Abstract
The nuclear fraction of the ProteoExtract subcellular fractionation kit was assessed using frozen rat liver and heart tissue. Fractionation was evaluated by western blot using protein markers for various subcellular compartments and followed up with LC/MS/MS analysis of the nuclear fractions. Of the proteins identified, nuclear proteins were in the minority (less than 15%) and there was poor representation of the various nuclear substructures when compared to liver nuclear isolations using a classical density-based centrifugation protocol. The ProteoExtract kit demonstrated poor specificity for the nucleus and offers limited promise for proteomics investigations of the nuclear subproteome in frozen tissue samples.
Keywords: Fractionation, ProteoExtract, cardiac muscle, liver
Prefractionation strategies allow for in-depth analyses by isolating or enriching a subset of proteins (subproteome) that may otherwise be below the threshold of detection and/or obscured by a complex proteome [1-4]. This is particularly useful when dealing with samples that have a broad dynamic range, where differences of up to 6-10 orders of magnitude can exist between the most and least abundant proteins, such as in serum or cardiac tissue [5, 6]. A major consideration when designing a prefractionation strategy for a proteomic investigation is a protocol's capacity to enrich or purify a particular subproteome as these parameters reflect the extent to which contamination can be tolerated and establishes the limits of data interpretation. Therefore, any new protocols, including those commercially available, must be evaluated to establish the degree of purification and reproducibility of the method for a given sample type.
Several fractionation strategies exist for separating and enriching the various cellular structures and classes of proteins for proteomic investigation [1]. In the case of subcellular fractionation, classical methods involving differential or isopycnic centrifugation have been used to separate and isolate individual organellar or soluble components [7]. Alternatively, a chemical or solubility-based separation can be used to produce fractions of differing protein composition [8-10]. In this study, we evaluate the enrichment and quality of the nuclear subproteome derived from frozen liver and heart tissue samples using the commercially available ProteoExtract subcellular fractionation kit (Calbiochem-EMD Biosciences, San Diego CA, USA). Successive extraction buffers are proposed to isolate four distinct subproteomes - cytosolic, membrane/organelle, nuclear and cytoskeletal - an approach indicative of that used by other extraction-based kits. Our goal was to assess the suitability of the nuclear fraction for use in subsequent proteomic studies. As a reference, proposed nuclear fractions were compared with a well-characterized classic density-based isolation of intact liver nuclei [11, 12].
Tissue extractions were carried out using the ProteoExtract kit on 25-50 mg of frozen rat liver (n=3) or heart (n=3) tissue (Pel-Freez Biologicals, Rogers AR, USA) following the manufacturer's protocol. In brief, fragmented tissue was mixed with 1 ml of cold Extraction Buffer 1 including protease inhibitors and incubated at 4°C for 10 min (all incubations were performed on an end-over-end shaker). Insoluble material was sedimented at 1000×g at 4°C for 10 min and the resulting supernatant, the cytosolic subproteome, was removed and stored. The pellet was mixed with 1 ml of cold Extraction Buffer 2 and incubated for 30 min at 4°C. The insoluble material was sedimented at 6000×g at 4°C for 10 min. The supernatant, the membrane/organelle subproteome, was removed and the pellet mixed with 500 μl of cold Extraction Buffer 3 including 1.5 μl Benzoase to digest DNA. Following 10 min of incubation the insoluble material was sedimented at 7000×g at 4°C for 10 min and the supernatant, the nuclear fraction, was removed. The final fraction, the cytoskeletal subproteome, was obtained by resuspension of the remaining pellet in 500 μl of 25°C Extraction Buffer 4. Fractions were aliquoted and stored at -80°C until further use.
Intact nuclei were isolated from rat liver tissue following the protocol of Jung et al. [11]. 3 g of frozen tissue (n=3) were fragmented in liquid nitrogen and mixed with 40 mL of homogenization buffer A (60 mM KCl, 15 mM NaCl, 0.15 mM spermine, 0.5 mM spermidine, 15 mM HEPES) plus 0.3 M sucrose, 0.2%(v/v) IPEGAL 680, and protease inhibitors then homogenized with 10 strokes in a 40 mL Dounce homogenizer (Kontes, Vineland NJ, USA). The homogenate was passed twice through a 0.9 M sucrose cushion by centrifugation at 2000×g for 10 min at 4°C in a swinging bucket rotor (Eppendorf, Hamburg Germany). The resulting pellet was resuspended and passed though a 1.8 M sucrose cushion by centrifugation for 1 hour at 100000×g. The final nuclei pellet was collected, aliquoted and stored at -80°C until further analysis. For detailed methods see online supplement.
The tissue fractionation was characterized by 1DE western blot probing for protein markers of various subcellular compartments present in both heart and liver tissue. Nuclear enrichment and integrity were tracked using antibodies against histone H3 (H3), a DNA binding protein, and lamin associated polypeptide 1A (LAP1A), a protein associated with the inner nuclear membrane [13, 14]. Non-nuclear compartments were assessed using antibodies to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) a cytosolic protein, ATP synthase subunit β (ATP-β), a mitochondrial protein and caveolin-1 (liver) or 3 (heart) (Cav-1/3), a membrane protein [15-17]. In the case of heart tissue, the myofilament subproteome was assessed using an antibody against myosin heavy chain (MHC) due to the dominance of this subproteome in striated muscle [18]. Western blot signal intensities of proposed nuclear fractions (Figure 1B lanes 9-Density or 3-ProteoExtract) were quantitatively measured and normalized to whole tissue (wt) homogenate lanes for each marker (Figure 1A and B; Table S1 for summary of quantitative data). The relative distribution and reproducibility for each protein marker across the fractions was also determined (Figure S1 and Table S2 in online supplement).
Figure 1. Western blot evaluation of density-based (liver) and ProteoExtract (liver and heart) fractionation.
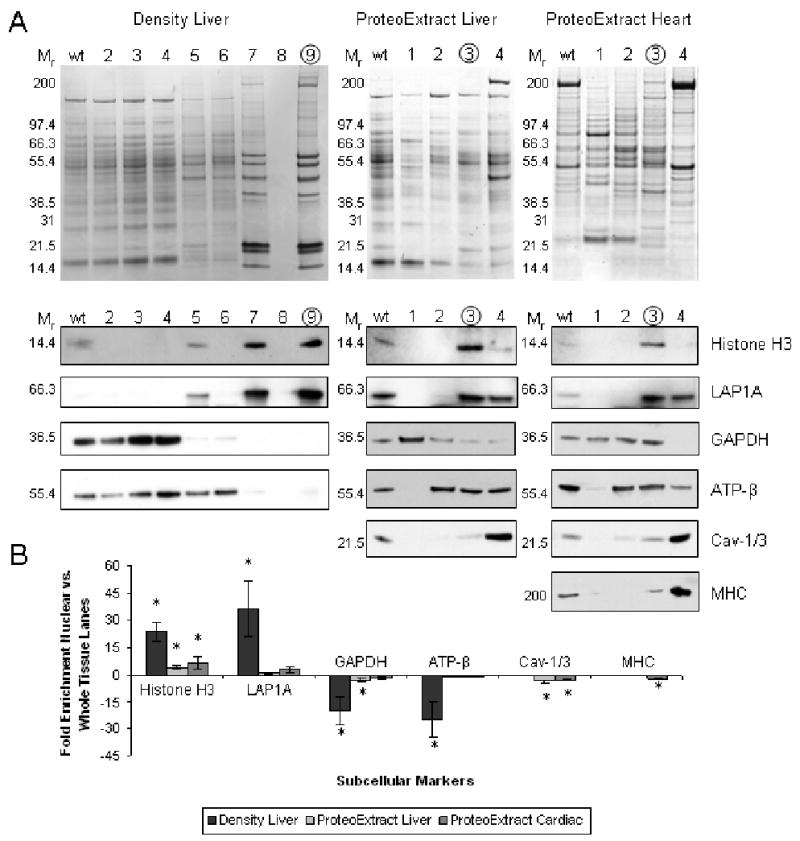
Panel A shows representative 1D Coomassie stained gel of whole tissue (wt) and each of the steps during fractionation protocol (5 μg/lane). Proposed nuclear fractions are circled. Below are equivalent western blots indicating the relative distribution of various subcellular localization markers (1-10 μg/lane): H3 and LAP1A (nuclear), GAPDH (cytosolic), ATP-β (mitochondrial), Cav-1/3 (membrane) and MHC (myofilament). Panel B is the summary of average fold difference between western blot signals for wt and nuclear fraction (n=3). Error bars are ± one standard deviation and * = P<0.05. See Table S1 in online supplement.
Liver nuclear isolations based on organelle density were enriched for the nuclear markers (23.8±5 fold H3, 36.6±15 fold LAP1A) and of equal importance, were depleted in the markers of other cellular compartments (-20±8 fold GAPDH, -25±9 fold ATP-β). However, the ProteoExtract kit was found to have only a modest 4.3±0.8 fold (liver) and a 6.6±3.5 fold (heart) enrichment of H3 in the nuclear fraction. No significant nuclear enrichment was observed for LAP1A and it was found to be distributed between fractions three and four. The presence of LAP1A and Cav-1/3 in the final proposed cytoskeletal fraction may indicate protein interactions persisting throughout the sequential extraction protocol [10]. Both LAP1A (potentially via its associated nuclear lamina) and Cav-1/3 are known to make connections with the cytoskeleton possibly resisting earlier solubilization [15, 19]. The cytosolic and mitochondrial markers were found to be distributed across multiple fractions.
To further characterize the constituents of these subproteomes, a one dimensional electrophoresis (1DE) and LC/MS/MS analysis was performed on the nuclear fractions obtained from the ProteoExtract (liver and heart) and density-based (liver) protocols. Large format (18cm) 1DE was chosen to separate the proteins in the fractions because of its ability to resolve abundant, hydrophobic, high molecular weight and very basic proteins [20] (Figure 2A, gel map; Table S3 in online supplement for protein identifications per band and MS data). Gel bands or regions were excised across gels from all three extracts to provide a survey of the proteins present in each fraction. Dominant protein bands present in the density-based isolation of liver nuclei were used as a guide to ensure that molecular weight ranges of known nuclear proteins were included. Subcelluar localizations were assigned to each protein following MS/MS analysis. This was largely determined by annotation in the Uniprot database (www.uniprot.org). In some cases proteins were annotated with multiple subcellular localizations in the database including proteins known to translocate in and out of the nucleus. Any protein with a nuclear localization was assigned to the nucleus. This was done to ensure a maximum number of potential nuclear proteins would be considered and to mitigate any possible bias associated with the enrichment of any particular subset of nuclear proteins by either of the methods. For all other proteins, assignment was based on the first listing present in the database. If no localization was available, a determination was made based on consensus from the subcellular localization prediction programs WoLF PSORT [21], pTARGET [22] and LOCSVMpsi [23] (see online supplement and Figure S2 for prediction schema).
Figure 2. 1DE and LC/MS/MS analysis of density-based (liver) and ProteoExtract (liver and heart) derived nuclear extracts.
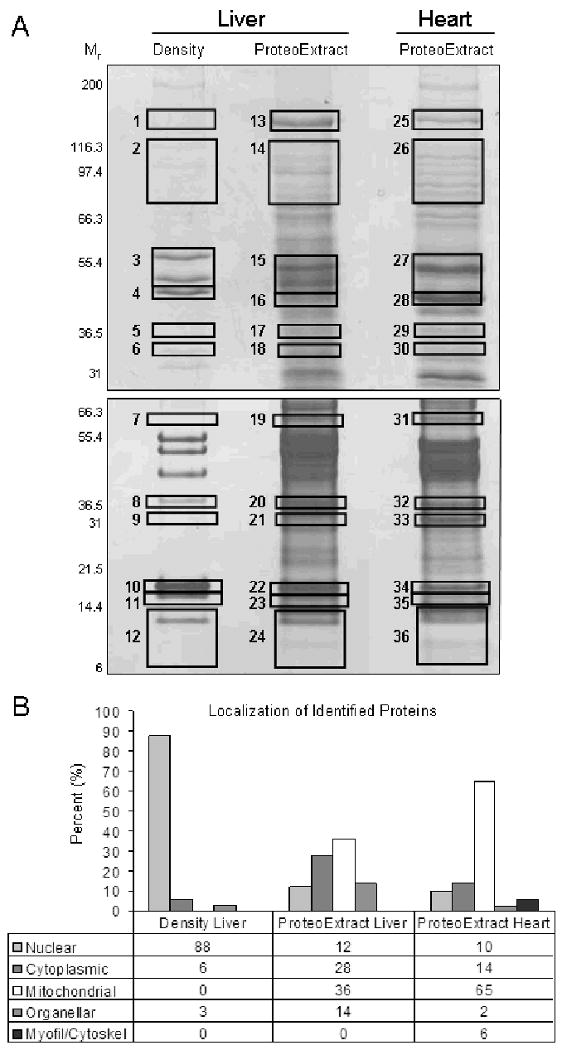
Panel A is the 1DE gel map, 100 μg of each nuclear extract was separated by an 8% (upper) and a 4-12% (lower) acrylamide gel (n=2). Boxes indicate gel bands/regions which were analyzed by LC/MS/MS, see Table S3 in online supplement for list of protein identifications per gel band. Panel B is a summary of the subcellular localization assignment for non-redundant proteins identified in each fractionation, see online supplement for details on subcellular assignment.
In liver nuclear isolations based on organelle density, 88% (29 of 33) of the proteins identified were of known nuclear origin (Figure 2B). This included proteins from a variety of nuclear substructures, including DNA binding proteins, nuclear lamina, nucleolus and the splicing factors (Table 1). The same was not true for either tissue using the ProteoExtract kit where the nuclear enrichment was found to be limited and incomplete. The majority of proteins identified were either mitochondrial or cytosolic and only 12% (6 of 50) of liver and 10% (4 of 43) of cardiac proteins identified were determined to reside in the nucleus. Of the nuclear proteins identified, many were found to be DNA binding histone proteins while other nuclear components were poorly represented. A possible explanation for these results is the lack of any physical homogenization in the protocol. Without any significant disruption, portions of cellular compartments can be shielded from the various detergents during the solubilization steps. In that case, carry over would occur between extractions producing imprecise fractionation.
Table 1. Summary of nuclear proteins identified from liver and heart tissue nuclear isolations by 1DE/MS/MS analysis.
| Nuclear Protein | Nuclear compartment | Liver | Heart | |
|---|---|---|---|---|
| Density | PE.a | PE. | ||
| Histone H1 | DNA binding | ✓b | ✓ | ✓ |
| Histone H2B | DNA binding | ✓ | ✓ | ✓ |
| Histone H2A | DNA binding | ✓ | -c | ✓ |
| Histone H4 | DNA binding | ✓ | ✓ | ✓ |
| Histone H3 | DNA binding | ✓ | - | - |
| DNA-directed RNA polymerases III | DNA binding | ✓ | - | - |
| DNA helicase II | DNA binding | ✓ | - | - |
| Lamin A | nuclear matrix | ✓ | - | - |
| Lamin B1 | nuclear matrix | ✓ | - | - |
| Cytokeratin 8 | nuclear matrix | ✓ | - | - |
| Cytokeratin 18 | nuclear matrix | ✓ | - | - |
| Brix domain containing 1 | nucleolus | ✓ | - | - |
| Fibrillarin | nucleolus | ✓ | - | - |
| hnRNP A2/B1 | hnRNPs/splicing factor | - | ✓ | - |
| hnRNP C | hnRNPs/splicing factor | ✓ | - | - |
| hnRNP M | hnRNPs/splicing factor | ✓ | - | - |
| Small nuclear ribonucleoprotein B | hnRNPs/splicing factor | ✓ | - | - |
| Splicing factor 3b, subunit 1 | hnRNPs/splicing factor | ✓ | - | - |
PE. - ProteoExtract kit.
‘✓’indicates that the protein is present in fraction.
‘-’ indicates that protein was not detected in fraction
The primary goal of prefractionation is to isolate or enrich a subset of proteins, often from a particular organelle. The more specific the prefractionation, i) the higher the probability will be for observing low abundance proteins, and ii) the more confidence in being able to associate a given protein with a particular organelle. Although there is no minimum standard for prefractionation, the quality and reproducibility of the method needs to be established for different tissues. In this study, we have assessed the commercially available ProteoExtract protocol for the isolation of nuclear subproteome from frozen liver and found it to be lacking in specificity compared to the well established centrifugation method. We further investigated the kit using frozen heart tissue which was equally unsuccessful. The ProteoExtract subcellular fractionation kit was found to display a very limited enrichment of nuclear proteins and those present gave an incomplete representation of the nuclear components. It is likely the nuclear membrane, structural and matrix proteins were lost to the other fractions based on alternate solubility properties resulting in a proposed nuclear fraction that consisted of only a minority of DNA binding histones (released by DNA digestion with benozoase) amid an assortment of contaminating proteins. Given these findings, we conclude that this kit displays poor specificity for the nucleus and therefore offers little promise for in-depth proteomic analysis of nuclear subproteome from frozen tissue samples.
Supplementary Material
Acknowledgments
The authors would like to thank the following funding sources: JVE: The National Heart Lung Blood Institute Proteomic Initiative (contract NO-HV-28120), the Donald P. Amos Family Foundation, and the NIH (grants P01HL081427 and P01HL077180). We would also like to thank Bob Cole and Dawn Chen at the Johns Hopkins Mass Spectrometry and Proteomic Facility for their assistance.
References
- 1.Yates JR, 3rd, Gilchrist A, Howell KE, Bergeron JJ. Proteomics of organelles and large cellular structures. Nat Rev Mol Cell Biol. 2005;6:702–714. doi: 10.1038/nrm1711. [DOI] [PubMed] [Google Scholar]
- 2.Stasyk T, Huber LA. Zooming in: fractionation strategies in proteomics. Proteomics. 2004;4:3704–3716. doi: 10.1002/pmic.200401048. [DOI] [PubMed] [Google Scholar]
- 3.Huber LA, Pfaller K, Vietor I. Organelle proteomics: implications for subcellular fractionation in proteomics. Circ Res. 2003;92:962–968. doi: 10.1161/01.RES.0000071748.48338.25. [DOI] [PubMed] [Google Scholar]
- 4.Dreger M. Subcellular proteomics. Mass Spectrom Rev. 2003;22:27–56. doi: 10.1002/mas.10047. [DOI] [PubMed] [Google Scholar]
- 5.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 6.Corthals GL, Wasinger VC, Hochstrasser DF, Sanchez JC. The dynamic range of protein expression: a challenge for proteomic research. Electrophoresis. 2000;21:1104–1115. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1104::AID-ELPS1104>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 7.Foster LJ, de Hoog CL, Zhang Y, Xie X, et al. A mammalian organelle map by protein correlation profiling. Cell. 2006;125:187–199. doi: 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 8.Abdolzade-Bavil A, Hayes S, Goretzki L, Kroger M, et al. Convenient and versatile subcellular extraction procedure, that facilitates classical protein expression profiling and functional protein analysis. Proteomics. 2004;4:1397–1405. doi: 10.1002/pmic.200300710. [DOI] [PubMed] [Google Scholar]
- 9.Neverova I, Van Eyk JE. Application of reversed phase high performance liquid chromatography for subproteomic analysis of cardiac muscle. Proteomics. 2002;2:22–31. [PubMed] [Google Scholar]
- 10.Ramsby ML, Makowski GS, Khairallah EA. Differential detergent fractionation of isolated hepatocytes: biochemical, immunochemical and two-dimensional gel electrophoresis characterization of cytoskeletal and noncytoskeletal compartments. Electrophoresis. 1994;15:265–277. doi: 10.1002/elps.1150150146. [DOI] [PubMed] [Google Scholar]
- 11.Jung E, Hoogland C, Chiappe D, Sanchez JC, et al. The establishment of a human liver nuclei two-dimensional electrophoresis reference map. Electrophoresis. 2000;21:3483–3487. doi: 10.1002/1522-2683(20001001)21:16<3483::AID-ELPS3483>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 12.Blobel G, Potter VR. Nuclei from rat liver: isolation method that combines purity with high yield. Science. 1966;154:1662–1665. doi: 10.1126/science.154.3757.1662. [DOI] [PubMed] [Google Scholar]
- 13.Maison C, Pyrpasopoulou A, Theodoropoulos PA, Georgatos SD. The inner nuclear membrane protein LAP1 forms a native complex with B-type lamins and partitions with spindle-associated mitotic vesicles. Embo J. 1997;16:4839–4850. doi: 10.1093/emboj/16.16.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Usachenko SI, Bradbury EM. Histone-DNA contacts in structure/function relationships of nucleosomes as revealed by crosslinking. Genetica. 1999;106:103–115. doi: 10.1023/a:1003785031470. [DOI] [PubMed] [Google Scholar]
- 15.Thomsen P, Roepstorff K, Stahlhut M, van Deurs B. Caveolae are highly immobile plasma membrane microdomains, which are not involved in constitutive endocytic trafficking. Mol Biol Cell. 2002;13:238–250. doi: 10.1091/mbc.01-06-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanley WC, Recchia FA, Lopaschuk GD. Myocardial substrate metabolism in the normal and failing heart. Physiol Rev. 2005;85:1093–1129. doi: 10.1152/physrev.00006.2004. [DOI] [PubMed] [Google Scholar]
- 17.Gaballo A, Zanotti F, Papa S. Structures and interactions of proteins involved in the coupling function of the protonmotive F(o)F(1)-ATP synthase. Curr Protein Pept Sci. 2002;3:451–460. doi: 10.2174/1389203023380558. [DOI] [PubMed] [Google Scholar]
- 18.Kostin S, Hein S, Arnon E, Scholz D, et al. The cytoskeleton and related proteins in the human failing heart. Heart Fail Rev. 2000;5:271–280. doi: 10.1023/A:1009813621103. [DOI] [PubMed] [Google Scholar]
- 19.Houben F, Ramaekers FC, Snoeckx LH, Broers JL. Role of nuclear lamina-cytoskeleton interactions in the maintenance of cellular strength. Biochim Biophys Acta. 2007;1773:675–686. doi: 10.1016/j.bbamcr.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 20.Lopez JL. Two-dimensional electrophoresis in proteome expression analysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2006 doi: 10.1016/j.jchromb.2006.11.049. [DOI] [PubMed] [Google Scholar]
- 21.Horton P, Park KJ, Obayashi T, Nakai K. the 4th Anual Asian Bioinformatics Conference APBC06. 2006:39–48. [Google Scholar]
- 22.Guda C, Subramaniam S. pTARGET [corrected] a new method for predicting protein subcellular localization in eukaryotes. Bioinformatics. 2005;21:3963–3969. doi: 10.1093/bioinformatics/bti650. [DOI] [PubMed] [Google Scholar]
- 23.Xie D, Li A, Wang M, Fan Z, et al. LOCSVMPSI: a web server for subcellular localization of eukaryotic proteins using SVM and profile of PSI-BLAST. Nucleic Acids Res. 2005;33:W105–110. doi: 10.1093/nar/gki359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


