe or change the optode couplings to the head, the signal levels before and after the motion may need to be corrected. This can be a major issue, since it requires a renormalization of the measured signal or in a worse case could mean that measurements represent a different brain region. In some cases, this may require the data to be analyzed in a piecewise fashion to properly compute normalized intensity in the expression for optical density changes [Eqs. (1)] before and after the motion.
B. Eigenvector-Based Reduction of Motion Artifacts
Since motion artifacts arise from the physical displacement of the optical probe from the surface of the subject’s head, motion artifacts often exhibit statistically high covariance across multiple source–detector pairs and across optical wavelengths. This feature may be used to remove the motion artifact by using filtering methods based on principal component analysis (PCA). PCA (or truncated singular value decomposition) filters are based on the subtraction of the eigenvector components exhibiting high (or low) covariance in the data. This approach was first demonstrated by Wilcox and co-workers and was shown to reduce motion artifacts in a study of infant brain function [64]. The principal components of the data covariance are calculated by using a standard procedure for singular value decomposition (SVD). For removing motion, the covariance of the data should be calculated from all source–detector measurement pairs and all wavelengths and thus represent should the covariance across the entire set of measurements. The spatial eigenvectors of this covariance matrix are defined by the singular value factorization
| (5a) |
where
| (5b) |
Since motion artifacts will typically have a highly covariant structure, motion is generally captured in the first (strongest) eigenvector components. This allows these artifacts to be subtracted from the time series of the optical density measurements by using linear regression. The filtered time course of the data is given by
| (6a) |
where the PCA filter component is defined by
| (6b) |
with the eigenvalue weights (wi) given by
| (6c) |
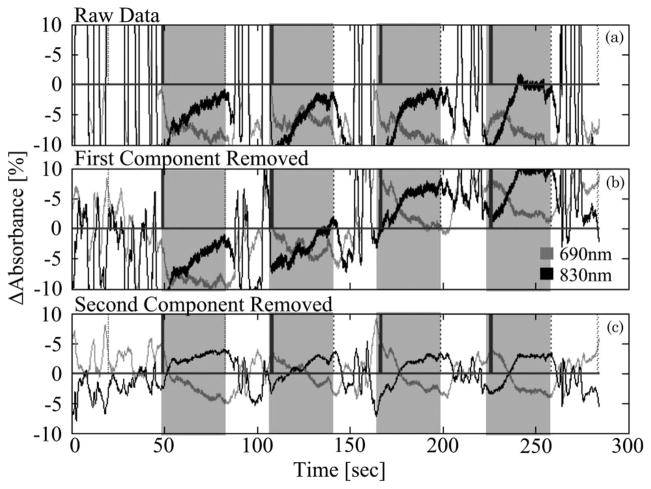
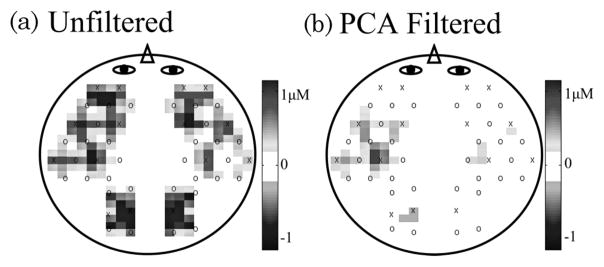
w1 ≥ w2 ≥ wN, and i is summed from 1 to the number of components to be removed. The vectors σi and υi represent the ith columns of matrices Σ and V, respectively. For motion correction, typically as many as 1 or 2 components may be removed, depending on the total number of measurements in the data and the structure of the covariance matrix (e.g., [64]). An example of this PCA filter is demonstrated in Fig. 4. Here we show an example data set taken as part of a NIRS study on object recognition in infants as described in [64]. The raw data, shown in Fig. 4(a), demonstrate very strong motion artifacts, which dominate the optical signals. In addition, these motion artifacts were covariant across most of the NIRS probe, making analysis of this data virtually impossible. When this PCA filter is used to remove the first and second principal components, the motion artifact is greatly reduced [Figs. 4(b) and 4(c)].
Fig. 4.
Example of motion artifact removal by principal component (PCA) filtering. The raw data shown in (a) is experimental data measured in the inferior temporal cortex in an infant presented with an object recognition task [64]. The data were dominated by several motion artifacts that resulted from the movement of the probe on the head’s surface. These motion artifacts were highly covariant across the entire probe. (b) and (c) show the filtered data after the removal of the first and second principal components. After reduction of the covariance in the data by the removal of these components, the motion artifacts were greatly reduced. The gray shading indicates the presentation of stimuli.
This motion correction algorithm exploits the property that a motion artifact has an intense and correlated effect over a large area of the NIRS probe. This is a general characteristic of motion, but does not entirely encompass all motion artifacts. This algorithm works best for reducing motion that affects areas much larger than the functional region. For a small number of source–detector pairs or a probe with a spatial coverage comparable with the extent of the functional region, PCA can negatively affect the estimate of the hemodynamic response. If used too strongly, PCA filters can remove brain activation signals, which typically also exhibit spatial covariance. Statistical metrics, such as the goodness-of-fit metric of the model (e.g., a partial F test [111]) or an effects test (e.g., T test [95]) for the brain activation signal can help to decide the optimal number of principal components to remove.
Alternative approaches to motion correction including wavelet filtering [112], Wiener filtering [113], and autoregression models have been proposed. These methods work on the basis of exploiting temporal signatures of the motion artifact. In comparison, PCA, as described above, is a filter based on removing spatial covariance structures.
C. Reduction of Physiological Interference
After optical signals have been checked and corrected for motion artifacts, there will still be several other sources of noise in the data. Physiological systemic noise arising from the cardiac cycle, respiration, systemic blood pressure, and Mayer wave fluctuations are a common component of signal interference in NIRS measurements [3,69,70,106,107,109,114]. In fact, the absence of cardiac and other fluctuations can be a sign that data quality is low (perhaps because of poor coupling of the optical probe with the head) and often serve as a quality assurance test to indicate that functional analysis will not be possible. Background physiological signals have been explored and may in fact reveal interesting details about vascular physiology [69]. However, in most studies, attempts are made to reduce background physiological signals. Generally, these techniques are designed around specific characteristics of physiology based on either (i) known approximate frequency content of signals, (ii) the spatial covariance of physiology, or (iii) the ability to remotely measure physiological signals from areas away from the functional activation signal either from a measurement pair in a different brain region, a measurement pair with a short separation distance, or direct measurements of physiology at extracranial locations. Several of these techniques will be discussed in the following sections.
D. Bandpass Filtering Techniques
Bandpass filtering, which removes specific frequency content from the measured signals, can provide a method for effectively removing many sources of noise in the data. Low-pass filters can be used to remove high-frequency instrument noise and the fast cardiac oscillations. High-pass filters, which remove slow, low-frequency content, can be used to remove physiological noise such as blood pressure oscillations, which are generally found between 0.08–0.12 Hz, or cardiac pulsation (around 1 Hz, 60 × beats/min). Although bandpass filtering can be used to remove extreme low- and high-frequency noise, these approaches must be used carefully to avoid accidentally removing frequencies associated with the functional hemodynamic response. Thus, bandpass filtering can generally not be used to remove respiratory signals and some components of the blood pressure signal, since these contain frequency bands that overlap with the typical content of the hemodynamic response. As a result, to remove these specific effects, bandpass filtering cannot be used without also compromising the stimulus response frequency bands. Alternatives to conventional bandpass filtering, such as the use of wavelets [115,116] or adaptive filtering [118,119], have also been used in optical and fMRI to remove systemic physiology from specific frequency bands while preserving the evoked response.
E. Eigenvector-Based Reduction of Spatial Covariance
Often a feature of systemic physiology is its spatial covariance. For example, signal changes due to the breathing cycle fluctuate across the brain in a specific and repeated spatial pattern. Similar to its use for motion correction, PCA may also be used to remove physiological noise by taking advantage of this spatial structure. PCA-based physiological filtering methods have been previously described by Zhang and co-workers [70]. Similar to motion artifacts, systemic fluctuations are covariant between the NIRS measurements from different regions of the head. Thus, the reduction of this covariance can be used to filter physiology of systemic origin from the measurements. In comparison with the motion correction, for this PCA filter, the spatial covariance in Eqs. (5) and (6) is calculated from a separate baseline (rest state) data set, rather than from the functional data themselves. By using a separate baseline time course to derive the spatial structure associated with background systemic physiology, the principal component filter can be used to remove covariance from functional data that have spatial patterns similar to the baseline data. Since systemic signals are different for oxyhemoglobin and deoxyhemoglobin, the spatial covariance used in this PCA analysis [Eq. (6)] is calculated and applied separately for the data corresponding to each of the two hemoglobin species by computing and applying PCA to separate covariance matrices for oxyhemoglobin and deoxyhemoglobin.
In Fig. 5, we demonstrate how the reduction of spatial covariance allows the better localization hemodynamic activation associated with a functional motor task by the removal of global, systemic changes. This data, which was previously described in Franceschini and co-workers [69], demonstrates that in addition to the regional cerebral changes, functional tasks may result in additional global systemic changes, which must be accounted for. During the finger-tapping task, shown in Fig. 5, changes in heart rate, respiration, and blood pressure resulted in a task-associated, global hemodynamic response. These changes result in the appearance of a functional activation over nearly the entire head of the subject, as shown in Fig. 5(a). By using the PCA filter to project the strongest principal components calculated from a separate baseline recording, the resulting filtered response was localized to the contralateral motor cortex [Fig. 5(b)]. Similar to the use of PCA for motion correction, the baseline derived eigenvectors can either be removed from the data in a preprocessing step as described by Zhang et al. ([70,124]) or used as additional regressors of the data within the general linear model as part of the estimation of functional activation.
Fig. 5.
Removing systemic physiological noise by PCA: NIRS measurements are often sensitive to systemic fluctuations arising from blood pressure changes, respiration, or the cardiac cycle. As a complication, this physiology may change during the performance of intense stimulus tasks, such as motor activity. As is shown in (a), these changes can result in a systemic response giving the appearance of global functional activation. (b) When the PCA filter described in the text is used to remove this systemic effect by reducing this covariance, the activation region is localized to the motor area. These data were previously published in [69].
F. Empirically Derived Filters of Systemic Physiology
A final method for removing systemic physiological noise that we will discuss is the use of auxiliary measurements of systemic physiology to design specific filters for the optical measurements. As demonstrated by Diamond and co-workers [108], the linear model can also be used to remove systemic physiological signals by incorporating external measurements of cardiac, respiratory, or blood pressure oscillations as model regressors. Similar methods have also been proposed for reducing noise in fMRI signals [120,121]. In this approach, it is assumed that the observed hemodynamic signal can be expressed as a linear sum of the contributions of both the functional stimulus and the additional systemic physiological terms, i.e.,
| (7) |
In Eq. (7), U is the regression variable. The stimulus regressor (Ustim) describes the timing of the experimental task. ⊗ is the convolution operator. The systemic regressors for the cardiac, blood pressure (BP), or respiration signals are externally measured signals such as from a dynamic blood pressure cuff, pulse oximetry, or an electrocardiogram instrument. The impulse response function (β) for each of these regressors describes the transfer function from the regression variable to the effect on the observation. The convolution of the regressor and the individual transfer function (U ⊗ β) can describe the identified component in the observations. This approach is very similar to the methods termed RET-ROICOR for analysis of functional MRI developed by Glover and colleagues [120]. One important difference between the application of these methods to fMRI versus optical data is that the higher temporal resolution of the optical data allows a high-temporal-resolution transfer function to be estimated to relate the recorded external physiology and the optical signals. In comparison, for removal of systemic signals from fMRI, which is often sampled at only once every 1–3 s, only a single temporal shift of the systemic physiology is used to filter the data.
A demonstration of this approach as used in the HomER program is provided in Fig 6. In this figure, we show the analysis of simulated evoked hemodynamic responses with experimentally measured baseline physiological signals from a finger pulse oximeter and beat-to-beat blood pressure cuff (low-pass filtered) added on top of the response. We used both of these signals as regressors in a linear model as given by Eq. (7) to separate the raw signal (left) into three components (right panels) corresponding to the models of the evoked response, cardiac-related, and blood-pressure-related signals.
Fig. 6.
Linear filtering of systemic physiology based on auxiliary measurements. Here we demonstrate the use of auxiliary measurements to improve the estimate of the functional hemodynamic response. Without the removal of these signals by bandpass filtering, the calculated hemodynamic response is heavily corrupted by these fluctuations. When the cardiac cycle and blood pressure fluctuations are used as additional regression variables, the functional hemodynamic response is more clearly separated from the effects of these systemic variables. The regression of this data with the external physiological measurements allows the separation of the data into the functional and systemic contributions. The panels on the right (b)–(d) show the separated system components for the evoked response (b), the cardiac related response (c) and the blood pressure component (d) composing the raw data (a).
6. Estimation of functional signals in NIRS
Since the CW NIRS technique is capable of measuring only hemodynamic changes and is unable to report baseline values, the most common use of this technique is to assess the brain’s functional response to stimulation. In general, most analysis of functional hemodynamic changes in neuroimaging techniques such as NIRS or fMRI is based on an assumption of the linear addition of hemodynamic changes. In the linear model, the effects of hemodynamic variations are additive with the individual components summing linearly to give the measured total signal change. For example, the hemodynamic responses associated with multiple types of stimuli or overlapping responses are assumed to add to give the overall measured signal. In this model, the observations of hemodynamic changes over time can be expressed as a series of linear equations, described by the equations
| (8a) |
| (8b) |
In this notation, Y is the recorded time series of observations of a hemodynamic variable. β represents the parameter vector, for which we wish to solve. The operator ⊗ represents convolution of the regression variable (i.e., the timing of stimuli) with the impulse response to that regressor (β, i.e., the hemodynamic response). In the matrix form of Eq. (8b), the matrix G is typically termed the design matrix. When multiplied by the impulse response functions (β), this matrix performs the convolution of the regression parameters and the response functions. The effects of individual responses are summed to reproduce the observations. Last, the final term in Eqs. (8), e, is the error in each measurement. This is generally assumed to be normally distributed, zero-mean, random noise.
To estimate the mean effect of each individual functional response, we must perform a deconvolution of Eqs. (8) to estimate β. The minimum least-squares solution to this linear equation is provided by the equation [122]
| (9) |
The superscript t represents the transpose matrix operation. The efficiency of this deconvolution equation is optimized by the careful experimental design of the timing of stimulus presentation [99–102]. Inappropriate stimulus timing can result in cross talk between components of β or create an ill-posed structure for the matrix G, which prevents inversion or introduces numerical imprecision. In addition, the nonlinear effects of short interstimulus intervals should also be considered, which may affect the assumption of the linear model.
7. Canonical Temporal Functions in Optical Imaging
As previously mentioned, in fMRI analysis, a typical a priori assumption is made about the temporal shape of the hemodynamic response. For example, a canonical temporal basis set, which model the response as a combination of one or more Γ-variant or Gaussian functions (e.g., [123]), is often used as an assumption to describe the shape of the response and to provide a support vector for modeling the evoked response. In other words, an assumption is made that the evoked response may be modeled as a linear combination of these functions (e.g., see [95]). For the analysis of brain activation signals, this approach has several advantages. First, it reduces the number of unknowns in the model, thereby improving the statistical power of certain statistical metrics (for example, an F test). Second, canonical basis functions impose a priori assumptions on the model; for example, using a basis set of spline functions or multiple Gaussian curves can impose a specific level of temporal smoothness on the evoked response. This approach can offer vast improvements to the detection efficiency of the evoked response and greatly simplify statistical testing. Because the evoked response is reduced to one or a few numerical values, it is more straightforward to perform statistical tests of whether this value is nonzero, thereby rejecting the null hypothesis that the signal did not change. Reduction of the number of parameters tested also reduces the possibility of finding a false positive because of multiple comparisons. In contrast, traditional deconvolution methods provide an estimation of the full time course of the evoked response by estimating each point independently. This provides a model-free estimate of the shape of the evoked response but adds the complexity of having to choose a temporal window for defining the statistical test and introduces multiple comparison concerns from the many more parameters that could be tested.
While canonical basis functions offer several advantages for the analysis of NIRS data (e.g., [94,124]), there are still several issues to consider. First, the specific hypothesis that will be tested will influence whether a canonical basis is appropriate and, if so, determine which basis should be used. As discussed previously, one of the advantages of optical imaging is the higher temporal resolution in comparison with fMRI. This resolution allows for an additional dimension in which to pose a hypothesis. That is, we can test on the basis of not only magnitude, but also timing differences. For example, a hypothesis might be that a specific form of cognitive impairment increases the duration of the evoked oxyhemoglobin response. We can also ask questions about relative timing parameters; for example, increased cerebral vascular reserve will delay the onset of deoxyhemoglobin response relative to the oxyhemoglobin response (there is evidence of this from hypercapnia studies, e.g., [125]). In using a canonical basis set to test timing-based hypotheses, it is important to choose the basis functions appropriately so that uniform sensitivity to different dynamics of the evoked response can be achieved. Work by Zhang et al. [124] and Diamond et al. [109] both used a large number of Gaussian basis functions to capture the temporal dynamics of the evoked response. Similarly Boverman et al. used a similar basis set based on spline functions to model dynamics in optical mammography [126]. An alternative is to use a nonlinear estimation procedure [127–130]. In this approach, an underlying temporal function, such as a Γ-variant functional form or biological model, may be assumed, but the timing parameters are unknowns estimated by using a minimization routine. This allows additional hypothesis to be statistically tested based on the recovered values and uncertainty of the timing parameters.
A second issue that must be addressed concerning the use of canonical functions in NIRS analysis is the uniformity of sensitivity to oxyhemoglobin and deoxyhemoglobin changes. Unlike fMRI, which typically reports a single type of functional contrast, NIRS measures two distinct signals (oxyhemoglobin and deoxyhemoglobin), and each of these is expected to have a slightly different timing (e.g., [10]). Thus, the use of a single common set of canonical basis functions to model both of the hemoglobin responses will give rise to nonuniform detection efficiencies of the oxyhemoglobin and deoxyhemoglobin changes. The canonical basis functions must be carefully chosen not to sacrifice the spectroscopic resolution of the NIRS measurements. For example, if a canonical function that was optimized to model the deoxyhemoglobin response were used (i.e., taken from the fMRI literature, e.g., [131]), this would give good sensitivity to detect significant deoxyhemoglobin changes, but would not guarantee the same sensitivity to estimate oxyhemoglobin changes, which can have different timing [10]. Because of the assumption that the expected experimental response can be modeled by the canonical response, the sensitivity of this approach is influenced by the proper choice of the canonical model. Using a model optimized to describe the timing of deoxyhemoglobin may not be able to model oxyhemoglobin as well and would result in an underestimation of the significance of oxyhemoglobin changes in comparison with deoxyhemoglobin changes. Therefore, any interpretation of relative oxyhemoglobin and deoxyhemoglobin changes would depend on the basis functions chosen. Care must be exercised in choosing these basis functions correctly to provide uniform sensitivity to both hemoglobin species. The recent work by Plichta and co-workers compared the model-based and model-free analysis of the NIRS-measured hemodynamic response to graded visual stimuli [132]. In this work, Plichta and co-workers demonstrated the use of a Gaussian temporal basis function along with its first and second derivatives in order to linearly model both the oxyhemoglobin and deoxyhemoglobin responses. Although they showed that this canonical model was able to model both hemoglobin signals significantly, the correlation between the model-based and model-free estimates was consistently lower for the deoxyhemoglobin model that for the oxyhemoglobin model.
Although canonical models improve the contrast-to-noise ratio and statistical power of the resulting hemodynamic responses with NIRS [107,124,133], more investigation is needed to verify and choose basis functions that will not compromise the temporal and spectroscopic resolutions of NIRS, which are amongst the strengths of this technique. In comparison with fMRI, NIRS instruments generally have better temporal resolutions (fMRI can typically achieve 2–3 s). This temporal resolution allows the potential to distinguish responses based on temporal characteristics such as onset or peak times. Which of these parameters or hemoglobin species is most informative in analysis and hypothesis testing is not clear.
8. HomER—Graphical Interface for Functional Near-Infrared Spectroscopy Signals
The HomER program is based on a flexible data architecture, which allows data to be imported with limited preparatory processing from virtually any currently existing commercial NIRS system. The analysis of this data can be tailored to allow the users complete versatility to specify the details of their experiment, including NIRS probe geometry and layout, optical wavelength selection, stimulus design, and other instrumental and acquisition parameters. A screen shot of the HomER program is shown in Fig 7. Example files and scripts are provided along with the download of the program demonstrating the preconditioning of data for import into HomER. Although HomER is principally designed for CW NIRS measurements, both time-domain [83,135] and frequency-domain [136] data can be analyzed with additional preprocessing. The HomER program has been made freely available and is available from the Photon Migration Imaging Laboratory at the Massachusetts General Hospital ([6]). This program is available in both its native MATLAB format as open source code and as a compiled standalone executable binary program. The standalone executable is distributed with the MATLAB Runtime Compiler and will run under Windows NT or higher. The open-source MATLAB code is written for MATLAB version 7.0 or higher.
Fig. 7.
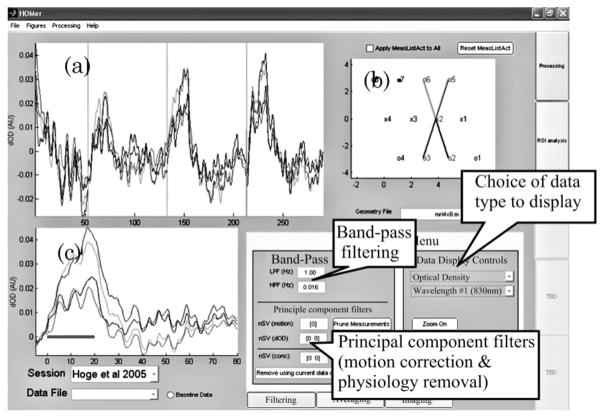
Screen shot of the HomER program. The layout of the HomER program is based around an interactive graphical display of the NIRS probe, shown in the upper right (b). The user specifies this probe geometry within the data file imported into HomER as described in the text. By selecting source (displayed as “x”) or detector (‘o”) positions on this probe layout, the user navigates through the display of their data. The original data are shown in (a) and the average evoked response is shown in (c). The data presented are described in [134] and were recorded during a 20 s finger-tapping task. Data shown are from a single run of one of the subjects.
The structure of the HomER program is based on three levels of data processing as shown in Fig 8. (i) At the first level, individual data files are represented. This level contains the data for a single experimental run, for example, a single acquisition scan. Scan-level processing allows the data from a single experimental run to be interactively visualized, time-series data filtered, and analyzed for functional responses. For a typical experiment, multiple such data files will be recorded within a single experimental session. (ii) The session level represents the second organizing level in HomER. This level includes the joint processing of one or more individual data files for a single experimental subject, recorded during a single session. At the session level, response averages and effects analysis can be calculated from multiple data runs. First-level statistical tests can be performed to investigate the significance of functional changes (i.e., the F test and T test). In addition, images of the changes in optical absorption or hemoglobin concentrations can be reconstructed from these session averages. Session-level data should have consistent optical probe placement and geometry to allow analysis between scans. (iii) At the final level of processing, multisession or group information is represented. This allows group averaging from multiple experimental subjects or multiple sessions from the same subject. Data processing at this level is allowed for region-of-interest comparisons to avoid intersubject registration issues. At all of these three levels, data analysis, visualization, and data export are provided for MATLAB and ASCII formats.
Fig. 8.
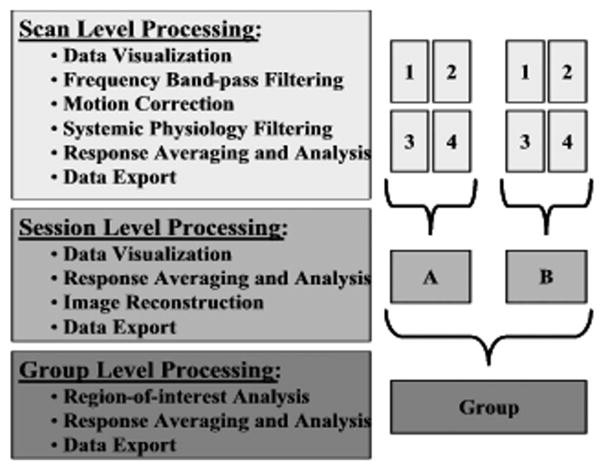
Levels of analysis in HomER. The HomER program architecture is based on three levels of analysis and processing: a single experimental scan, a session (single subject), and group analysis. At each level, various options for processing, visualization, and data management are offered.
The primary purpose of the design of the HomER program is to provide the users with easy, quick, and productive interaction and visualization of their data. The program has been designed such that the user should be able to interact with their data from the level of a general overview, handling large data sets, down to the detailed management of individual time courses from a single measurement between a source and detector pair. In HomER, the detailed management of individual measurements is facilitated by a visual layout of the NIRS probe created from user-specified probe geometry and a list of data measurements in the raw data file. By interactively selecting source or detector positions from the NIRS probe, the user is able to toggle the display of different measurements. The time-series data display is color-coded with lines drawn between the corresponding source and detector positions on the NIRS probe. Data can be discarded from analysis, exported to file, or copied to the clipboard function by simply clicking on one of these lines. This probe image allows the user to interact with the data for both the individual data file and session levels of processing. The user can interactively navigate through their data with ease by clicking on the probe geometry exactly as it was set up during data acquisition, allowing the results of an experiment to be easily viewed. The probe also allows users a means to prune measurements from the analysis for subjective reasons, such as the presence of motion artifacts, or objectively based on a low signal-to-noise ratio or source–detector separation criterion.
In addition to the interaction across the spatial dimensions of the probe, HomER allows the user to visually interact with the time-series data by enabling the user to remove subject motion or other artifacts from the data. Regions of time can be highlighted and discarded from functional averaging or deconvolution of the hemodynamic response. These regions can also be discarded from the calculation of the data covariance used in the PCA filtering as described earlier in this paper. In addition to selecting blocks of time, individual stimulus epochs can also be removed prior to functional analysis. These two features allow the user to manually select regions of time that are affected by motion or other artifacts. Several statistical based plots, such as studentized residual analysis for outlier detection, F tests, and other analysis of variance analyses, are also included in HomER for the analysis of functional data.
For filtering, HomER offers both bandpass and PCA-based filtering modules. Bandpass filtering is done by using an finite-impulse response (FIR) model. The default FIR filter used in HomER is a fourth-order Butterworth filter and filters twice in a forward and inverse pass of the data using the MATLAB function filtfilt. This back-and-forth filter reduces accumulation of phase in the data and offers approximately a twofold better rejection in the stop band. Additional options can be selected to specify the characteristics of the filter model (stop band, order, and type of filter). Three options are given for PCA analysis to allow (i) motion correction (see [64] for an example), (ii) physiological correction using a separate baseline file (see [117]), and (iii) physiological correction using components derived and applied to the current file.
In HomER, functional responses can be calculated by using either block averaging or a deconvolution model based on the estimation of an FIR impulse response. These methods use an ordinary least-squares fit of the data. The estimation of the evoked response is performed subsequent to filtering and motion correction. Responses are calculated for both the change in optical density and the change in oxyhemoglobin and deoxyhemoglobin. Individual files (scans) are processed first and then averaged together to calculate the average response for a session. Selected regions of interest can also be averaged within and across subjects. Regions of interest are specified based on either T statistics calculated from the effects of the functional model or from F tests describing the ability of the model to fit the data. Joint probabilities across specific wavelengths or hemoglobin types are not currently calculated.
Finally, HomER also includes basic tools for image reconstruction. HomER currently supports calculations of the optical forward model (sensitivity matrix) from a homogeneous, semi-infinite, slab geometry, using the analytic form given in [11]. The user is given options to specify the absorption and scattering properties of the medium at each wavelength. Currently only CW forward models are supported. Images are reconstructed by using either the back-projection method (default) or via a Tikhonov regularized inverse. For reconstruction of images of hemoglobin, HomER uses a spectral prior incorporated into the inverse problem as described in [137,138]. Movies of the functional response can also be reconstructed and saved by using HomER.
9. Conclusions and Future Directions
The NIRS technology will continue to make advances toward the understanding of the functioning human brain. Along with the ever-increasing applications for this technology, innovations will continue to be made in the methods used to analysis this data. In the future, it is hoped that the HomER program will provide the framework to disseminate these advances. By providing HomER as a free resource, it is our hope that this program will continue to grow and advance along with the NIRS field.
Acknowledgments
The authors thank Danny Joseph and all the current users of this program, whose comments and software debugging have led to improvements in HomER. We also acknowledge Dana Brooks, Rick Gaudette, Tom Gaudette, Eric Miller, Quan Zhang, and Jonathan Stott, who contributed to the original PMI toolbox. We also thank Rick Hoge, Teresa Wilcox, and Heather Bortfeld, who contributed data for the examples used in this text. This work was supported by the National Institutes of Health (NIH, P41-RR14075).
Footnotes
OCIS codes: 170.2655, 170.3880, 300.0300, 070.0070.
References
- 1.Villringer A, Chance B. Non-invasive optical spectroscopy and imaging of human brain function. Trends Neurosci. 1997;20:435–442. doi: 10.1016/s0166-2236(97)01132-6. [DOI] [PubMed] [Google Scholar]
- 2.Yodh A, Boas D, editors. Functional Imaging with Diffusing Light. CRC Press; 2003. [Google Scholar]
- 3.Boas DA, Dale AM, Franceschini MA. Diffuse optical imaging of brain activation: approaches to optimizing image sensitivity, resolution, and accuracy. NeuroImage. 2004;23(Suppl 1):S275–S288. doi: 10.1016/j.neuroimage.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Gibson AP, Hebden JC, Arridge SR. Recent advances in diffuse optical imaging. Phys Med Biol. 2005;50:R1–R43. doi: 10.1088/0031-9155/50/4/r01. [DOI] [PubMed] [Google Scholar]
- 5.Gratton E, Toronov V, Wolf U, Wolf M, Webb A. Measurement of brain activity by near-infrared light. J Biomed Opt. 2005;10:011008. doi: 10.1117/1.1854673. [DOI] [PubMed] [Google Scholar]
- 6.PMI Lab—HomER. http://www.nmr.mgh.harvard.edu/PMI/resources/homer/home.htm.
- 7.Jöbsis FF. Noninvasive, infrared monitoring of cerebral and myocardial oxygen sufficiency and circulatory parameters. Science. 1977;198:1264–1267. doi: 10.1126/science.929199. [DOI] [PubMed] [Google Scholar]
- 8.Wyatt JS, Cope M, Delpy DT, Wray S, Reynolds EO. Quantification of cerebral oxygenation and haemodynamics in sick newborn infants by near infrared spectrophotometry. Lancet. 1986;2:1063–1066. doi: 10.1016/s0140-6736(86)90467-8. [online ed. Lancet 328, 1063–1066 2003. [DOI] [PubMed] [Google Scholar]
- 9.Chance B, Leigh JS, Miyake H, Smith DS, Nioka S, Greenfeld R, Finander M, Kaufmann K, Levy W, Young M. Comparison of time-resolved and -unresolved measurements of deoxyhemoglobin in brain. Proc Natl Acad Sci USA. 1988;85:4971–4975. doi: 10.1073/pnas.85.14.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huppert TJ, Hoge RD, Diamond SG, Franceschini MA, Boas DA. A temporal comparison of BOLD, ASL, and NIRS hemodynamic responses to motor stimuli in adult humans. NeuroImage. 2006;29:368–382. doi: 10.1016/j.neuroimage.2005.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huppert T, Franceschini M, Boas D. Non-invasive imaging of cerebral activation with diffuse optical tomography. In: Frostig R, editor. In Vivo Optical Imaging of Brain Function. 2. CRC Press; 2009. [PubMed] [Google Scholar]
- 12.Fukui Y, Ajichi Y, Okada E. Monte Carlo prediction of near-infrared light propagation in realistic adult and neonatal head models. Appl Opt. 2003;42:2881–2887. doi: 10.1364/ao.42.002881. [DOI] [PubMed] [Google Scholar]
- 13.Okada E, Delpy DT. Near-infrared light propagation in an adult head model. II. Effect of superficial tissue thickness on the sensitivity of the near-infrared spectroscopy signal. Appl Opt. 2003;42:2915–2922. doi: 10.1364/ao.42.002915. [DOI] [PubMed] [Google Scholar]
- 14.Okada E, Delpy DT. Near-infrared light propagation in an adult head model. I. Modeling of low-level scattering in the cerebrospinal fluid layer. Appl Opt. 2003;42:2906–2914. doi: 10.1364/ao.42.002906. [DOI] [PubMed] [Google Scholar]
- 15.Cope M, Delpy DT, Reynolds EO, Wray S, Wyatt J, van der Zee P. Methods of quantitating cerebral near infrared spectroscopy data. Adv Exp Med Biol. 1988;222:183–189. doi: 10.1007/978-1-4615-9510-6_21. [DOI] [PubMed] [Google Scholar]
- 16.Boas D, Culver J, Stott J, Dunn A. Three dimensional Monte Carlo code for photon migration through complex heterogeneous media including the adult human head. Opt Express. 2002;10:159–170. doi: 10.1364/oe.10.000159. [DOI] [PubMed] [Google Scholar]
- 17.Huppert T, Hoge RD, Dale AM, Franceschini MA, Boas DA. A quantitative spatial comparison of diffuse optical imaging with BOLD- and ASL-based fMRI. J Biomed Opt. 2006;11:064018. doi: 10.1117/1.2400910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heekeren HR, Obrig H, Wenzel R, Eberle K, Ruben J, Villringer K, Kurth R, Villringer A. Cerebral haemoglobin oxygenation during sustained visual stimulation—a near-infrared spectroscopy study. Philos Trans R Soc London Ser B. 1997;352:743–750. doi: 10.1098/rstb.1997.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meek JH, Elwell CE, Khan MJ, Romaya J, Wyatt JS, Delpy DT, Zeki S. Regional changes in cerebral haemodynamics as a result of a visual stimulus measured by near infrared spectroscopy. Proc R Soc London Ser B. 1995;261:351–356. doi: 10.1098/rspb.1995.0158. [DOI] [PubMed] [Google Scholar]
- 20.Ruben J, Wenzel R, Obrig H, Villringer K, Bernarding J, Hirth C, Heekeren H, Dirnagl U, Villringer A. Haemoglobin oxygenation changes during visual stimulation in the occipital cortex. Adv Exp Med Biol. 1997;428:181–187. doi: 10.1007/978-1-4615-5399-1_26. [DOI] [PubMed] [Google Scholar]
- 21.Sakatani K, Chen S, Lichty W, Zuo H, Wang YP. Cerebral blood oxygenation changes induced by auditory stimulation in newborn infants measured by near infrared spectroscopy. Early Hum Dev. 1999;55:229–236. doi: 10.1016/s0378-3782(99)00019-5. [DOI] [PubMed] [Google Scholar]
- 22.Chen S, Sakatani K, Lichty W, Ning P, Zhao S, Zuo H. Auditory-evoked cerebral oxygenation changes in hypoxic-ischemic encephalopathy of newborn infants monitored by near infrared spectroscopy. Early Hum Dev. 2002;67:113–121. doi: 10.1016/s0378-3782(02)00004-x. [DOI] [PubMed] [Google Scholar]
- 23.Zaramella P, Freato F, Amigoni A, Salvadori S, Marangoni P, Suppjei A, Schiavo B, Chiandetti L. Brain auditory activation measured by near-infrared spectroscopy (NIRS) in neonates. Pediatr Res. 2001;49:213–219. doi: 10.1203/00006450-200102000-00014. [DOI] [PubMed] [Google Scholar]
- 24.Franceschini MA, Fantini S, Thompson JH, Culver JP, Boas DA. Hemodynamic evoked response of the sensorimotor cortex measured noninvasively with near-infrared optical imaging. Psychophysiology. 2003;40:548–560. doi: 10.1111/1469-8986.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obrig H, Wolf T, Doge C, Hulsing JJ, Dirnagl U, Villringer A. Cerebral oxygenation changes during motor and somatosensory stimulation in humans, as measured by near-infrared spectroscopy. Adv Exp Med Biol. 1996;388:219–224. doi: 10.1007/978-1-4613-0333-6_27. [DOI] [PubMed] [Google Scholar]
- 26.Obrig H, Hirth C, Junge-Hulsing JG, Doge C, Wolf T, Dirnagl U, Villringer A. Cerebral oxygenation changes in response to motor stimulation. J Appl Physiol. 1996;81:1174–1183. doi: 10.1152/jappl.1996.81.3.1174. [DOI] [PubMed] [Google Scholar]
- 27.Colier WN, Quaresima V, Oeseburg B, Ferrari M. Human motor-cortex oxygenation changes induced by cyclic coupled movements of hand and foot. Exp Brain Res. 1999;129:457–461. doi: 10.1007/s002210050913. [DOI] [PubMed] [Google Scholar]
- 28.Hirth C, Obrig H, Villringer K, Thiel A, Bernarding J, Muhlnickel W, Flor H, Dirnagl U, Villringer A. Non-invasive functional mapping of the human motor cortex using near-infrared spectroscopy. NeuroReport. 1996;7:1977–1981. doi: 10.1097/00001756-199608120-00024. [DOI] [PubMed] [Google Scholar]
- 29.Kleinschmidt A, Obrig H, Requardt M, Merboldt KD, Dirnagl U, Villringer A, Frahm J. Simultaneous recording of cerebral blood oxygenation changes during human brain activation by magnetic resonance imaging and near-infrared spectroscopy. J Cereb Blood Flow Metab. 1996;16:817–826. doi: 10.1097/00004647-199609000-00006. [DOI] [PubMed] [Google Scholar]
- 30.Kameyama M, Fukuda M, Yamagishi Y, Sato T, Uehara T, Ito M, Suto T, Mikuni M. Frontal lobe function in bipolar disorder: a multichannel near-infrared spectroscopy study. NeuroImage. 2006;29:172–184. doi: 10.1016/j.neuroimage.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 31.Nagamitsu S, Nagano M, Yamashita Y, Takashima S, Matsuishi T. Prefrontal cerebral blood volume patterns while playing video games–a near-infrared spectroscopy study. Brain Dev. 2006;28:315–321. doi: 10.1016/j.braindev.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Okamoto M, Matsunami M, Dan H, Kohata T, Kohyama K, Dan I. Prefrontal activity during taste encoding: an fNIRS study. NeuroImage. 2006;31:796–806. doi: 10.1016/j.neuroimage.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 33.Sumitani S, Tanaka T, Tayoshi S, Ota K, Kameoka N, Morimune M, Shibuya-Tayoshi S, Kinouchi S, Ueno S, Ohmori T. Hemodynamic changes in the prefrontal cortex during mental works as measured by multi channel near-infrared spectroscopy (NIRS) J Med Invest. 2005;52(Suppl):302–303. doi: 10.2152/jmi.52.302. [DOI] [PubMed] [Google Scholar]
- 34.Sato H, Takeuchi T, Sakai KL. Temporal cortex activation during speech recognition: an optical topography study. Cognition. 1999;73:B55–66. doi: 10.1016/s0010-0277(99)00060-8. [DOI] [PubMed] [Google Scholar]
- 35.Adelson PD, Nemoto E, Scheuer M, Painter M, Morgan J, Yonas H. Noninvasive continuous monitoring of cerebral oxygenation periictally using near-infrared spectroscopy: a preliminary report. Epilepsia (1909–1915, 1937–1950, 1952–1955, 1959-) 1999;40:1484–1489. doi: 10.1111/j.1528-1157.1999.tb02030.x. [DOI] [PubMed] [Google Scholar]
- 36.Sokol DK, Markand ON, Daly EC, Luerssen TG, Malkoff MD. Near infrared spectroscopy (NIRS) distinguishes seizure types. Seizure: the Journal of the British Epilepsy Association. 2000;9:323–327. doi: 10.1053/seiz.2000.0406. [DOI] [PubMed] [Google Scholar]
- 37.Steinhoff BJ, Herrendorf G, Kurth C. Ictal near infrared spectroscopy in temporal lobe epilepsy: a pilot study. Seizure: Eur J Epilepsy. 1996;5:97–101. [PubMed] [Google Scholar]
- 38.Watanabe E, Maki A, Kawaguchi F, Yamashita Y, Koizumi H, Mayanagi Y. Noninvasive cerebral blood volume measurement during seizures using multichannel near infrared spectroscopic topography. J Biomed Opt. 2000;5:287–290. doi: 10.1117/1.429998. [DOI] [PubMed] [Google Scholar]
- 39.Buchheim K, Obrig H, Pannwitz Wv, Muller A, Heekeren H, Villringer A, Meierkord H. Decrease in haemoglobin oxygenation during absence seizures in adult humans. Neurosci Lett Suppl. 2004;354:119–122. doi: 10.1016/j.neulet.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe E, Nagahori Y, Mayanagi Y. Focus diagnosis of epilepsy using near-infrared spectroscopy. Epilepsia (1909–1915, 1937–1950, 1952–1955, 1959-) 2002;43(Suppl 9):50–55. doi: 10.1046/j.1528-1157.43.s.9.12.x. [DOI] [PubMed] [Google Scholar]
- 41.Fallgatter AJ, Roesler M, Sitzmann L, Heidrich A, Mueller TJ, Strik WK. Loss of functional hemispheric asymmetry in Alzheimer’s dementia assessed with near-infrared spectroscopy. Brain Res Cognit Brain Res. 1997;6:67–72. doi: 10.1016/s0926-6410(97)00016-5. [DOI] [PubMed] [Google Scholar]
- 42.Hanlon EB, Itzkan I, Dasari RR, Feld MS, Ferrante RJ, McKee AC, Lathi D, Kowall NW. Near-infrared fluorescence spectroscopy detects Alzheimer’s disease in vitro. Photochem Photobiol. 1999;70:236–242. [PubMed] [Google Scholar]
- 43.Hock C, Villringer K, Muller-Spahn F, Hofmann M, Schuh-Hofer S, Heekeren H, Wenzel R, Dirnagl U, Villringer A. Near infrared spectroscopy in the diagnosis of Alzheimer’s disease. Ann NY Acad Sci. 1996;777:22–29. doi: 10.1111/j.1749-6632.1996.tb34397.x. [DOI] [PubMed] [Google Scholar]
- 44.Rolfe P, Wickramasinghe YA, Thorniley MS, Faris F, Houston R, Kai Z, Yamakoshi K, O’Brien S, Doyle M, Palmer K, Spencer S. Fetal and neonatal cerebral oxygen monitoring with NIRS: theory and practice. Early Hum Dev. 1992;29:269–273. doi: 10.1016/0378-3782(92)90163-b. [DOI] [PubMed] [Google Scholar]
- 45.Liebert A, Wabnitz H, Steinbrink J, Moller M, Macdonald R, Rinneberg H, Villringer A, Obrig H. Bedside assessment of cerebral perfusion in stroke patients based on optical monitoring of a dye bolus by time-resolved diffuse reflectance. NeuroImage. 2005;24:426–435. doi: 10.1016/j.neuroimage.2004.08.046. [DOI] [PubMed] [Google Scholar]
- 46.Orihashi K, Sueda T, Okada K, Imai K. Near-infrared spectroscopy for monitoring cerebral ischemia during selective cerebral perfusion. Eur J Cardiothorac Surg. 2004;26:907–911. doi: 10.1016/j.ejcts.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 47.Terborg C, Bramer S, Harscher S, Simon M, Witte OW. Bedside assessment of cerebral perfusion reductions in patients with acute ischaemic stroke by near-infrared spectroscopy and indocyanine green. J Neurol Neurosurg Psychiatry. 2004;75:38–42. [PMC free article] [PubMed] [Google Scholar]
- 48.Thiagarajah JR, Papadopoulos MC, Verkman AS. Noninvasive early detection of brain edema in mice by near-infrared light scattering. J Neorosci Res. 2005;80:293–299. doi: 10.1002/jnr.20439. [DOI] [PubMed] [Google Scholar]
- 49.Chen WG, Li PC, Luo QM, Zeng SQ, Hu B. Hemodynamic assessment of ischemic stroke with near-infrared spectroscopy. Space Med Med Eng (Beijing) 2000;13:84–89. [PubMed] [Google Scholar]
- 50.Nemoto EM, Yonas H, Kassam A. Clinical experience with cerebral oximetry in stroke and cardiac arrest. Crit Care Med. 2000;28:1052–1054. doi: 10.1097/00003246-200004000-00023. [DOI] [PubMed] [Google Scholar]
- 51.Saitou H, Yanagi H, Hara S, Tsuchiya S, Tomura S. Cerebral blood volume and oxygenation among poststroke hemiplegic patients: effects of 13 rehabilitation tasks measured by near-infrared spectroscopy. Arch Phys Med Rehabil. 2000;81:1348–1356. doi: 10.1053/apmr.2000.9400. [DOI] [PubMed] [Google Scholar]
- 52.Vernieri F, Rosato N, Pauri F, Tibuzzi F, Passarelli F, Rossini PM. Near infrared spectroscopy and transcranial Doppler in monohemispheric stroke. Eur Neurol. 1999;41:159–162. doi: 10.1159/000008041. [DOI] [PubMed] [Google Scholar]
- 53.Eschweiler GW, Wegerer C, Schlotter W, Spandl C, Stevens A, Bartels M, Buchkremer G. Left prefrontal activation predicts therapeutic effects of repetitive transcranial magnetic stimulation (rTMS) in major depression. Psychiatry Res. 2000;99:161–172. doi: 10.1016/s0925-4927(00)00062-7. [DOI] [PubMed] [Google Scholar]
- 54.Matsuo K, Kato T, Fukuda M, Kato N. Alteration of hemoglobin oxygenation in the frontal region in elderly depressed patients as measured by near-infrared spectroscopy. J Neuropsychiatry Clin Neurosci. 2000;12:465–471. doi: 10.1176/jnp.12.4.465. [DOI] [PubMed] [Google Scholar]
- 55.Okada F, Takahashi N, Tokumitsu Y. Dominance of the ‘nondominant’ hemisphere in depression. J Affect Disord. 1996;37:13–21. doi: 10.1016/0165-0327(95)00040-2. [DOI] [PubMed] [Google Scholar]
- 56.Matsuo K, Kato N, Kato T. Decreased cerebral haemodynamic response to cognitive and physiological tasks in mood disorders as shown by near-infrared spectroscopy. Psychol Med. 2002;32:1029–1037. doi: 10.1017/s0033291702005974. [DOI] [PubMed] [Google Scholar]
- 57.Suto T, Fukuda M, Ito M, Uehara T, Mikuni M. Multichannel near-infrared spectroscopy in depression and schizophrenia: cognitive brain activation study. Biol Psychiatry. 2004;55:501–511. doi: 10.1016/j.biopsych.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 58.Fallgatter AJ, Strik WK. Reduced frontal functional asymmetry in schizophrenia during a cued continuous performance test assessed with near-infrared spectroscopy. Schizophr Bull. 2000;26:913–919. doi: 10.1093/oxfordjournals.schbul.a033505. [DOI] [PubMed] [Google Scholar]
- 59.Okada F, Tokumitsu Y, Hoshi Y, Tamura M. Impaired interhemispheric integration in brain oxygenation and hemodynamics in schizophrenia. Eur Arch Psychiatry Clin Neurosci. 1994;244:17–25. doi: 10.1007/BF02279807. [DOI] [PubMed] [Google Scholar]
- 60.Kubota Y, Toichi M, Shimizu M, Mason RA, Coconcea CM, Findling RL, Yamamoto K, Calabrese JR. Prefrontal activation during verbal fluency tests in schizophrenia—a near-infrared spectroscopy (NIRS) study. Schizophr Res. 2005;77:65–73. doi: 10.1016/j.schres.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 61.Shinba T, Nagano M, Kariya N, Ogawa K, Shinozaki T, Shimosato S, Hoshi Y. Near-infrared spectroscopy analysis of frontal lobe dysfunction in schizophrenia. Biol Psychiatry. 2004;55:154–164. doi: 10.1016/s0006-3223(03)00547-x. [DOI] [PubMed] [Google Scholar]
- 62.Watanabe A, Kato T. Cerebrovascular response to cognitive tasks in patients with schizophrenia measured by near-infrared spectroscopy. Schizophr Bull. 2004;30:435–444. doi: 10.1093/oxfordjournals.schbul.a007090. [DOI] [PubMed] [Google Scholar]
- 63.Elwell CE, Henty JR, Leung TS, Austin T, Meek JH, Delpy DT, Wyatt JS. Measurement of CMRO2 in neonates undergoing intensive care using near infrared s pectroscopy. Adv Exp Med Biol. 2005;566:263–268. doi: 10.1007/0-387-26206-7_35. [DOI] [PubMed] [Google Scholar]
- 64.Wilcox T, Bortfeld H, Woods R, Wruck E, Boas DA. Using near-infrared spectroscopy to assess neural activation during object processing in infants. J Biomed Opt. 2005;10:11010. doi: 10.1117/1.1852551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sowell ER, Thompson PM, Leonard CM, Welcome SE, Kan E, Toga AW. Longitudinal mapping of cortical thickness and brain growth in normal children. J Neurosci. 2004;24:8223–8231. doi: 10.1523/JNEUROSCI.1798-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perrey S. Non-invasive NIR spectroscopy of human brain function during exercise. Methods. 2008;45:289–299. doi: 10.1016/j.ymeth.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 67.Ide K, Secher NH. Cerebral blood flow and metabolism during exercise. Prog Neurobiol (Oxford) 2000;61:397–414. doi: 10.1016/s0301-0082(99)00057-x. [DOI] [PubMed] [Google Scholar]
- 68.Mehagnoul-Schipper DJ, Colier WN, Jansen RW. Reproducibility of orthostatic changes in cerebral oxygenation in healthy subjects aged 70 years or older. Clin Physiol. 2001;21:77–84. doi: 10.1046/j.1365-2281.2001.00290.x. [DOI] [PubMed] [Google Scholar]
- 69.Franceschini MA, Joseph DK, Huppert TJ, Diamond SG, Boas DA. Diffuse optical imaging of the whole head. J Biomed Opt. 2006;11:054007. doi: 10.1117/1.2363365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, Brooks DH, Franceschini MA, Boas DA. Eigenvector-based spatial filtering for reduction of physiological interference in diffuse optical imaging. J Biomed Opt. 2005;10:011014. doi: 10.1117/1.1852552. [DOI] [PubMed] [Google Scholar]
- 71.Selb J, Stott JJ, Franceschini MA, Sorensen AG, Boas DA. Improved sensitivity to cerebral hemodynamics during brain activation with a time-gated optical system: analytical model and experimental validation. J Biomed Opt. 2005;10:011013. doi: 10.1117/1.1852553. [DOI] [PubMed] [Google Scholar]
- 72.Gratton E, Fantini S, Franceschini MA, Gratton G, Fabiani M. Measurements of scattering and absorption changes in muscle and brain. Philos Trans R Soc London Ser B. 1997;352:727–735. doi: 10.1098/rstb.1997.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang L, Jacques SL, Zheng L. MCML—Monte Carlo modeling of light transport in multi-layered tissues. Comput Methods Programs Biomed. 1995;47:131–146. doi: 10.1016/0169-2607(95)01640-f. [DOI] [PubMed] [Google Scholar]
- 74.Hiraoka M, Firbank M, Essenpreis M, Cope M, Arridge SR, van der Zee P, Delpy DT. A Monte Carlo investigation of optical pathlength in inhomogeneous tissue and its application to near-infrared spectroscopy. Phys Med Biol. 1993;38:1859–1876. doi: 10.1088/0031-9155/38/12/011. [DOI] [PubMed] [Google Scholar]
- 75.Flock ST, Patterson MS, Wilson BC, Wyman DR. Monte Carlo modeling of light propagation in highly scattering tissue—I: Model predictions and comparison with diffusion theory. IEEE Trans Biomed Eng. 1989;36:1162–1168. doi: 10.1109/tbme.1989.1173624. [DOI] [PubMed] [Google Scholar]
- 76.Flock ST, Wilson BC, Patterson MS. Monte Carlo modeling of light propagation in highly scattering tissues—II: Comparison with measurements in phantoms. IEEE Trans Biomed Eng. 1989;36:1169–1173. doi: 10.1109/10.42107. [DOI] [PubMed] [Google Scholar]
- 77.Custo A, Wells WM, III, Barnett AH, Hillman EM, Boas DA. Effective scattering coefficient of the cerebral spinal fluid in adult head models for diffuse optical imaging. Appl Opt. 2006;45:4747–4755. doi: 10.1364/ao.45.004747. [DOI] [PubMed] [Google Scholar]
- 78.Barbour RL, Graber HL, Chang J, Barbour SLS, Koo PC, Aronson R. MRI-guided optical tomography: prospectives and computation for a new imaging method. IEEE Comput Sci Eng. 1995;2:63–77. [Google Scholar]
- 79.Zhang X, Toronov VY, Webb AG. Spatial and temporal hemodynamic study of human primary visual cortex using simultaneous functional MRI and diffuse optical tomography. 27th Annual International Conference of the Engineering in Medicine and Biology Society, 2005. IEEE-EMBS 2005; IEEE; 2005. pp. 727–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chang J, Graber HL, Koo PC, Aronson R, Barbour SL, Barbour RL. Optical imaging of anatomical maps derived from magnetic resonance images using time-independent optical sources. IEEE Trans Med Imaging. 1997;16:68–77. doi: 10.1109/42.552056. [DOI] [PubMed] [Google Scholar]
- 81.Huppert TJ, Diamond SG, Boas DA. Direct estimation of evoked hemoglobin changes by multimodality fusion imaging. J Biomed Opt. 2008;13:054031. doi: 10.1117/1.2976432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Buxton RB, Uludag K, Dubowitz DJ, Liu TT. Modeling the hemodynamic response to brain activation. Neuro-Image. 2004;23(Suppl 1):S220–S233. doi: 10.1016/j.neuroimage.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 83.Delpy DT, Cope M, van der Zee P, Arridge S, Wray S, Wyatt J. Estimation of optical pathlength through tissue from direct time of flight measurement. Phys Med Biol. 1988;33:1433–1442. doi: 10.1088/0031-9155/33/12/008. [DOI] [PubMed] [Google Scholar]
- 84.Sassaroli A, Fantini S. Comment on the modified Beer–Lambert law for scattering media. Phys Med Biol. 2004;49:N255–N257. doi: 10.1088/0031-9155/49/14/n07. [DOI] [PubMed] [Google Scholar]
- 85.Kocsis L, Herman P, Eke A. The modified Beer–Lambert law revisited. Phys Med Biol. 2006;51:N91–N98. doi: 10.1088/0031-9155/51/5/N02. [DOI] [PubMed] [Google Scholar]
- 86.Strangman G, Franceschini MA, Boas DA. Factors affecting the accuracy of near-infrared spectroscopy concentration calculations for focal changes in oxygenation parameters. NeuroImage. 2003;18:865–879. doi: 10.1016/s1053-8119(03)00021-1. [DOI] [PubMed] [Google Scholar]
- 87.Yalavarthy PK, Pogue BW, Dehghani H, Paulsen KD. Weight-matrix structured regularization provides optimal generalized least-squares estimate in diffuse optical tomography. Med Phys. 2007;34:2085–2098. doi: 10.1118/1.2733803. [DOI] [PubMed] [Google Scholar]
- 88.Hoshi Y. Functional near-infrared optical imaging: utility and limitations in human brain mapping. Psychophysiology. 2003;40:511–520. doi: 10.1111/1469-8986.00053. [DOI] [PubMed] [Google Scholar]
- 89.Boas DA, Gaudette T, Strangman G, Cheng X, Marota JJ, Mandeville JB. The accuracy of near infrared spectroscopy and imaging during focal changes in cerebral hemodynamics. NeuroImage. 2001;13:76–90. doi: 10.1006/nimg.2000.0674. [DOI] [PubMed] [Google Scholar]
- 90.Kohl M, Nolte C, Heekeren HR, Horst S, Scholz U, Obrig H, Villringer A. Determination of the wavelength dependence of the differential pathlength factor from near-infrared pulse signals. Phys Med Biol. 1998;43:1771–1782. doi: 10.1088/0031-9155/43/6/028. [DOI] [PubMed] [Google Scholar]
- 91.Boas DA, Dale AM. Simulation study of magnetic resonance imaging-guided cortically constrained diffuse optical tomography of human brain function. Appl Opt. 2005;44:1957–1968. doi: 10.1364/ao.44.001957. [DOI] [PubMed] [Google Scholar]
- 92.Okui N, Okada E. Wavelength dependence of crosstalk in dual-wavelength measurement of oxy- and deoxyhemoglobin. J Biomed Opt. 2005;10:11015. doi: 10.1117/1.1846076. [DOI] [PubMed] [Google Scholar]
- 93.Sato H, Kiguchi M, Kawaguchi F, Maki A. Practicality of wavelength selection to improve signal-to-noise ratio in near-infrared spectroscopy. NeuroImage. 2004;21:1554–1562. doi: 10.1016/j.neuroimage.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 94.Cohen-Adad J, Chapuisat S, Doyon J, Rossignol S, Lina JM, Benali H, Lesage F. Activation detection in diffuse optical imaging by means of the general linear model. Med Image Anal. 2007;11:616–629. doi: 10.1016/j.media.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 95.Friston KJ. Statistical Parametric Mapping: the Analysis of Functional Brain Images. Elsevier/Academic; 2007. [Google Scholar]
- 96.Miyai I, Tanabe HC, Sase I, Eda H, Oda I, Konishi I, Tsunazawa Y, Suzuki T, Yanagida T, Kubota K. Cortical mapping of gait in humans: a near-infrared spectroscopic topography study. NeuroImage. 2001;14:1186–1192. doi: 10.1006/nimg.2001.0905. [DOI] [PubMed] [Google Scholar]
- 97.Joseph DK, Huppert TJ, Franceschini MA, Boas DA. Design and validation of a time division multiplexed continuous wave diffuse optical tomography (DOT) system optimized for brain function imaging. Appl Opt. 2006;45:8142–8151. doi: 10.1364/ao.45.008142. [DOI] [PubMed] [Google Scholar]
- 98.Boas DA, Chen K, Grebert D, Franceschini MA. Improving the diffuse optical imaging spatial resolution of the cerebral hemodynamic response to brain activation in humans. Opt Lett. 2004;29:1506–1508. doi: 10.1364/ol.29.001506. [DOI] [PubMed] [Google Scholar]
- 99.Dale AM. Optimal experimental design for event-related fMRI. Hum Brain Mapp. 1999;8:109–114. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<109::AID-HBM7>3.0.CO;2-W. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Josephs O, Henson RN. Event-related functional magnetic resonance imaging: modelling, inference and optimization. Philos Trans R Soc London Ser B. 1999;354:1215–1228. doi: 10.1098/rstb.1999.0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Friston KJ. Experimental design and statistical issues. In: Mazziotta JC, Toga AW, Frackowiak RSJ, editors. Brain Mapping: The Disorders. Academic; 2000. pp. 33–58. [Google Scholar]
- 102.Wager TD, Nichols TE. Optimization of experimental design in fMRI: a general framework using a genetic algorithm. NeuroImage. 2003;18:293–309. doi: 10.1016/s1053-8119(02)00046-0. [DOI] [PubMed] [Google Scholar]
- 103.Cannestra AF, Pouratian N, Shomer MH, Toga AW. Refractory periods observed by intrinsic signal and fluorescent dye imaging. J Neurophysiol. 1998;80:1522–1532. doi: 10.1152/jn.1998.80.3.1522. [DOI] [PubMed] [Google Scholar]
- 104.Sato H, Kiguchi M, Maki A, Fuchino Y, Obata A, Yoro T, Koizumi H. Within-subject reproducibility of near-infrared spectroscopy signals in sensorimotor activation after 6 months. J Biomed Opt. 2006;11:014021. doi: 10.1117/1.2166632. [DOI] [PubMed] [Google Scholar]
- 105.Sato H, Fuchino Y, Kiguchi M, Katura T, Maki A, Yoro T, Koizumi H. Intersubject variability of near-infrared spectroscopy signals during sensorimotor cortex activation. J Biomed Opt. 2005;10:44001. doi: 10.1117/1.1960907. [DOI] [PubMed] [Google Scholar]
- 106.Obrig H, Neufang M, Wenzel R, Kohl M, Steinbrink J, Einhaupl K, Villringer A. Spontaneous low frequency oscillations of cerebral hemodynamics and metabolism in human adults. NeuroImage. 2000;12:623–639. doi: 10.1006/nimg.2000.0657. [DOI] [PubMed] [Google Scholar]
- 107.Toronov V, Franceschini MA, Filiaci M, Fantini S, Wolf M, Michalos A, Gratton E. Near-infrared study of fluctuations in cerebral hemodynamics during rest and motor stimulation: temporal analysis and spatial mapping. Med Phys. 2000;27:801–815. doi: 10.1118/1.598943. [DOI] [PubMed] [Google Scholar]
- 108.Diamond SG, Huppert TJ, Kolehmainen V, Franceschini MA, Kaipio JP, Arridge SR, Boas DA. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2005, Vol. 3750 of Lecture Notes in Computer Science. Springer; 2005. Physiological system identification with the Kalman filter in diffuse optical tomography; pp. 649–656. [DOI] [PubMed] [Google Scholar]
- 109.Diamond SG, Huppert TJ, Kolehmainen V, Franceschini MA, Kaipio JP, Arridge SR, Boas DA. Dynamic physiological modeling for functional diffuse optical tomography. NeuroImage. 2006;30:88–101. doi: 10.1016/j.neuroimage.2005.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.von Siebenthal K, Beran J, Wolf M, Keel M, Dietz V, Kundu S, Bucher HU. Cyclical fluctuations in blood pressure, heart rate and cerebral blood volume in preterm infants. Brain Dev. 1999;21:529–534. doi: 10.1016/s0387-7604(99)00062-5. [DOI] [PubMed] [Google Scholar]
- 111.Montgomery DC, Peck EA, Vining GG. Introduction to Linear Regression Analysis. Wiley-Interscience; 2006. [Google Scholar]
- 112.Sato H, Tanaka N, Uchida M, Hirabayashi Y, Kanai M, Ashida T, Konishi I, Maki A. Wavelet analysis for detecting body-movement artifacts in optical topography signals. NeuroImage. 2006;33:580–587. doi: 10.1016/j.neuroimage.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 113.Izzetoglu M, Devaraj A, Bunce S, Onaral B. Motion artifact cancellation in NIR spectroscopy using Wiener filtering. IEEE Trans Biomed Eng. 2005;52:934–938. doi: 10.1109/TBME.2005.845243. [DOI] [PubMed] [Google Scholar]
- 114.Strangman G, Boas DA, Sutton JP. Non-invasive neuroimaging using near-infrared light. Biol Psychiatry. 2002;52:679–693. doi: 10.1016/s0006-3223(02)01550-0. [DOI] [PubMed] [Google Scholar]
- 115.Long C, Brown EN, Manoach D, Solo V. Spatiotemporal wavelet analysis for functional MRI. NeuroImage. 2004;23:500–516. doi: 10.1016/j.neuroimage.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 116.Shimizu Y, Barth M, Windischberger C, Moser E, Thurner S. Wavelet-based multifractal analysis of fMRI time series. NeuroImage. 2004;22:1195–1202. doi: 10.1016/j.neuroimage.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 117.Baillet S, Garnero L, Marin G, Hugonin JP. Combined MEG and EEG source imaging by minimization of mutual information. IEEE Trans Biomed Eng. 1999;46:522–534. doi: 10.1109/10.759053. [DOI] [PubMed] [Google Scholar]
- 118.Prince S, Kolehmainen V, Kaipio JP, Franceschini MA, Boas D, Arridge SR. Time-series estimation of biological factors in optical diffusion tomography. Phys Med Biol. 2003;48:1491–1504. doi: 10.1088/0031-9155/48/11/301. [DOI] [PubMed] [Google Scholar]
- 119.Luo H, Puthusserypady S. fMRI data analysis with nonstationary noise models: a Bayesian approach. IEEE Trans Biomed Eng. 2007;54:1621–1630. doi: 10.1109/TBME.2007.902591. [DOI] [PubMed] [Google Scholar]
- 120.Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn Reson Med. 2000;44:162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 121.Birn RM, Diamond JB, Smith MA, Bandettini PA. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. NeuroImage. 2006;31:1536–1548. doi: 10.1016/j.neuroimage.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 122.Devore JL. Probability and Statistics for Engineering and the Sciences. Wadsworth; 1995. Simple linear regression and correlation; pp. 474–522. [Google Scholar]
- 123.Chen H, Yao D, Liu Z. A comparison of gamma and Gaussian dynamic convolution models of the fMRI BOLD response. Magn Reson Imaging. 2005;23:83–88. doi: 10.1016/j.mri.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 124.Zhang Y, Brooks DH, Boas DA. A haemodynamic response function model in spatio-temporal diffuse optical tomography. Phys Med Biol. 2005;50:4625–4644. doi: 10.1088/0031-9155/50/19/014. [DOI] [PubMed] [Google Scholar]
- 125.Cohen ER, Ugurbil K, Kim SG. Effect of basal conditions on the magnitude and dynamics of the blood oxygenation level-dependent fMRI response. J Cereb Blood Flow Metab. 2002;22:1042–1053. doi: 10.1097/00004647-200209000-00002. [DOI] [PubMed] [Google Scholar]
- 126.Boverman G, Fang Q, Carp SA, Miller EL, Brooks DH, Selb J, Moore RH, Kopans DB, Boas DA. Spatio-temporal imaging of the hemoglobin in the compressed breast with diffuse optical tomography. Phys Med Biol. 2007;52:3619–3641. doi: 10.1088/0031-9155/52/12/018. [DOI] [PubMed] [Google Scholar]
- 127.Deneux T, Faugeras O. Using nonlinear models in fMRI data analysis: model selection and activation detection. NeuroImage. 2006;32:1669–1689. doi: 10.1016/j.neuroimage.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 128.Riera JJ, Watanabe J, Kazuki I, Naoki M, Aubert E, Ozaki T, Kawashima R. A state-space model of the hemodynamic approach: nonlinear filtering of BOLD signals. NeuroImage. 2004;21:547–567. doi: 10.1016/j.neuroimage.2003.09.052. [DOI] [PubMed] [Google Scholar]
- 129.Friston KJ, Mechelli A, Turner R, Price CJ. Nonlinear responses in fMRI: the Balloon model, Volterra kernels, and other hemodynamics. NeuroImage. 2000;12:466–477. doi: 10.1006/nimg.2000.0630. [DOI] [PubMed] [Google Scholar]
- 130.Huppert TJ, Allen MS, Diamond SG, Boas DA. Estimating cerebral oxygen metabolism from fMRI with a dynamic multicompartment Windkessel model. Hum Brain Mapp. 2008 doi: 10.1002/hbm.20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Glover GH. Deconvolution of impulse response in event-related BOLD fMRI. NeuroImage. 1999;9:416–429. doi: 10.1002/hbm.20628. [DOI] [PubMed] [Google Scholar]
- 132.Plichta MM, Heinzel S, Ehlis AC, Pauli P, Fallgatter AJ. Model-based analysis of rapid event-related functional near-infrared spectroscopy (NIRS) data: a parametric validation study. NeuroImage. 2007;35:625–634. doi: 10.1016/j.neuroimage.2006.11.028. [DOI] [PubMed] [Google Scholar]
- 133.Schroeter ML, Bucheler MM, Muller K, Uludag K, Obrig H, Lohmann G, Tittgemeyer M, Villringer A, von Cramon DY. Towards a standard analysis for functional near-infrared imaging. NeuroImage. 2004;21:283–290. doi: 10.1016/j.neuroimage.2003.09.054. [DOI] [PubMed] [Google Scholar]
- 134.Hoge RD, Franceschini MA, Covolan RJ, Huppert T, Mandeville JB, Boas DA. Simultaneous recording of task-induced changes in blood oxygenation, volume, and flow using diffuse optical imaging and arterial spin-labeling MRI. NeuroImage. 2005;25:701–707. doi: 10.1016/j.neuroimage.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 135.Steinbrink J, Wabnitz H, Obrig H, Villringer A, Rinneberg H. Determining changes in NIR absorption using a layered model of the human head. Phys Med Biol. 2001;46:879–896. doi: 10.1088/0031-9155/46/3/320. [DOI] [PubMed] [Google Scholar]
- 136.Wolf M, Franceschini MA, Paunescu LA, Toronov V, Michalos A, Wolf U, Gratton E, Fantini S. Absolute frequency-domain pulse oximetry of the brain: methodology and measurements. Adv Exp Med Biol. 2003;530:61–73. doi: 10.1007/978-1-4615-0075-9_7. [DOI] [PubMed] [Google Scholar]
- 137.Brooksby B, Srinivasan S, Jiang S, Dehghani H, Pogue BW, Paulsen KD, Weaver J, Kogel C, Poplack SP. Spectral priors improve near-infrared diffuse tomography more than spatial priors. Opt Lett. 2005;30:1968–1970. doi: 10.1364/ol.30.001968. [DOI] [PubMed] [Google Scholar]
- 138.Li A, Zhang Q, Culver JP, Miller EL, Boas DA. Reconstructing chromosphere concentration images directly by continuous-wave diffuse optical tomography. Opt Lett. 2004;29:256–258. doi: 10.1364/ol.29.000256. [DOI] [PubMed] [Google Scholar]