Abstract
The rotation of the earth on its axis influences the physiology of all organisms. A highly conserved set of genes encoding the “core circadian regulatory proteins” (CCRP) has evolved across species. The CCRP acts through transcriptional and post-transcriptional mechanisms to direct the oscillatory expression of genes essential for key metabolic events. In addition to the light:dark cycle, the CCRP expression can be entrained by changes in feeding and physical activity patterns. While mammalian CCRP were originally associated with the central clock located within the suprachiasmatic nucleus of the brain, there is a growing body of evidence documenting the presence of the CCRP in peripheral tissues. It is now evident that the CCRP play a role in regulating the proliferation, differentiation, and function of adult stem cells in multiple organs. This concise review highlights findings concerning the role of the CCRP in modulating the adult stem cell activities. Although the manuscript focuses on hematopoietic stem cells (HSCs), bone marrow-derived mesenchymal stem cells (BMSCs), adipose-derived stem cells (ASCs) and cancer stem cells, it is likely that the contribution of the CCRP merits consideration and evaluation in all stem cell pathways.
Keywords: Adipose-derived Stem Cell, Bone Marrow-derived Mesenchymal Stem Cell, Cancer Stem Cell, Circadian, Hematopoietic Stem Cell
Introduction
Why ask questions about the role of circadian biology in the regulation of adult stem cell function?
Biochemical and molecular tools, model organisms, and transgenic and knock out gene technology have promoted rapid gains in our understanding of adult stem cells. With these approaches, we have gained insight into fundamental questions relating to cell development, migration, and differentiation fate. Nevertheless, the majority of adult stem cell studies have been performed under static conditions. By necessity, cells are examined in a single point in time, rather than in a kinetic manner. As a result, it is possible that important information concerning stem cell dynamics and their regulatory pathways have been overlooked.
Advances from the field of circadian biology may provide the tools necessary to address this question. Using a variety of model organisms, sleep researchers and circadian biologists have defined a set of core circadian regulatory proteins (CCRP) that are highly conserved throughout the phylogenetic tree [Hirota and Fukada, 2004; Lowrey and Takahashi, 2004]. The levels of these proteins and their activities oscillate rhythmically over a 24 hour period [Lowrey and Takahashi, 2004]. Although the mammalian CCRPs were first defined in the context of the body’s central clock, the suprachiasmic nucleus of brain, they have since been detected in peripheral tissues as well [Yoo et al., 2004]. This concise review highlights the growing body of literature linking the molecular components of the body’s circadian clock to adult stem cell physiology and pathology. Currently, we focus on growth factors, receptors, and signal transduction pathways as mechanisms for manipulating adult stem cell proliferation, differentiation, and function; however, if timing really does matter, we may be able to exploit components of the body’s own molecular clock to achieve these same goals.
Overview of Common Circadian Regulatory Proteins (CCRP)
What drives the circadian clock?
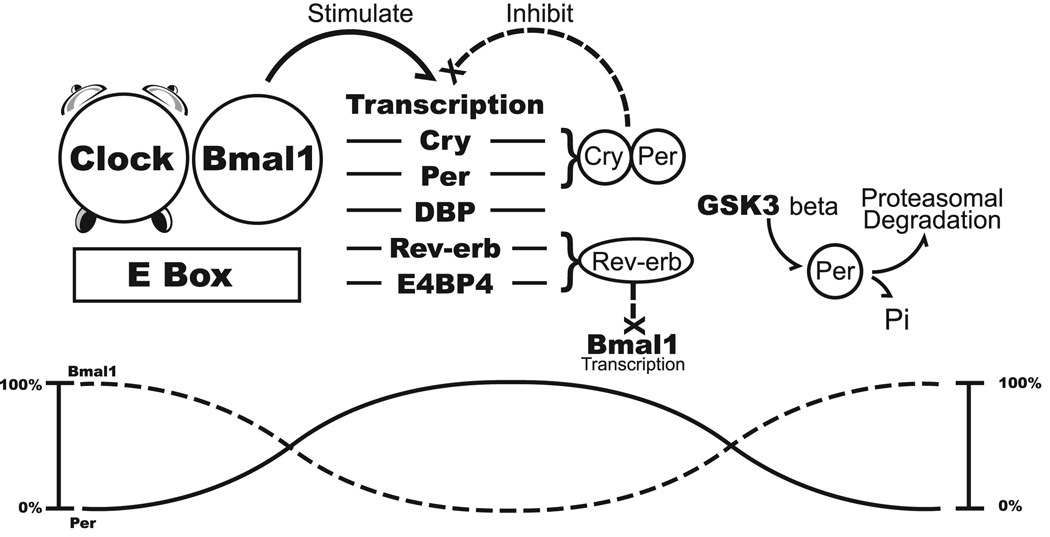
Molecular biological studies of circadian mutations in plants, Drosophila, and mammalian systems have identified the biochemical components of the central circadian oscillator (Figure 1) [Hirota and Fukada, 2004; Lowrey and Takahashi, 2004]. Transcriptional regulatory proteins belonging to the basic helix-loop-helix/Per-ARNT-Singleminded (bHLH-PAS) domain family interact with each other and downstream target proteins to create a self-perpetuating, rhythmic pattern of gene transcription (Table 1) [Hirota and Fukada, 2004; Lowrey and Takahashi, 2004]. The proteins CLOCK (or its ortholog, neural PAS domain 2 or NPAS2) and brain and muscle ARNT-like 1 (BMAL1) form heterodimers that act as positive transcriptional regulators. In contrast, heterodimers of the PERIOD (Per1, Per2, Per3) and CRYPTOCHROME (Cry1, Cry2) family members serve in a negative feedback capacity. As a result of these self-contained feedback loops, the circadian protein levels oscillate in a rhythmic manner.
Figure 1.
Table 1.
Circadian Rhythm Regulatory Genes/Proteins
| bHLH-PAS Family Members (basic Helix Loop Helix – Period/Arnt/Simpleminded) |
bHLH-PAS Family Post-Translational Modifiers and/or Putative Downstream Targets |
|---|---|
| BMAL 1 & 2 (Bone and Muscle Arnt- Like) |
CK1ε (Casein Kinase 1 ε) |
| Clock | Cry 1,2 (Cryptochrome) |
| DEC 1 & 2 (Differentially Expressed in Chondrocytes) |
DBP (Albumin D site binding protein) |
| EPAS 1 (Endothelial PAS) | E4BP4 |
| NPAS1 & 2 (Neuronal PAS) | GSK3β (Glycogen synthase kinase 3β) |
| Per 1,2,3 (Period) (PAS domain only) | Rev-Erb α & β |
The positive transcriptional regulators direct expression of immediate downstream targets, such as the albumin D site binding protein (DBP) which exerts a further transcriptional activating role. In contrast, related downstream targets such as Rev-erbα, Rev-erbβ, and Retinoid Orphan Receptor α (RORα), or other transcription factor families (E4BP4/NFIL3), repress transcription [Hirota and Fukada, 2004; Lowrey and Takahashi, 2004] [Duez and Staels, 2008; Yin and Lazar, 2005; Yin et al., 2006]. The serine/threonine kinases, casein kinase Iε (CK1 ε) and glycogen synthase kinase 3β (GSK3 β), phosphorylate BMAL1, PER, and other proteins. Once modified, the proteins are targeted to the ubiquitin/proteasomal pathway for degradation [Ko and Edery, 2005; Lee et al., 2008].
Circadian intersections with the stem cell regulatory highway
The CCRP intersects with a number of identified adult stem cell regulatory pathways. For example, the PAS domain family is notable for other protein members relevant to stem cell biology. The hypoxia inducible factor 1 (HIF-1) belongs to the PAS domain family and heterodimerizes with the aryl hydrocarbon receptor nuclear translocator (ARNT) to form a transcriptional regulatory complex. Under hypoxic conditions, HIF-1 induces expression of vascular endothelial growth factor (VEGF), stromal derived factor-1, and related factors known to modulate adult stem cell migration and differentiation. There is a growing body of literature evaluating the relationship between tissue oxygen tension, stem cell number, and function. Indeed, it appears that hypoxic conditions within a stem cell’s microenvironment or niche may be critical to the preservation of the stem cell state [D'Ippolito et al., 2006; Fehrer et al., 2007; Grayson et al., 2006]. The shared protein structure between circadian and hypoxic transcriptional regulators, together with the recognized importance of hypoxia in stem cell biology, suggests that a similar link remains to be defined between circadian and stem cell biology.
It is well established that the nuclear hormone receptor family, acting through the glucocorticoid, retinoid, estrogen, and related receptors, plays an influential role in directing and determining adult stem cell fate. Furthermore, the expression of the mRNAs encoding these receptor exhibits circadian rhythmicity [Lemberger et al., 1996] [Yang et al., 2006]. The CCRP involves the NHR family members Rev-erb and ROR [Akashi and Takumi, 2005; Duez and Staels, 2008; Triqueneaux et al., 2004; Yin et al., 2006]. These proteins exert circadian modulating effects in what may prove to be a ligand dependent manner. Although Rev-erb had been characterized as an orphan receptor, recent studies have determined that heme is its natural ligand [Burris, 2008; Raghuram et al., 2007; Rogers et al., 2008; Yin et al., 2007]. This ubiquitous iron containing compound is capable of activating Rev-erb dependent transcription in in vitro models. Indeed, heme injection into mice has profound effects on the circadian biology of peripheral tissues, causing a phase shift in the expression of both the positive and negative CCRP genes in the liver [Kaasik and Lee, 2004].
The advent of induced pluripotent stem cells has underscored the importance of epigenetic mechanisms in adult stem cell biology. The introduction of transcription factors such as Oct 4, Sox2, Myc, and KLF4 have endowed adult stem cells with pluripotential properties similar to those demonstrated by embryonic stem cells [Takahashi et al., 2007; Wernig et al., 2007]. This has been associated with altered levels of histone acetyl transferase activity. Recently, valproic acid and related small molecule inhibitors of histone deacetylases (HDACs) have used to substitute for or complement these transgenic methods with success [Huangfu et al., 2008]. At least one CCRP protein, Clock, has been shown to possess histone acetyl transferase activity [Doi et al., 2006]. This chromatin modifying activity is an essential feature of the clock protein’s circadian function [Doi et al., 2006]. Furthermore, recent studies have determined that the NAD+ dependent deacetylase, SIRT1, is responsible for the deacetylation of Period 2 [Asher et al., 2008]. This histone deacetylase enzyme plays a prominent role in regulating the oscillatory expression profile of multiple CCRP genes [Nakahata et al., 2008]. Likewise, the disruption of HDAC interaction with the nuclear receptor co-repressor (NCoR) has been found to disrupt circadian oscillations and metabolic events in murine models [Alenghat T, 2008]. Together, these studies demonstrate a close relationship between chromatin remodeling and circadian mechanisms.
Finally, GSK3β has profound effects on stem cell biology through its role in the Wnt signal transduction pathway [Baksh et al., 2007; Baksh and Tuan, 2007; Etheridge et al., 2004; Gregory et al., 2005; Nemeth and Bodine, 2007; Sato et al., 2004]. Studies have demonstrated that GSK3β inhibition and subsequent modification of β-catenin phosphorylation modulate bone marrow hematopoietic and mesenchymal stem cell differentiation and function [Trowbridge et al., 2006]. Likewise, GSK3β is responsible for phosphorylation and turnover of Period and related CCRP proteins [Akashi et al., 2002]. Inhibition of GSK3 using lithium chloride has been shown to lengthen the circadian period in animal studies [Iwahana et al., 2004; Padiath et al., 2004]. Thus, the CCRP intersects with multiple established adult stem cell regulatory pathways at the biochemical and protein level.
Stem Cell Dysfunction in CCRP Mutant Mice
Murine models with mutations or deficiencies in critical CCRP genes have revealed important insights into circadian biology [Antoch et al., 2008; King et al., 1997; Kondratov et al., 2006; Turek et al., 2005]. In many of these models, gene alterations are systemic and not limited to a single organ or tissue type. Consequently, they cannot always be used to distinguish between central versus peripheral circadian mechanisms. Nevertheless, these animals have provided valuable experimental tools. Among the best studied models are the Clock mutant mice which display arrhythmic circadian biology based on activity and biomarker evaluation [King et al., 1997; Turek et al., 2005]. These mice are prone to abnormalities directly or indirectly related to metabolism and adipose tissue function. Clock deficient mice are prone to hyperphagia, hyperinsulinemia, hyperglycemia, dyslipidemia, and obesity [Turek et al., 2005]. In response to radiation damage, the Clock mutant mice exhibit a phenotype of accelerated aging associated with reduced growth rates, cataract development, graying of the hair, and altered cell cycle dynamics [Antoch et al., 2008]. Bmal1 deficient mice exhibit a similar phenotype. Relative to wild type controls, these mice exhibit a shorter life span and increased atrophy of the bone, fat, and skeletal muscle with advancing age [Kondratov et al., 2006]. The mice show evidence of tendon calcification, consistent with abnormalities in the function and differentiation of the tendon stem cells [Bi et al., 2007; Kondratov et al., 2006]. Further evidence from Period deficient mice will be presented in the Cancer Stem Cell section below.
CCRP Regulation of Adult Stem Cell Models
Hematopoietic Stem Cells (HSC)
Studies from the 1950’s and earlier first provided evidence of circadian rhythmicity in hematopoietic cell numbers [Halberg et al., 1953a; Halberg and Ulstrom, 1952; Halberg and Visscher, 1950; Halberg et al., 1953b; Halberg F, 1953]. Investigations by Halberg and others demonstrated that the number of circulating eosinophils in multiple murine strains, canine models, and human infants varied as a function of time of day. These findings were extended by Sharkis et al.[Sharkis et al., 1971; Sharkis et al., 1974], Aardal, Abrahamsen, Laerum, and their colleagues in Bergen, Norway [Aardal, 1984; Aardal and Laerum, 1983; Abrahamsen et al., 1997; Sletvold and Laerum, 1988a; Sletvold et al., 1991; Smaaland et al., 2002; Tsinkalovsky et al., 2006; Tsinkalovsky et al., 2005; Tsinkalovsky et al., 2007], Quesenberry [D'Hondt et al., 2004], and others [Baudoux et al., 1998; Bourin et al., 2002; Chen et al., 2000; Haus et al., 1983; Haus and Smolensky, 1999; Wood et al., 1998] who applied flow cytometric analyses, colony forming unit (CFU), and related assays to monitor circulating hematopoietic cell numbers and cell cycle. The circadian oscillation of hematopoietic stem cell-derived erythroid, myeloid, and lymphoid populations were demonstrated as well as those of circulating endothelial progenitor cells [Thomas et al., 2008]. Svetvold, Laerum, and colleagues demonstrated that the circadian oscillation of hematopoietic lineages was age dependent [Sletvold and Laerum, 1988b; Sletvold et al., 1988a; Sletvold et al., 1988b]. While young mice (3 months of age) displayed robust rhythmicity of splenic-CFU and granulocyte-monocyte-CFU, independent of season, the amplitude and peak levels declined in aged mice (26 months of age) [Sletvold and Laerum, 1988b; Sletvold et al., 1988a; Sletvold et al., 1988b]. Correlation between cell numbers and cell cycle indicated that common mechanisms regulated hematopoietic events in the marrow microenvironment. These observations were followed at the molecular level by direct measurements of CCRP gene expression profiles in circulating blood cells [Chen et al., 2000; Fukuya et al., 2007; Sun et al., 2006; Tsinkalovsky et al., 2006]. These studies indicated that the Cry and Per genes exhibited a robust circadian oscillation in peripheral blood cells. Analyses were extended to the examination of bone marrow CD34+ cells obtained [Tsinkalovsky et al., 2005; Tsinkalovsky et al., 2007]. The CD34+ cells displayed a peak level in early morning hours and their levels of Cry and Per genes exhibited a robust circadian oscillation; in contrast, Bmal1 did not. A similar profile has been reported in peripheral blood leukocytes [Chen et al., 2000; Fukuya et al., 2007]. Consistent with these observations, circulating levels of hematopoietic growth factors such as Granulocyte-Colony Stimulating Factor, Granulocyte-Monocyte Colony Stimulating Factor, Tumor Necrosis Factor, and Interleukins 2, 6, and 10 were also observed to display circadian oscillations [Abdelaal et al., 2000; Dincol et al., 2000; Sothern et al., 1995; Young et al., 1995].
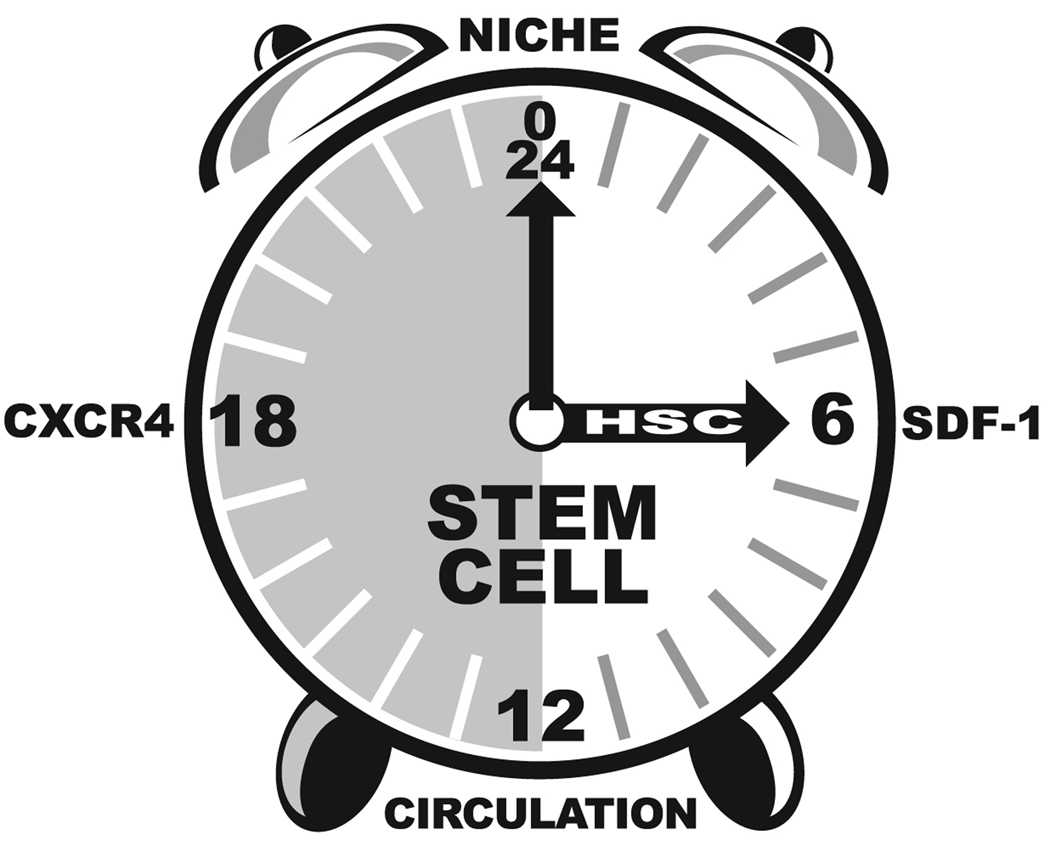
The circadian dynamics of bone marrow hematopoiesis have been linked to the signaling of neural and bone marrow cell derived catecholamines [Maestroni et al., 1998]. Recently, the circadian oscillation in circulating hematopoietic stem cell number have been correlated with circulating levels of stromal derived factor 1 (SDF-1 or CXCL12) in murine models (Figure 2) [Lucas et al., 2008; Mendez-Ferrer et al., 2008]. This dynamic oscillation in HSC levels was absent in Bmal1−/− mice [Mendez-Ferrer et al., 2008]. Based in part on this observation, Mendez-Ferrer et al. hypothesize that the suprachiasmatic nucleus transmit signals to the bone marrow microenvironment via the β-adrenergic system to directly regulate SDF-1 levels and subsequent HSC release [Mendez-Ferrer et al., 2008]. It is of interest that this phenomenon appears to be species dependent [Lucas et al., 2008]. In contrast to nocturnal mice, the timing of the HSC release and SDF-1 expression in humans is reversed relative to the light dark cycle [Lucas et al., 2008; Mendez-Ferrer et al., 2008]. Indeed, SDF-1 and its receptor CXCR4 are likely to play a critical role in determining the circadian dynamics of HSCs; however, in light of the fact that up to 25% of the transcriptome in peripheral tissues displays a circadian oscillation [Ptitysn AA, 2006; Zvonic et al., 2006], it is likely that the expression profile of multiple growth factors, receptors, and related metabolic enzymes will exhibit circadian characteristics.
Figure 2.
Bone Marrow-derived Mesenchymal Stem Cells (BMSC)
Circadian mechanisms influence the other stem cell constituent of the bone marrow microenvironment, the bone mesenchymal stem cell (BMSC), and its progeny, the adipocytes, osteoblasts, and stromal cells. The BMSCs serve in multiple capacities. In the context of hematopoiesis, the BMSCs express surface adhesion proteins such as β1 intergrin (CD29), hyaluronate receptor (CD44), and vascular cell adhesion molecule (VCAM-1) that sequester the HSCs and their lineage committed progeny [Kincade, 1991; Kincade et al., 1989; Simmons et al., 1992; Yin and Li, 2006]. By anchoring the HSCs, the BMSCs serve as a stromal feeder layer or nurse cell for hematopoietic events [Kincade et al., 1989]. Locally, the BMSCs release cytokines and growth factors that can maintain the HSCs in a stem-like state, stimulate proliferation, or promote differentiation along the various hematopoietic lineage pathways [Gimble et al., 1989; Kincade, 1991; Kincade et al., 1989; Yin and Li, 2006]. Systemically, the circulating levels of these growth factors fluctuate in a circadian manner. Well established examples include Granulocyte Colony Stimulating Factor and Interleukin 6 which oscillate in a time dependent manner [Abdelaal et al., 2000; Dincol et al., 2000; Sothern et al., 1995; Young et al., 1995]. Furthermore, the MSCs are themselves multipotent and can differentiate into adipocytes, chondrocytes, and osteoblasts as well as other lineages [Gimble et al., 1990; Gimble et al., 1996; Gimble et al., 2006].
Schibler and his colleagues performed pioneering work demonstrating that adherent rat fibroblast cells in culture contain an intact and operational molecular circadian clock [Balsalobre et al., 2000a; Balsalobre et al., 1998; Balsalobre et al., 2000b; Brown et al., 2005]. Their initial studies found that the CCRP in confluent and quiescent fibroblasts was synchronized following exposure to a 2 hr dexamethasone or 30% serum shock [Balsalobre et al., 1998]. Over the next 48 hrs, the levels of Cry, Per, DBP, and Rev-Erb mRNAs oscillated with a period of ~24 hrs. When human dermal fibroblasts were transduced with viral vectors containing a circadian gene promoter/luciferase reporter construct, the luciferase activity levels could be entrained in vitro [Brown et al., 2005; Brown et al., 2008]. Following dexamethasone stimulation, the human cells expressed luciferase under the control of the murine Per2 promoter with a period of 23 to 26 hrs. Indeed, the period length could be correlated with the reported sleep/awakening patterns of the fibroblast cell donors [Brown et al., 2008].
The BMSCs display a pattern of CCRP synchronization similar to fibroblast models [Wu X, 2008]. When human or murine BMSCs were synchronized with dexamethasone, multiple CCRP genes displayed an oscillatory expression profile [Wu X, 2008]. These included Bmal1, Cry 1, DBP, Per 2/3, and Rev-erbα/β. Furthermore, the presence of the GSK3β inhibitor, lithium chloride, shifted the acrophase or time of peak expression for all genes by ~4 hrs [Wu X, 2008]. For BMSCs obtained from male mice, lithium chloride significantly increased the length of the period of gene expression; otherwise, no gender related differences were noted [Wu X, 2008]. In vivo studies of bone and related in vitro analyses of osteoblasts suggest that BMSCs and their progeny exhibit circadian dependent functionality [Fu et al., 2005; Meyer et al., 2000; Zvonic et al., 2007]. In transcriptomic analyses of the murine calvarial bone, ~25% of the expressed mRNAs exhibited a circadian oscillation [Zvonic et al., 2007]. This included a number of gene families directly associated with osteoblast function. Furthermore, murine models deficient in the Cry or Per genes exhibited increased bone mass relative to their controls [Fu et al., 2005]. This was attributed to centrally mediated clock effects on sympathetic signaling and osteoblast proliferation [Fu et al., 2005]. Likewise, mice deficient in RORα expression display skeletal abnormalities [Meyer et al., 2000]. Together, these studies indicate that both central and peripheral clocks directly regulate the biology of BMSCs.
Adipose-derived Stem Cells (ASC)
Adipose tissue contains adipose-derived stem cells (ASCs) that resemble BMSC with respect to differentiation potential, immunophenotype, immunogenicity, proteome, and transcriptome [Gimble and Guilak, 2003; Gimble et al., 2007]. In the undifferentiated state, the expression of CCRP genes by ASCs was similar to that of BMSCs [Wu X, 2007]. In vitro, the ASC profiles of Bmal1, Cry, Per, DBP, and Rev-erb were synchronized following exposure to dexamethasone, thiazolidinedione, or serum shock [Wu X, 2007]. Furthermore, the presence of lithium chloride increased the period of Rev-erbα and β gene expression by 4 hrs in the ASCs [Wu X, 2007]. While the adipocyte differentiated ASCs displayed an overall CCRP response profile similar to the undifferentiated cells, the period of expression was prolonged by 1 to 6 hrs, depending on the synchronizing agent [Wu X, 2007]. Consistent with these observations, individual CCRP genes have been implicated directly in the adipocyte transcriptional regulatory pathway [Shimba et al., 2005; Shimba et al., 2004]. In 3T3-L1 murine pre-adipocytes, knock down of the Bmal1 mRNAs by RNAi inhibited adipogenesis based on morphological criteria [Shimba et al., 2005]. Complimentary gain of function studies demonstrated that over-expression of Bmal1 increased expression of multiple adipogenic and lipogenic genes [Shimba et al., 2005]. Furthermore, transfection studies demonstrated that Bmal1 drove transcription from both an SREBP1α and Rev-erbα promoter construct [Shimba et al., 2005]. The related CCRP, EPAS1, displayed similar adipogenic regulatory function [Shimba et al., 2004]. Over expression of EPAS1 promotes adipogenesis in 3T3 fibroblasts which, unlike 3T3-L1 cells, are poorly able to differentiate along the adipocyte pathway [Shimba et al., 2004]. In contrast, the CCRP Dec 1 has been found to inhibit adipogenesis by modulating the association of the transcription factor, C/EBPβ, with a histone deacetylase [Gulbagci et al., 2008]. This , in turn, down regulates the expression of adipogenic master-regulators, C/EBPα and PPARγ2, which both lie downstream of C/EBPβ [Gulbagci et al., 2008]. Finally, both Rev-erbα and Rev-erbβ were initially identified as late markers gene markers of adipogenesis and have since been shown to regulate adipogenesis at the transcriptional level [Chawla and Lazar, 1993; Yin and Lazar, 2005; Yin et al., 2006; Yin et al., 2007]. Recent studies have demonstrated that heme functions as Rev-erb ligand [Burris, 2008; Raghuram et al., 2007; Rogers et al., 2008; Yin et al., 2007]. In light of the fact that heme induces adipogenesis in the 3T3-L1 model, this suggests that Rev-erb acts as a ligand-dependent transcription factor controlling adipogenic differentiation while likewise serving as a CCRP [Chen and London, 1981].
In vivo findings complement these in vitro observations [Ando et al., 2005; Bray and Young, 2007; Ptitysn AA, 2006; Zvonic et al., 2006]. Transcriptomic studies have shown that between 20–25% of the mRNA transcripts in both white and brown murine adipose tissues exhibit a circadian expression profile [Ptitysn AA, 2006; Zvonic et al., 2006]. These include multiple genes induced with ASC adipogenesis, including those in the oxidative phosphorylation, glucose and lipid metabolic, steroidogenic, and heat shock/chaperone families [Zvonic et al., 2006]. In vivo, treatment of animals or human subjects with the lithium chloride leads to weight gain, increased adiposity, and obesity [Baptista et al., 1995; Zvonic S, 2006]. Paradoxically, the presence of lithium chloride inhibits adipogenesis in both human ASCs and murine 3T3-L1 cells in vitro [Aratani et al., 1987; Ross et al., 2000; Wu et al., 2007]. Despite this discrepancy, the weight of the evidence supports a direct role of CCRPs in regulating the ASC adipogenic program and lineage selection.
Cancer Derived Cell Models
There is a wealth of in vivo and in vitro evidence implicating the CCRP in the growth of tumors and, by extension, cancer stem/progenitor cells. The Period genes have been associated with a number of tumors, including myeloid leukemia, breast, and lung cancers [Fu et al., 2002; Gery et al., 2005; Gery and Koeffler, 2007; Gery et al., 2006]. In a number of human tumors, the levels of the Period genes were reduced [Gery et al., 2006]. In mice deficient for Per2, the animals showed an increased risk of tumor development following exposure to ionizing radiation [Fu et al., 2002]. This resulted in an increased incidence of salivary gland hyperplasia, lymphoma, angiosarcoma, and premature graying of the hair in Per2−/− mice relative to wild type controls [Fu et al., 2002]. Furthermore, thymocytes in the Per2−/− mice were relatively resistant to the apoptotic effects of ionizing radiation [Fu et al., 2002]. Consistent with this observation, over expression of Period in human cancer cell lines increased their sensitivity to DNA damage and apoptosis; in contrast, down regulation of Period was associated with protection against ionizing radiation induced apoptosis [Gery et al., 2006]. Both the CCRP proteins Period and Bmal1 regulate components of the cell cycle, such as the cell cycle inhibitor p21WAF1/CIP1 [Fu et al., 2002; Grechez-Cassiau et al., 2008]. The levels of p21WAF/CIP1 and related cell cycle genes displayed a robust circadian oscillation in the liver of wild type mice; however, in Bmal1−/− animals, all cell cycle genes were expressed at relatively constant levels [Grechez-Cassiau et al., 2008]. The transcription of the p21WAF/CIP1 promoter was positively regulated by expression of Clock and Bmal1 in co-transfection assay; the further addition of Rev-erbα/β or RORα/γ antagonized the actions of Clock and Bmal1 [Grechez-Cassiau et al., 2008]. It is noteworthy that disruption of Clock and Bmal1 genes in mice was not associated with increased carcinogenesis [Antoch et al., 2008; Kondratov et al., 2006]. When exposed to ionizing radiation, these mice show accelerated aging and increased evidence of lymphoid apoptosis but do not exhibit tumors [Antoch et al., 2008; Kondratov et al., 2006]. One interpretation of this finding is that the negative arm of the CCRP (Cry, Per), as opposed to the positive arm (Clock, Bmal1) plays a more direct role in suppressing tumor formation.
Dysregulation of the CCRP may account for related changes in downstream features of tumor cells. In murine breast cancer models, while the anti-apoptotic gene Bcl2 displayed a robust circadian oscillation in normal tissues, this rhythmicity was absent in the tumor itself [Granda et al., 2005]. The involvement of the CCRP in tumor development has substantial implications with respect to chemotherapy [Gery and Koeffler, 2007; Gorbacheva et al., 2005; Granda et al., 2005; Levi, 2006; Levi et al., 2007]. The time of day when drugs are delivered to patients can profoundly impact both their benefits and adverse effects. Murine and human clinical studies have determined that there is an optimal time of day for the delivery of chemotherapy [Gery and Koeffler, 2007; Gorbacheva et al., 2005; Granda et al., 2005; Levi, 2006; Levi et al., 2007; Sahar and Sassone-Corsi, 2007]. For example, the administration of cyclin dependent kinase inhibitors to tumor bearing animals has been shown to enhance the rhythmicity of CCRP genes in the tumors themselves [Iurisci et al., 2006]. This has been associated with improved outcomes as evidenced by >50% reduction in the extent of tumor growth [Iurisci et al., 2006]. Oncologists have begun to incorporate the emerging concepts of chronotherapy into their strategies for dosage administration [Levi, 2006; Levi et al., 2007]. These findings merit continued evaluation and research to better define the underlying mechanisms of circadian biology in cancer and cell proliferation.
Future Directions
Again, why ask these questions? Because:
Timing is a variable that we can readily manipulate. As we develop cell based therapies that use exogenous, culture expanded stem cells or target the endogenous stem cells and their associated microenvironment, it is critical to maximize the returns to the patient. Studies of the CCRP in the context of stem cell biology will allow us to optimize the time when stem cell-based treatments are administered. Furthermore, this pathway will provide a molecular read out that can be readily monitored with appropriate diagnostic tools.
The CCRP provides an under-explored and novel regulatory pathway for potential direct interventions. The recent identification of heme as a direct ligand for Rev-erb proteins underscores this point. Since Rev-erb plays a significant role in adipogenic differentiation by ASCs, heme analogues have potential benefits for the prevention or treatment of obesity. Likewise, related nuclear hormone receptor ligands and substrates of critical rate limiting metabolic enzymes may also prove to be productive targets of opportunity. One example is the recent identification of cyclic AMP and its signaling pathway as a critical component of the circadian oscillatory network [O'Neill et al., 2008]. Furthermore, similar studies in model organisms have implicated cyclic adenosine diphosphate ribose and calcium as feedback components of the circadian oscillations [Dodd et al., 2007; O'Neill et al., 2008]. We hypothesize that these small molecules capable of entraining the CCRP apparatus can be used as drug discovery targets to manipulate adult stem cell lineage differentiation and function.
Conclusions
Modifying the period and amplitude of circadian oscillations in stem cell number and in stem cell metabolic activity is feasible using small molecule or genetic agents. Directing increased attention to these mechanisms has the potential to improve our understanding of stem cell biology and subsequently, our ability to modulate stem cell proliferation, differentiation, and function. The outcomes of such studies will directly impact our future capability to manipulate stem cell physiology and to prevent stem cell pathology.
Acknowledgements
This work was partially supported by a CNRU Center Grant # 1P30 DK072476 entitled “Nutritional Programming: Environmental and Molecular Interactions” sponsored by NIDDK (JMG), the Pennington Biomedical Research Foundation (JMG), the COBRE Center Grant P20-RR021945 from the National Center for Research Resources at NIH (ZEF), a grant from the NIH/NCRR P50RR00164, and a grant from eth Louisiana Gene Therapy Research Consortium (BAB).
The authors thank Mr. Lary Loupe of Loupe Graphics (lploupe@yahoo.com) for the providing the cartoons for this manuscript and their colleagues in the Stem Cell Laboratory at the Pennington Biomecical Research Center and the Center for Gene Therapy for their discussion and support of this work.
References
- Aardal NP. Circannual variations of circadian periodicity in murine colony-forming cells. Exp Hematol. 1984;12:61–67. [PubMed] [Google Scholar]
- Aardal NP, Laerum OD. Circadian variations in mouse bone marrow. Exp Hematol. 1983;11:792–801. [PubMed] [Google Scholar]
- Abdelaal MA, Hashim IA, Zawawi TH, Felimban SK, Sobhi EM, Jeje O, Oni GA. Circadian rhythm of granulocyte-macrophage colony-stimulating factor in normal subjects and neutropenic hospitalised patients. Ir J Med Sci. 2000;169:55–57. doi: 10.1007/BF03170487. [DOI] [PubMed] [Google Scholar]
- Abrahamsen JF, Smaaland R, Sothern RB, Laerum OD. Circadian cell cycle variations of erythro- and myelopoiesis in humans. Eur J Haematol. 1997;58:333–345. doi: 10.1111/j.1600-0609.1997.tb01680.x. [DOI] [PubMed] [Google Scholar]
- Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- Akashi M, Tsuchiya Y, Yoshino T, Nishida E. Control of intracellular dynamics of mammalian period proteins by casein kinase I epsilon (CKIepsilon) and CKIdelta in cultured cells. Mol Cell Biol. 2002;22:1693–1703. doi: 10.1128/MCB.22.6.1693-1703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alenghat TMK, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, Bućan M, Ahima RS, Kaestner KH, Lazar MA. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando H, Yanagihara H, Hayashi Y, Obi Y, Tsuruoka S, Takamura T, Kaneko S, Fujimura A. Rhythmic mRNA Expression of Clock Genes and Adipocytokines in Mouse Visceral Adipose Tissue. Endocrinology. 2005 doi: 10.1210/en.2005-0771. [DOI] [PubMed] [Google Scholar]
- Antoch MP, Gorbacheva VY, Vykhovanets O, Toshkov IA, Kondratov RV, Kondratova AA, Lee C, Nikitin AY. Disruption of the circadian clock due to the Clock mutation has discrete effects on aging and carcinogenesis. Cell Cycle. 2008;7:1197–1204. doi: 10.4161/cc.7.9.5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aratani Y, Sugimoto E, Kitagawa Y. Lithium ion reversibly inhibits inducer-stimulated adipose conversion of 3T3-L1 cells. FEBS Lett. 1987;218:47–51. doi: 10.1016/0014-5793(87)81015-3. [DOI] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Baksh D, Boland GM, Tuan RS. Cross-talk between Wnt signaling pathways in human mesenchymal stem cells leads to functional antagonism during osteogenic differentiation. J Cell Biochem. 2007;101:1109–1124. doi: 10.1002/jcb.21097. [DOI] [PubMed] [Google Scholar]
- Baksh D, Tuan RS. Canonical and non-canonical wnts differentially affect the development potential of primary isolate of human bone marrow mesenchymal stem cells. J Cell Physiol. 2007;212:817–826. doi: 10.1002/jcp.21080. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000a;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Marcacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr Biol. 2000b;10:1291–1294. doi: 10.1016/s0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- Baptista T, Teneud L, Contreras Q, Alastre T, Burguera JL, de Burguera M, de Baptista E, Weiss S, Hernandez L. Lithium and body weight gain. Pharmacopsychiatry. 1995;28:35–44. doi: 10.1055/s-2007-979586. [DOI] [PubMed] [Google Scholar]
- Baudoux E, Beguin Y, Cornu G, Brichard B, Debruyn C, De Bruyere M, De Hemptinne D, Delforge A, Deneys V, Fillet G, Germeau N, Joris I, Lefevre P, Massy M, Paulus JM, Raymakers N, Schaaps JP, Sondag D, Van Cauwenberge JR, Vermylen C, Strijckmans P. Circadian and seasonal variations of hematopoiesis in cord blood. Bone Marrow Transplant. 1998;22 Suppl 1:S12. [PubMed] [Google Scholar]
- Bi Y, Ehirchiou D, Kilts TM, Inkson CA, Embree MC, Sonoyama W, Li L, Leet AI, Seo BM, Zhang L, Shi S, Young MF. Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat Med. 2007;13:1219–1227. doi: 10.1038/nm1630. [DOI] [PubMed] [Google Scholar]
- Bourin P, Ledain AF, Beau J, Mille D, Levi F. In-vitro circadian rhythm of murine bone marrow progenitor production. Chronobiol Int. 2002;19:57–67. doi: 10.1081/cbi-120002677. [DOI] [PubMed] [Google Scholar]
- Bray MS, Young ME. Circadian rhythms in the development of obesity: potential role for the circadian clock within the adipocyte. Obes Rev. 2007;8:169–181. doi: 10.1111/j.1467-789X.2006.00277.x. [DOI] [PubMed] [Google Scholar]
- Brown SA, Fleury-Olela F, Nagoshi E, Hauser C, Juge C, Meier CA, Chicheportiche R, Dayer JM, Albrecht U, Schibler U. The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biol. 2005;3:e338. doi: 10.1371/journal.pbio.0030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Kunz D, Dumas A, Westermark PO, Vanselow K, Tilmann-Wahnschaffe A, Herzel H, Kramer A. Molecular insights into human daily behavior. Proc Natl Acad Sci U S A. 2008;105:1602–1607. doi: 10.1073/pnas.0707772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burris TP. Nuclear Hormone Receptors for Heme: REV-ERB{alpha} and REV-ERB{beta} Are Ligand-Regulated Components of the Mammalian Clock. Mol Endocrinol. 2008 doi: 10.1210/me.2007-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Lazar MA. Induction of Rev-ErbA alpha, an orphan receptor encoded on the opposite strand of the alpha-thyroid hormone receptor gene, during adipocyte differentiation. J Biol Chem. 1993;268:16265–16269. [PubMed] [Google Scholar]
- Chen JJ, London IM. Hemin enhances the differentiation of mouse 3T3 cells to adipocytes. Cell. 1981;26:117–122. doi: 10.1016/0092-8674(81)90039-8. [DOI] [PubMed] [Google Scholar]
- Chen YG, Mantalaris A, Bourne P, Keng P, Wu JH. Expression of mPer1 and mPer2, two mammalian clock genes, in murine bone marrow. Biochem Biophys Res Commun. 2000;276:724–728. doi: 10.1006/bbrc.2000.3536. [DOI] [PubMed] [Google Scholar]
- D'Hondt L, McAuliffe C, Damon J, Reilly J, Carlson J, Dooner M, Colvin G, Lambert JF, Hsieh CC, Habibian H, Stencel K, Quesenberry PJ. Circadian variations of bone marrow engraftability. J Cell Physiol. 2004;200:63–70. doi: 10.1002/jcp.20032. [DOI] [PubMed] [Google Scholar]
- D'Ippolito G, Diabira S, Howard GA, Roos BA, Schiller PC. Low oxygen tension inhibits osteogenic differentiation and enhances stemness of human MIAMI cells. Bone. 2006;39:513–522. doi: 10.1016/j.bone.2006.02.061. [DOI] [PubMed] [Google Scholar]
- Dincol D, Akbulut H, Buyukcelik A, Icli F. Diurnal variations of serum GM-CSF levels. Cytokine. 2000;12:1151–1155. doi: 10.1006/cyto.2000.0677. [DOI] [PubMed] [Google Scholar]
- Dodd AN, Gardner MJ, Hotta CT, Hubbard KE, Dalchau N, Love J, Assie JM, Robertson FC, Jakobsen MK, Goncalves J, Sanders D, Webb AA. The Arabidopsis circadian clock incorporates a cADPR-based feedback loop. Science. 2007;318:1789–1792. doi: 10.1126/science.1146757. [DOI] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Duez H, Staels B. Rev-erb alpha gives a time cue to metabolism. FEBS Lett. 2008;582:19–25. doi: 10.1016/j.febslet.2007.08.032. [DOI] [PubMed] [Google Scholar]
- Etheridge SL, Spencer GJ, Heath DJ, Genever PG. Expression profiling and functional analysis of wnt signaling mechanisms in mesenchymal stem cells. Stem Cells. 2004;22:849–860. doi: 10.1634/stemcells.22-5-849. [DOI] [PubMed] [Google Scholar]
- Fehrer C, Brunauer R, Laschober G, Unterluggauer H, Reitinger S, Kloss F, Gully C, Gassner R, Lepperdinger G. Reduced oxygen tension attenuates differentiation capacity of human mesenchymal stem cells and prolongs their lifespan. Aging Cell. 2007;6:745–757. doi: 10.1111/j.1474-9726.2007.00336.x. [DOI] [PubMed] [Google Scholar]
- Fu L, Patel MS, Bradley A, Wagner EF, Karsenty G. The molecular clock mediates leptin-regulated bone formation. Cell. 2005;122:803–815. doi: 10.1016/j.cell.2005.06.028. [DOI] [PubMed] [Google Scholar]
- Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- Fukuya H, Emoto N, Nonaka H, Yagita K, Okamura H, Yokoyama M. Circadian expression of clock genes in human peripheral leukocytes. Biochem Biophys Res Commun. 2007;354:924–928. doi: 10.1016/j.bbrc.2007.01.063. [DOI] [PubMed] [Google Scholar]
- Gery S, Gombart AF, Yi WS, Koeffler C, Hofmann WK, Koeffler HP. Transcription profiling of C/EBP targets identifies Per2 as a gene implicated in myeloid leukemia. Blood. 2005;106:2827–2836. doi: 10.1182/blood-2005-01-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gery S, Koeffler HP. The role of circadian regulation in cancer. Cold Spring Harb Symp Quant Biol. 2007;72:459–464. doi: 10.1101/sqb.2007.72.004. [DOI] [PubMed] [Google Scholar]
- Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Gimble J, Guilak F. Adipose-derived adult stem cells: isolation, characterization, and differentiation potential. Cytotherapy. 2003;5:362–369. doi: 10.1080/14653240310003026. [DOI] [PubMed] [Google Scholar]
- Gimble JM, Dorheim MA, Cheng Q, Medina K, Wang CS, Jones R, Koren E, Pietrangeli C, Kincade PW. Adipogenesis in a murine bone marrow stromal cell line capable of supporting B lineage lymphocyte growth and proliferation: biochemical and molecular characterization. Eur J Immunol. 1990;20:379–387. doi: 10.1002/eji.1830200222. [DOI] [PubMed] [Google Scholar]
- Gimble JM, Katz AJ, Bunnell BA. Adipose-derived stem cells for regenerative medicine. Circ Res. 2007;100:1249–1260. doi: 10.1161/01.RES.0000265074.83288.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimble JM, Pietrangeli C, Henley A, Dorheim MA, Silver J, Namen A, Takeichi M, Goridis C, Kincade PW. Characterization of murine bone marrow and spleen-derived stromal cells: analysis of leukocyte marker and growth factor mRNA transcript levels. Blood. 1989;74:303–311. [PubMed] [Google Scholar]
- Gimble JM, Robinson CE, Wu X, Kelly KA. The function of adipocytes in the bone marrow stroma: an update. Bone. 1996;19:421–428. doi: 10.1016/s8756-3282(96)00258-x. [DOI] [PubMed] [Google Scholar]
- Gimble JM, Zvonic S, Floyd ZE, Kassem M, Nuttall ME. Playing with bone and fat. J Cell Biochem. 2006;98:251–266. doi: 10.1002/jcb.20777. [DOI] [PubMed] [Google Scholar]
- Gorbacheva VY, Kondratov RV, Zhang R, Cherukuri S, Gudkov AV, Takahashi JS, Antoch MP. Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the CLOCK/BMAL1 transactivation complex. Proc Natl Acad Sci U S A. 2005;102:3407–3412. doi: 10.1073/pnas.0409897102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granda TG, Liu XH, Smaaland R, Cermakian N, Filipski E, Sassone-Corsi P, Levi F. Circadian regulation of cell cycle and apoptosis proteins in mouse bone marrow and tumor. FASEB J. 2005;19:304–306. doi: 10.1096/fj.04-2665fje. [DOI] [PubMed] [Google Scholar]
- Grayson WL, Zhao F, Izadpanah R, Bunnell B, Ma T. Effects of hypoxia on human mesenchymal stem cell expansion and plasticity in 3D constructs. J Cell Physiol. 2006;207:331–339. doi: 10.1002/jcp.20571. [DOI] [PubMed] [Google Scholar]
- Grechez-Cassiau A, Rayet B, Guillaumond F, Teboul M, Delaunay F. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J Biol Chem. 2008;283:4535–4542. doi: 10.1074/jbc.M705576200. [DOI] [PubMed] [Google Scholar]
- Gregory CA, Gunn WG, Reyes E, Smolarz AJ, Munoz J, Spees JL, Prockop DJ. How Wnt signaling affects bone repair by mesenchymal stem cells from the bone marrow. Ann N Y Acad Sci. 2005;1049:97–106. doi: 10.1196/annals.1334.010. [DOI] [PubMed] [Google Scholar]
- Gulbagci NT, Li L, Ling B, Gopinadhan S, Walsh M, Rossner M, Nave KA, Taneja R. SHARP1/DEC2 inhibits adipogenic differentiation by regulating the activity of C/EBP. EMBO Rep. 2008 doi: 10.1038/embor.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halberg F, Halberg E, Wargo DC, Visscher MB. Eosinophil levels in dogs with surgically established arteriovenous anastomoses. Am J Physiol. 1953a;174:313–315. doi: 10.1152/ajplegacy.1953.174.2.313. [DOI] [PubMed] [Google Scholar]
- Halberg F, Ulstrom RA. Morning changes in number of circulating eosinophils in infants. Proc Soc Exp Biol Med. 1952;80:747–748. doi: 10.3181/00379727-80-19750. [DOI] [PubMed] [Google Scholar]
- Halberg F, Visscher MB. Regular diurnal physiological variation in eosinophil levels in five stocks of mice. Proc Soc Exp Biol Med. 1950;75:846–847. doi: 10.3181/00379727-75-18365. [DOI] [PubMed] [Google Scholar]
- Halberg F, Visscher MB, Bittner JJ. Eosinophil rhythm in mice: range of occurrence; effects of illumination, feeding, and adrenalectomy. Am J Physiol. 1953b;174:109–122. doi: 10.1152/ajplegacy.1953.174.1.109. [DOI] [PubMed] [Google Scholar]
- Halberg FVM, Bittner JJ. Eosinophil rhythm in mice: range of occurrence, effects of illumination, feeding and adrenalectomy. American Journal of Physiology. 1953;174:109–122. doi: 10.1152/ajplegacy.1953.174.1.109. [DOI] [PubMed] [Google Scholar]
- Haus E, Lakatua DJ, Swoyer J, Sackett-Lundeen L. Chronobiology in hematology and immunology. Am J Anat. 1983;168:467–517. doi: 10.1002/aja.1001680406. [DOI] [PubMed] [Google Scholar]
- Haus E, Smolensky MH. Biologic rhythms in the immune system. Chronobiol Int. 1999;16:581–622. doi: 10.3109/07420529908998730. [DOI] [PubMed] [Google Scholar]
- Hirota T, Fukada Y. Resetting mechanism of central and peripheral circadian clocks in mammals. Zoolog Sci. 2004;21:359–368. doi: 10.2108/zsj.21.359. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iurisci I, Filipski E, Reinhardt J, Bach S, Gianella-Borradori A, Iacobelli S, Meijer L, Levi F. Improved tumor control through circadian clock induction by Seliciclib, a cyclin-dependent kinase inhibitor. Cancer Res. 2006;66:10720–10728. doi: 10.1158/0008-5472.CAN-06-2086. [DOI] [PubMed] [Google Scholar]
- Iwahana E, Akiyama M, Miyakawa K, Uchida A, Kasahara J, Fukunaga K, Hamada T, Shibata S. Effect of lithium on the circadian rhythms of locomotor activity and glycogen synthase kinase-3 protein expression in the mouse suprachiasmatic nuclei. Eur J Neurosci. 2004;19:2281–2287. doi: 10.1111/j.0953-816X.2004.03322.x. [DOI] [PubMed] [Google Scholar]
- Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- Kincade PW. Molecular interactions between stromal cells and B lymphocyte precursors. Semin Immunol. 1991;3:379–390. [PubMed] [Google Scholar]
- Kincade PW, Lee G, Pietrangeli CE, Hayashi S, Gimble JM. Cells and molecules that regulate B lymphopoiesis in bone marrow. Annu Rev Immunol. 1989;7:111–143. doi: 10.1146/annurev.iy.07.040189.000551. [DOI] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, Turek FW, Takahashi JS. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko HW, Edery I. Analyzing the Degradation of PERIOD Protein by the Ubiquitin-Proteasome Pathway in Cultured Drosophila Cells. Methods Enzymol. 2005;393:394–408. doi: 10.1016/S0076-6879(05)93018-8. [DOI] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lee Y, Lee MJ, Park E, Kang SH, Chung CH, Lee KH, Kim K. Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Mol Cell Biol. 2008;28:6056–6065. doi: 10.1128/MCB.00583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemberger T, Saladin R, Vazquez M, Assimacopoulos F, Staels B, Desvergne B, Wahli W, Auwerx J. Expression of the peroxisome proliferator-activated receptor alpha gene is stimulated by stress and follows a diurnal rhythm. J Biol Chem. 1996;271:1764–1769. doi: 10.1074/jbc.271.3.1764. [DOI] [PubMed] [Google Scholar]
- Levi F. Chronotherapeutics: the relevance of timing in cancer therapy. Cancer Causes Control. 2006;17:611–621. doi: 10.1007/s10552-005-9004-7. [DOI] [PubMed] [Google Scholar]
- Levi F, Filipski E, Iurisci I, Li XM, Innominato P. Cross-talks between circadian timing system and cell division cycle determine cancer biology and therapeutics. Cold Spring Harb Symp Quant Biol. 2007;72:465–475. doi: 10.1101/sqb.2007.72.030. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas D, Battista M, Shi PA, Isola L, Frenette PS. Mobilized hematopoietic stem cell yield depends on species-specific circadian timing. Cell Stem Cell. 2008;3:364–366. doi: 10.1016/j.stem.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestroni GJ, Cosentino M, Marino F, Togni M, Conti A, Lecchini S, Frigo G. Neural and endogenous catecholamines in the bone marrow Circadian association of norepinephrine with hematopoiesis? Exp Hematol. 1998;26:1172–1177. [PubMed] [Google Scholar]
- Mendez-Ferrer S, Lucas D, Battista M, Frenette PS. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008;452:442–447. doi: 10.1038/nature06685. [DOI] [PubMed] [Google Scholar]
- Meyer T, Kneissel M, Mariani J, Fournier B. In vitro and in vivo evidence for orphan nuclear receptor RORalpha function in bone metabolism. Proc Natl Acad Sci U S A. 2000;97:9197–9202. doi: 10.1073/pnas.150246097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth MJ, Bodine DM. Regulation of hematopoiesis and the hematopoietic stem cell niche by Wnt signaling pathways. Cell Res. 2007 doi: 10.1038/cr.2007.69. [DOI] [PubMed] [Google Scholar]
- O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padiath QS, Paranjpe D, Jain S, Sharma VK. Glycogen synthase kinase 3beta as a likely target for the action of lithium on circadian clocks. Chronobiol Int. 2004;21:43–55. doi: 10.1081/cbi-120027981. [DOI] [PubMed] [Google Scholar]
- Ptitysn AAZS, Conrad SA, Scott LK, Mynatt ML, Gimble JM. Circadian Clocks are Resounding in Peripheral Tissues. PLoS Computational Biology. 2006;2:e16. doi: 10.1371/journal.pcbi.0020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers PM, Ying L, Burris TP. Relationship between circadian oscillations of Rev-erbalpha expression and intracellular levels of its ligand, heme. Biochem Biophys Res Commun. 2008;368:955–958. doi: 10.1016/j.bbrc.2008.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P. Circadian clock and breast cancer: a molecular link. Cell Cycle. 2007;6:1329–1331. doi: 10.4161/cc.6.11.4295. [DOI] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Sharkis SJ, Alexander P, Jr, Rakowitz F, LoBue J, Weitz-Hamburger A, Gordon AS. Circadian variation in mouse hematopoiesis I. Sex difference in cellularity of femoral diaphyseal marrow. Proc Soc Exp Biol Med. 1971;136:283–284. doi: 10.3181/00379727-136-35247. [DOI] [PubMed] [Google Scholar]
- Sharkis SJ, Palmer JD, Goodenough J, LoBue J, Gordon AS. Daily variation of marrow and splenic erythropoiesis, pinna epidermal cell mitosis and physical activity in C57B1-6J mice. Cell Tissue Kinet. 1974;7:381–387. doi: 10.1111/j.1365-2184.1974.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, Hayashi M, Wada T, Aoyagi T, Tezuka M. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci U S A. 2005;102:12071–12076. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimba S, Wada T, Hara S, Tezuka M. EPAS1 promotes adipose differentiation in 3T3-L1 cells. J Biol Chem. 2004;279:40946–40953. doi: 10.1074/jbc.M400840200. [DOI] [PubMed] [Google Scholar]
- Simmons PJ, Masinovsky B, Longenecker BM, Berenson R, Torok-Storb B, Gallatin WM. Vascular cell adhesion molecule-1 expressed by bone marrow stromal cells mediates the binding of hematopoietic progenitor cells. Blood. 1992;80:388–395. [PubMed] [Google Scholar]
- Sletvold O, Laerum OD. Alterations of cell cycle distribution in the bone marrow of aging mice measured by flow cytometry. Exp Gerontol. 1988a;23:43–58. doi: 10.1016/0531-5565(88)90019-8. [DOI] [PubMed] [Google Scholar]
- Sletvold O, Laerum OD. Multipotent stem cell (CFU-S) numbers and circadian variations in aging mice. Eur J Haematol. 1988b;41:230–236. doi: 10.1111/j.1600-0609.1988.tb01186.x. [DOI] [PubMed] [Google Scholar]
- Sletvold O, Laerum OD, Riise T. Age-related differences and circadian and seasonal variations of myelopoietic progenitor cell (CFU-GM) numbers in mice. Eur J Haematol. 1988a;40:42–49. doi: 10.1111/j.1600-0609.1988.tb00795.x. [DOI] [PubMed] [Google Scholar]
- Sletvold O, Laerum OD, Riise T. Rhythmic variations of different hemopoietic cell lines and maturation stages in aging mice. Mech Ageing Dev. 1988b;42:91–104. doi: 10.1016/0047-6374(88)90065-6. [DOI] [PubMed] [Google Scholar]
- Sletvold O, Smaaland R, Laerum OD. Cytometry and time-dependent variations in peripheral blood and bone marrow cells: a literature review and relevance to the chronotherapy of cancer. Chronobiol Int. 1991;8:235–250. doi: 10.3109/07420529109063929. [DOI] [PubMed] [Google Scholar]
- Smaaland R, Sothern RB, Laerum OD, Abrahamsen JF. Rhythms in human bone marrow and blood cells. Chronobiol Int. 2002;19:101–127. doi: 10.1081/cbi-120002594. [DOI] [PubMed] [Google Scholar]
- Sothern RB, Roitman-Johnson B, Kanabrocki EL, Yager JG, Roodell MM, Weatherbee JA, Young MR, Nenchausky BM, Scheving LE. Circadian characteristics of circulating interleukin-6 in men. J Allergy Clin Immunol. 1995;95:1029–1035. doi: 10.1016/s0091-6749(95)70104-4. [DOI] [PubMed] [Google Scholar]
- Sun Y, Yang Z, Niu Z, Peng J, Li Q, Xiong W, Langnas AN, Ma MY, Zhao Y. MOP3, a component of the molecular clock, regulates the development of B cells. Immunology. 2006;119:451–460. doi: 10.1111/j.1365-2567.2006.02456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thomas HE, Redgrave R, Cunnington MS, Avery P, Keavney BD, Arthur HM. Circulating endothelial progenitor cells exhibit diurnal variation. Arterioscler Thromb Vasc Biol. 2008;28:e21–e22. doi: 10.1161/ATVBAHA.107.160317. [DOI] [PubMed] [Google Scholar]
- Triqueneaux G, Thenot S, Kakizawa T, Antoch MP, Safi R, Takahashi JS, Delaunay F, Laudet V. The orphan receptor Rev-erb{alpha} gene is a target of the circadian clock pacemaker. J Mol Endocrinol. 2004;33:585–608. doi: 10.1677/jme.1.01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge JJ, Xenocostas A, Moon RT, Bhatia M. Glycogen synthase kinase-3 is an in vivo regulator of hematopoietic stem cell repopulation. Nat Med. 2006;12:89–98. doi: 10.1038/nm1339. [DOI] [PubMed] [Google Scholar]
- Tsinkalovsky O, Filipski E, Rosenlund B, Sothern RB, Eiken HG, Wu MW, Claustrat B, Bayer J, Levi F, Laerum OD. Circadian expression of clock genes in purified hematopoietic stem cells is developmentally regulated in mouse bone marrow. Exp Hematol. 2006;34:1249–1261. doi: 10.1016/j.exphem.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Tsinkalovsky O, Rosenlund B, Laerum OD, Eiken HG. Clock gene expression in purified mouse hematopoietic stem cells. Exp Hematol. 2005;33:100–107. doi: 10.1016/j.exphem.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Tsinkalovsky O, Smaaland R, Rosenlund B, Sothern RB, Hirt A, Steine S, Badiee A, Abrahamsen JF, Eiken HG, Laerum OD. Circadian variations in clock gene expression of human bone marrow CD34+ cells. J Biol Rhythms. 2007;22:140–150. doi: 10.1177/0748730406299078. [DOI] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wood PA, Hrushesky WJ, Klevecz R. Distinct circadian time structures characterize myeloid and erythroid progenitor and multipotential cell clonogenicity as well as marrow precursor proliferation dynamics. Exp Hematol. 1998;26:523–533. [PubMed] [Google Scholar]
- Wu XYG, Parks H, Hebert T, Goh BC, Dietrich MA, Pelled G, Izadpanah R, Gazit D, Bunnell BA, Gimble JM. Circadian mechanisms in murine and human bone marrow mesenchymal stem cells following dexamethasone exposure. Bone. 2008;42:861–870. doi: 10.1016/j.bone.2007.12.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XZS, Floyd ZE, Kilroy G, Goh BC, Hernandez TL, Eckel RH, Mynatt RL, Gimble JM. Circadian Gene Expression in Human Subcutaneous Adipose-derived Stem Cells: Induction by Dexamethasone, Serum, and Thiazolidinedione in the Undifferentiated and Adipogenic States. Obesity. 2007;15:2560–2570. doi: 10.1038/oby.2007.308. [DOI] [PubMed] [Google Scholar]
- Wu X, Zvonic S, Floyd ZE, Kilroy G, Goh BC, Hernandez TL, Eckel RH, Mynatt RL, Gimble JM. Induction of circadian gene expression in human subcutaneous adipose-derived stem cells. Obesity (Silver Spring) 2007;15:2560–2570. doi: 10.1038/oby.2007.308. [DOI] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Yin L, Lazar MA. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol. 2005;19:1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- Yin L, Wang J, Klein PS, Lazar MA. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science. 2006;311:1002–1005. doi: 10.1126/science.1121613. [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Yin T, Li L. The stem cell niches in bone. J Clin Invest. 2006;116:1195–1201. doi: 10.1172/JCI28568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, Takahashi JS. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MR, Matthews JP, Kanabrocki EL, Sothern RB, Roitman-Johnson B, Scheving LE. Circadian rhythmometry of serum interleukin-2, interleukin-10, tumor necrosis factor-alpha, and granulocyte-macrophage colony-stimulating factor in men. Chronobiol Int. 1995;12:19–27. doi: 10.3109/07420529509064496. [DOI] [PubMed] [Google Scholar]
- Zvonic SFZ, Mynatt RL, Gimble JM. Circadian Rhythms and the Regulation of Metabolic Tissue Function and Energy Homeostasis. Obesity. 2006 doi: 10.1038/oby.2007.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, Wu X, Goh BC, Mynatt RL, Gimble JM. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- Zvonic S, Ptitsyn AA, Kilroy G, Wu X, Conrad SA, Scott LK, Guilak F, Pelled G, Gazit D, Gimble JM. Circadian Oscillation of Gene Expression in Murine Calvarial Bone. J Bone Miner Res. 2007;22:357–365. doi: 10.1359/jbmr.061114. [DOI] [PubMed] [Google Scholar]