Abstract
We report here the results of an analysis of the regulatory range of the GacS/GacA two-component system in Pseudomonas aeruginosa. Using microarrays, we identified a large number of genes that are regulated by the system, and detected a near complete overlap of these genes with those regulated by two small RNAs (sRNAs), RsmY and RsmZ, suggesting that the expression of all GacA-regulated genes is RsmY/Z-dependent. Using genome-wide DNA-protein interaction analyses, we identified only two genomic regions that associated specifically with GacA, located upstream of the rsmY and rsmZ genes. These results demonstrate that in P. aeruginosa, the GacS/GacA system transduces the regulatory signals to downstream genes exclusively by directly controlling the expression of only two genes rsmY and rsmZ. These two sRNAs serve as intermediates between the input signals and the output at the level of mRNA stability, although additional regulatory inputs can influence the levels of these two riboregulators. We show that the A+T-rich DNA segment upstream of rsmZ is bound and silenced by MvaT and MvaU, the global gene regulators of the H-NS family. This work highlights the importance of post-transcriptional mechanisms involving sRNAs in controlling gene expression during bacterial adaptation to different environments.
INTRODUCTION
Two-component regulatory systems, consisting of sensor kinase and response-regulator pairs have emerged as key mediators of successful adaptation of microorganisms to changing environments. These systems are capable of assimilating a wide range of environmental stimuli into different outputs, ranging from gene expression and motility to enzymatic activity (Gao et al., 2007). The majority of responses are associated with the control of gene expression, utilizing the ability of phosphorylated (or de-phosphorylated) response-regulators to recognize and bind to specific DNA sequences located upstream of regulated genes and thus influence their transcription. As such, the two-component systems often participate in complex networks where they control the expression of other regulatory factors, including transcriptional regulators and small regulatory RNAs (sRNAs) (Valverde and Haas, 2008).
In a number of human and plant pathogens, a conserved two-component system regulates a broad range of virulence and stress-responsive genes (Lapouge et al., 2008). This family of regulatory systems is referred to as GacS (LemA)/GacA in Pseudomonas, Erwinia and Vibrio fisherii species, VarS/VarA in Vibrio cholerae, BarA/UvrY in E. coli and BarA/SirA in Salmonella, and LetS/LetA in Legionella. These systems regulate the expression of a variety of phenotypes including production of extracellular enzymes, secreted toxins, quorum sensing molecules, various metabolic functions and motility. Where examined, transcription of small trans-acting regulatory RNAs of the RsmY/Z CsrB/C family has been shown to depend on these regulators. Since these sRNAs interact with and modulate the activity of the translation repressors of the RsmA/CsrA family, the phenotypes regulated by the GacS/GacA system and its homologs could be controlled not only at the level of transcription initiation by the response-regulator but also through mRNA turnover influenced by the interaction of the sRNAs with RsmA. In many cases, deletion of the regulatory sRNAs results in phenotypes that are similar to those of mutants in the GacS/GacA system. Such observations have been reported for rsmY rsmZ and gacA mutants in P. aeruginosa (Kay et al., 2006), rsmB and gacA mutants in E. carotovora (Liu et al., 1998), csrB csrC and uvrY mutants in E. coli (Weilbacher et al., 2003), csrB csrC and sirA mutants in S. enterica (Fortune et al., 2006), rsmX rsmY rsmZ and gacA mutants in P. fluorescens (Kay et al., 2005), and most recently for rsmY rsmZ and letA mutants in Legionella pneumophila (Sahr et al., 2009). This suggests that the GacS/GacA system and its homologs control expression of their target genes mainly through the regulation of the expression of the sRNA genes. However, the possibility that the systems directly regulate other types of genes cannot be excluded. In fact, in L. pneumophila, LetA regulates expression of flagellar genes by a mechanism that appears to be independent of rsmY and rsmZ (Sahr et al., 2009).
In Pseudomonas aeruginosa the GacS/GacA two-component system controls the reciprocal expression of acute and chronic virulence determinants (Goodman et al., 2004). In this organism, the GacS/GacA signal-transduction pathway itself is regulated by opposing activities of two additional sensor kinases, RetS and LadS (Goodman et al., 2004; Ventre et al., 2006). We have recently shown that RetS forms inactive heterodimers with GacS, preventing phosphorylation of the response regulator GacA (Goodman et al., 2009). In a microarray analysis of transcript levels in P. aeruginosa rsmA mutant, we have identified hundreds of genes whose expression levels were affected by the absence of RsmA, and many of these included genes previously shown to be regulated by RetS and LadS (Brencic and Lory, 2009; Goodman et al., 2004; Ventre et al., 2006). This suggested that RetS/GacS/GacA and RsmY/RsmZ/RsmA may be part of a single signal-transduction pathway, where RsmY and RsmZ serve as the regulatory intermediates between the input signals and the regulatory output at the levels of mRNA stability, and therefore that the only direct targets of GacA regulation are the two sRNAs. To test this idea, we have undertaken a study to identify all genes that are controlled by GacA and, among those, to identify the ones whose transcription is controlled by GacA directly. We found that in P. aeruginosa, GacS/GacA directly regulates only two genes, rsmY and rsmZ, and therefore that the transcripts of the hundreds of genes that are regulated by the RetS/GacS/GacA system are regulated by RsmY- and RsmZ-dependent modulation of RsmA activity. We also present evidence that GacA regulates rsmY and rsmZ by directly binding to sequences upstream of these genes. Furthermore, we show that the H-NS family of DNA-binding proteins MvaT and MvaU associate with the chromosomal region corresponding to rsmZ and also influence rsmZ expression. In summary, this study shows that in P. aeruginosa and possibly in other bacteria that possess orthologs of the GacS/GacA system, the output of the signal transduction pathway mediated by this two-component system is entirely directed towards controlling translation initiation or mRNA stability.
RESULTS
Gene regulation by RetS requires GacS
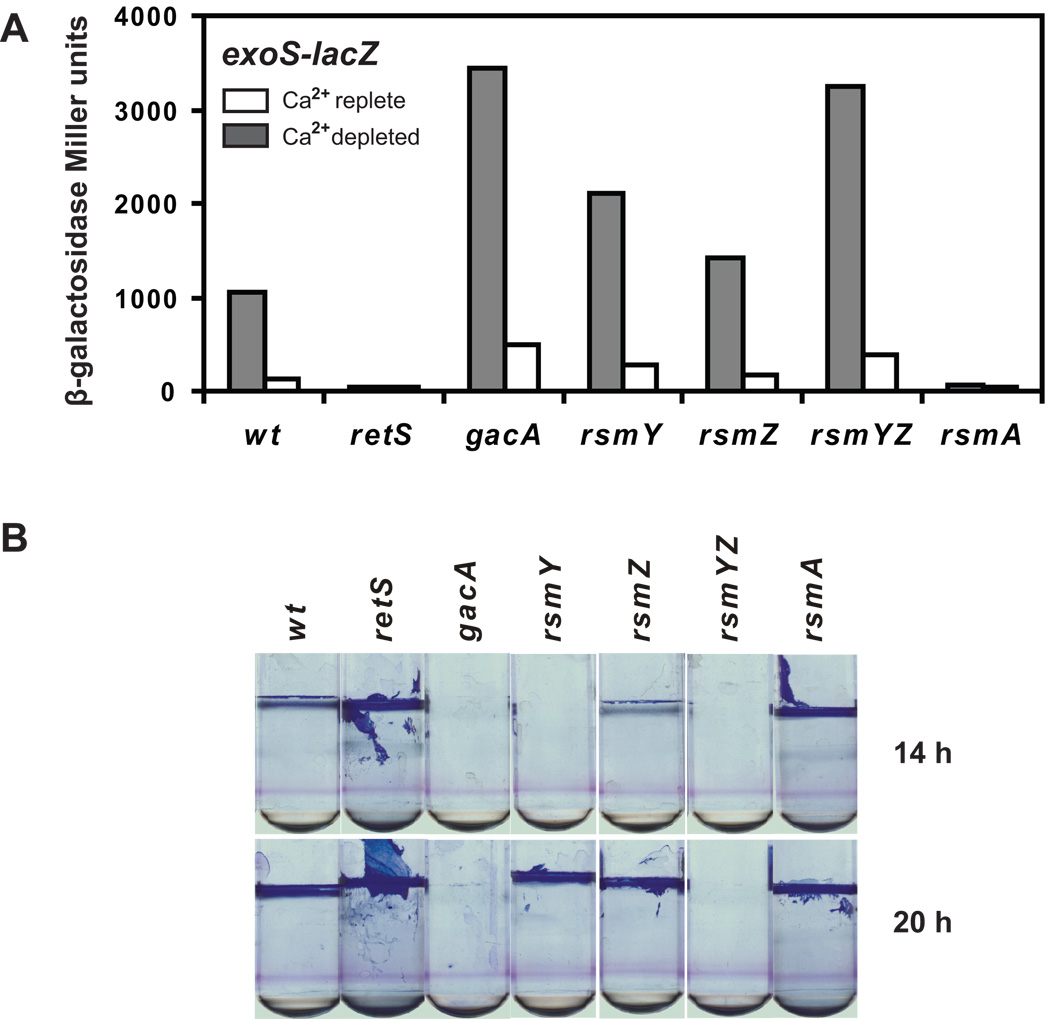
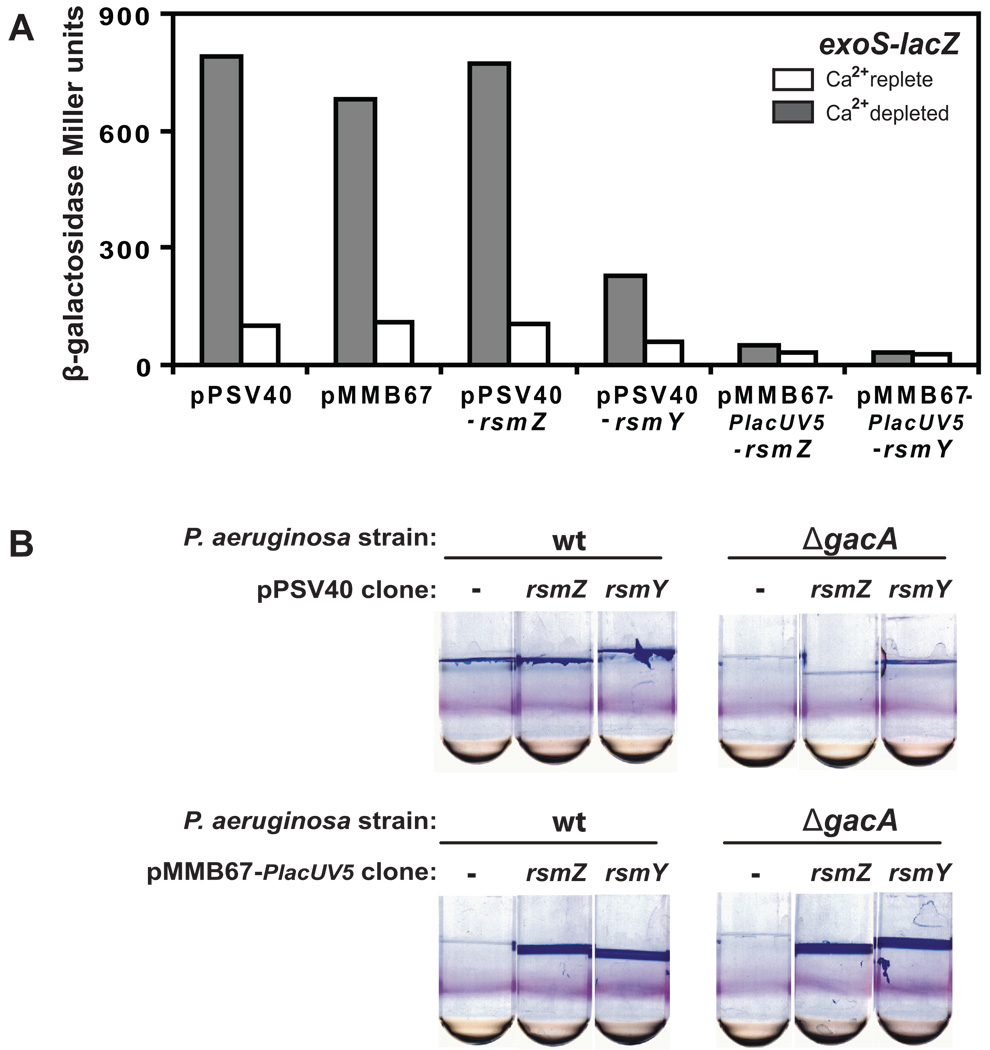
We have examined two known phenotypes regulated by RetS in a full range of mutants predicted to influence the downstream components of the RetS regulatory pathway (Figure 1). As predicted, a reciprocal regulation of transcription of the exoS gene, encoding a protein exported by the type III secretion (T3S) system, and formation of static biofilm was observed in strains with a deletion of the retS and rsmA genes compared to P. aeruginosa gacA, rsmY, rsmZ mutants, and to the rsmYZ double mutant. Examination of the phenotypes of the rsmY and rsmZ mutants showed that both mutations had an effect on exoS expression and on biofilm production (Figure 1). A somewhat stronger effect was observed in P. aeruginosa with a mutation in the rsmY gene, compared to the strain lacking rsmZ, however, a double rsmYZ deletion was required in order to observe a phenotype that equaled that of the gacA mutant. Similar results were previously reported for the RetS/GacS/A/RsmY/Z/A regulation of another T3S gene, exsD (Brencic and Lory, 2009). On the other hand, overexpression of either one of the small RNAs from a constitutively active exogenous promoter was sufficient to completely shut off the expression of exoS or to induce development of a robust biofilm that did not depend on the presence of gacA (Figure 2). When rsmY or rsmZ were expressed from a plasmid under the control of their respective native promoters, inhibition of exoS transcription was observed only with the rsmY construct, while in the same context, expression of rsmZ had no effect. Similarly, in the biofilm assay, expression of rsmY and rsmZ from their native promoters did not appear to significantly affect biofilm formation, although in the gacA mutant, the rsmY construct allowed for biofilm formation above the background. In general, rsmY and rsmZ seem to be redundant in function and deletion of both rsmY and rsmZ results in phenotypes that are similar to those of a gacA mutant. The stronger effect of rsmY on the two phenotypes is consistent with the higher levels of rsmY expression relative to those of rsmZ (Brencic and Lory, 2009; Kay et al., 2006).
Figure 1. Effect of mutations in the RetS/GacS/A/RsmY/Z/A pathway on expression of a T3S gene exoS and on development of biofilm.
A. Miller assays were used to measure expression of the exoS-lacZ fusion in wild type and mutant strains of P. aeruginosa. As induction of the T3S genes occurs under conditions of low calcium, cultures were grown in Ca2+-depleted (grey bars) and Ca2+-replete (white bars) LB media, at 37°C for 4 hours.
B. Biofilm phenotypes were measured in wild type and mutant strains of P. aeruginosa as the attachment of bacteria to tubes and formation of biofilm rings and pellicles after 14 and 20 h static incubation at 30°C.
Figure 2. Effect of RsmY and RsmZ overexpression on expression of the exoS gene and on biofilm development.
A. Expression of the exoS gene was measured using Miller assays in P. aeruginosa strains carrying an exoS-lacZ reporter fusion and rsmY or rsmZ cloned with their native promoter into a promoterless plasmid pPSV40, or under the control of a constitutively active PlacUV5 promoter in a derivative of the pMMB67 plasmid (See Experimental procedures). As induction of the T3S genes occurs under conditions of low calcium, cultures were grown in Ca2+-depleted (grey bars) and Ca2+-replete (white bars) LB media, at 37°C for 4 hours.
B. Biofilm phenotypes were measured in P. aeruginosa wild type and gacA mutant strains with rsmY or rsmZ cloned with their native promoter into a promoterless plasmid pPSV40, or under the control of a constitutively active PlacUV5 promoter in a derivative of the pMMB67 plasmid. Biofilms were scored as the attachment of P. aeruginosa to tubes and formation of the biofilm rings and pellicles after 20 h static incubation at 30°C.
These results are also consistent with the recently-proposed model in which RetS is an antagonist of the GacS-catalyzed phosphorylation of the response-regulator GacA, and where transcription of the sRNAs RsmY and RsmZ is activated by the GacS/GacA two-component system (Goodman et al., 2009). In addition to the T3S genes and the genes involved with biofilm production, RetS is known to regulate several hundred P. aeruginosa genes, many of which are also involved in virulence. We wondered whether or not all of these genes are regulated by RetS through the GacS/GacA pathway. We analyzed and compared transcription profiles of the P. aeruginosa retS gacS double mutant to the previously published profiles of the wild type and the retS mutant strains (Goodman et al., 2004). Comparison of the transcript levels in the retS and retS gacS mutant strains to the levels in the wild type strain showed that the 218 genes that were up-regulated in the retS mutant, were down-regulated in the retS gacS double mutant (Figure S1, Table S1). On the other hand, the 275 that were down-regulated in the retS strain were up-regulated in the retS gacS strain. This result confirms that GacS acts downstream of RetS and suggests that RetS acts exclusively through its control of the GacS/A system. The fact that the effect of the retS gacS double mutation was less pronounced compared to the effect of the retS mutation (Figure S1, Table S1), suggests that the effect of GacS on gene expression under the conditions that were used in the experiment is not significant. This is not surprising; the experiment was conducted with cultures in the log phase (at OD 600 of 0.5) when rsmY and rsmZ are expressed at very low levels (Brencic and Lory, 2009). At that time point, a mutation in retS results in a dramatic increase in the levels of rsmY and rsmZ, while in the gacS mutant, the resulting lack of expression of the sRNAs is not very different from the low levels of expression of the two sRNAs in the wild type strain. This rationale was also considered in the design of the experiments with the gacA and rsmYZ mutants described below, which were consequently conducted with stationary phase cultures. In summary, the results of the microarray analysis provide strong evidence that the regulation of all RetS-regulated genes is gacS-dependent. Since GacS phosphorylates GacA (Goodman et al., 2009), and GacA controls transcription of the sRNAs RsmY and RsmZ, the entire RetS regulon is very likely regulated through RsmY and RsmZ.
Regulation of all GacA-controlled genes is dependent on RsmY and RsmZ
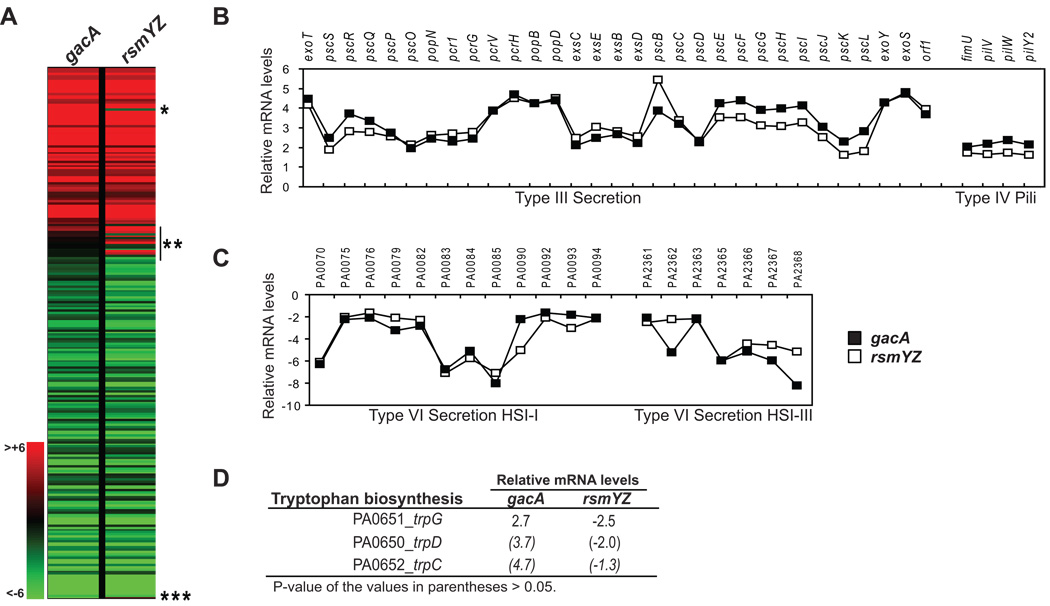
We next wondered whether or not the regulation of all of the GacA-controlled genes in P. aeruginosa is dependent on RsmY and RsmZ. Therefore, we examined the role of rsmY and rsmZ in the regulation of GacA-controlled genes by comparing mRNA levels of a wild type P. aeruginosa to those of a gacA mutant and of an rsmYZ double mutant (Figure 3, Table S2). A total of 241 genes showed changes in their transcipt levels by more than two-fold in at least one of the two mutants (Table S2). The affected genes included many virulence factors, including the T3S and the type IV pili genes, which were upregulated in the two mutants, and genes located on the type VI secretion islands HSI-I and HSI-III, which were downregulated (Figure 3B–C). Overall, the transcriptomes of the two strains were almost indistinguishable (Figure 3A), suggesting that all of the GacA-regulated genes are controlled through the RsmY/Z system.
Figure 3. Comparison of mRNA levels relative to wild type between P. aeruginosa gacA and rsmYZ mutants.
A. Heatmap of the 241 genes whose mRNA levels changed more than two fold relative to wild type in at least one of the two mutants (See text and Table S1 for detail). Red color represents genes that were upregulated relative to wild type, green represents genes that were downregulated relative to wild type (see color scale on the left). * trpC gene, which was upregulated in the gacA strain but was not affected in the rsmYZ strain (See text for detail); ** genes whose expression was affected in the rsmYZ strain only (See text and Table S3 for detail); *** gacA gene; downregulation in the gacA strain is artificial as this gene was deleted in this strain.
B–D. mRNA levels of selected genes in the gacA and rsmYZ mutants relative to wild type. B. Type III secretion genes and Type IV pili genes were upregulated in both gacA and rsmYZ mutants. C. Genes encoded in the Type VI secretion islands HSI-I and HSI-III were downregulated in both gacA and rsmYZ mutants. Open symbols – gacA mutant; Closed symbols – rsmYZ mutant. D. Genes encoded in the trp operon were upregulated in the gacA mutant and downregulated in the rsmYZ mutant (See text for detail).
There were 13 genes whose transcript levels were not affected in the gacA mutant, yet they were significantly different from the wild type levels in the rsmYZ mutant (Figure 3A, Table S3). For some of these genes, the standard deviation for the two replicates in the rsmYZ strain was rather large and this could explain this finding (Table S3 and data not shown). On the other hand, this result could suggest that the effect of RsmY and/or RsmZ on some of these genes can be influenced by factors other than GacA, such as the RNA chaperone Hfq, which is known to affect RsmY stability (Sonnleitner et al., 2006). The transcriptome analysis identified only one gene whose transcript concentration changed in the gacA mutant but was unaffected in the rsmYZ mutant (Figure 3A and 4D). Levels of trpC were increased in the gacA mutant, but did not change in the rsmYZ double mutant, suggesting that trpC is regulated by GacA independently of rsmY and rsmZ. The trpC gene is the last gene in a three-gene operon involved in tryptophan biosynthesis. The transcript levels for other two genes in the operon, trpG and trpD, were also increased in the gacA mutant and not affected in the rsmYZ double mutant, although because of their high P-value, these two genes did not pass our stringency cut-off. We constructed trpG-lacZ and trpC-lacZ reporter fusions and tested their expression in wild type and gacA and rsmYZ mutant backgrounds (Data not shown.). These fusions were expressed at the same levels in all three strains and therefore we conclude that the result is a false positive. In summary, our analysis found that the same genes are controlled by the GacS/GacA two-component system and by the RsmY/Z post-transcriptional mechanism. Therefore, it appears that regulation of all GacA-regulated genes is dependent on RsmY and RsmZ.
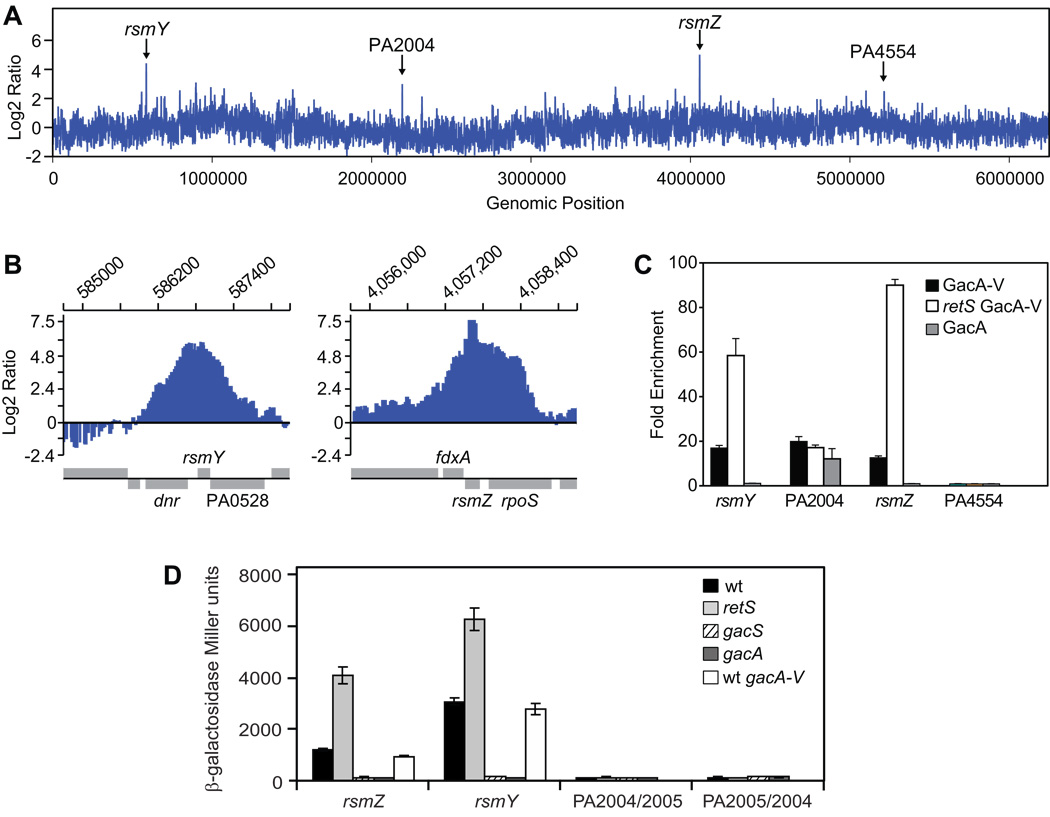
Figure 4. Identification of GacA binding sites in the P. aeruginosa genome.
A. Results of genome-wide ChIP-on-chip analysis using a P. aeruginosa retS strain with VSV-G-tagged GacA (GacA-V). Anti-VSV-G-tag antibody was used to capture GacA-V with bound DNA fragments, which were then used to analyze a high density DNA oligonucleotide microarray representing the P. aeruginosa genome. To obtain a genome-wide view of GacA-V-bound probes, data for the three biological replicates were averaged and were used to generate an enrichment map of fragments represented by probes corresponding to genes on the P. aeruginosa chromosome. Indicated are the four loci where enrichment of co-immunoprecipitated DNA fragments was observed.
B. Higher resolution of the rsmZ and rsmY genomic loci where the enrichment of DNA fragments was detected. Genes corresponding to the enriched regions are represented below the charts above the horizontal line if their orientation is left to right, and below the horizontal line if their orientation is right to left.
C. Chromatin immunoprecipitation (ChIP) using a wild type P. aeruginosa and a retS mutant expressing VSV-G tagged GacA (GacA-V), and a wild type strain expressing untagged GacA (mock ChIP). Quantitative PCR was used to quantify the co-immunoprecipitated DNA fragments using rsmZ-, rsmY-, PA2004/PA2005-, and PA4554-specific primers.
D. Activities of a lacZ reporter fused to DNA fragments that were enriched in the ChIP-on-chip experiment. The expression of the lacZ fusions was measured as β-galactosidase activity (in Miller units) in the P. aeruginosa wild type and retS, gacS, and gacA mutant strains. The PA2004/PA2005 intergenic region was tested in both orientations. For rsmY and rsmZ fusions, the expression was also measured in the strain expressing gacA with a carboxy-terminal VSV-G tag (gacA-V). The assays were conducted with cultures at 7 hours of growth.
GacA directly controls transcription of only two genes, rsmY and rsmZ
To confirm the above results and identify all P. aeruginosa genes that are directly regulated by GacA, we performed genome-wide ChIP-on-chip analysis using a P. aeruginosa retS mutant strain that expresses an epitope-tagged version of GacA from the native gacA locus. This strain has a fully active GacS/GacA phosphorelay pathway, and expresses GacA with a vesicular stomatitis virus glycoprotein (VSV-G) epitope tag fused to its carboxy terminus (Figure 4D). For the experiment, cells were grown for approximately 4 hours (at OD600 of 1.4). At this time point, RsmY and RsmZ begin to accumulate and it is therefore expected that rsmY and rsmZ are actively transcribed (Brencic and Lory, 2009). We used anti-VSV-G antibody-coated agarose beads to capture the VSV-G-tagged GacA (GacA-V) with bound DNA fragments, and these fragments were then hybridized to a high density DNA oligonucleotide microarray representing the P. aeruginosa (PAO1) genome. To obtain a genome-wide view of GacA-V-bound probes, data for three biological replicates were averaged and used to generate an enrichment map of fragments represented by probes corresponding to the P. aeruginosa chromosome. Using a stringent cut-off threshold of 99.9% (see Experimental Procedures), we identified a total of four putative binding regions present in all three replicates, and denoted by peaks in Figure 4A. Enrichment of DNA fragments at the rsmY and rsmZ loci was observed. In addition, regions corresponding to the intergenic region between PA2004 and PA2005, and the region corresponding to PA4554 were also enriched by the GacA-V immunoprecipitation.
To validate the enrichment of the specific regions identified by ChIP-on-chip, we carried out chromatin immunoprecipition (ChIP) with GacA-V in both the retS mutant, and in wild type P. aeruginosa; in the wild type strain phosphorylation and thus activation of the GacA response regulator is blocked by RetS. Although three of the regions that were enriched in the ChIP-on-chip analysis were detected in the ChIP experiment, only fragments containing the rsmY and rsmZ genes were bound in a RetS-responsive manner (Figure 4C). PA4554 was not enriched in the ChIP experiments, and represents a false positive result in the ChIP-on-chip data. Such a result is most likely due to an amplification and/or hybridization bias at this chromosomal location (Figure 4C). In order to further assess the specificity of the binding, we also carried out a mock ChIP experiment, where beads coated with antibody to the VSV-G tag were used to capture crosslinked DNA from P. aeruginosa expressing untagged GacA. In the mock experiment, significant enrichment of a fragment containing the PA2004-PA2005 intergenic region was detected, while no rsmY-or rsmZ-containing DNA fragments were detected (Figure 4C). Therefore the enrichment of the PA2004-PA2005 intergenic region in the ChIP-on-chip experiment is the result of a non-specific interaction between this region and reagents used in the IP reactions. Furthermore, when lacZ reporter fusions were constructed using promoter-containing fragments from each of the regions showing enhanced GacA binding, no difference in promoter activity was detected in the wild type, gacA or retS mutants using the DNA fragment from the PA2004-PA2005 intergenic region in either orientation (Figure 4D). On the other hand, transcription of rsmY and rsmZ was strongly dependent on gacA and enhanced by the retS mutation (Figure 4D). In summary, our analysis of GacA binding to the P. aeruginosa chromosome DNA suggests that the RetS/GacS/GacA pathway regulates global gene expression by directly controlling transcription of only two genes specifying the sRNAs RsmY and RsmZ.
Features of the rsmY and rsmZ regulatory regions and the role of MvaT and MvaU
In P. aeruginosa a conserved region upstream of the rsmY and rsmZ promoters has been identified, consisting of the sequences TGTAAGCATTAACTTACA and TGTAAGCCAAGGCTTACA, respectively (Heeb et al., 2002; Kay et al., 2006; Kulkarni et al., 2006). Variations of these sequences can be also found upstream of csrB/csrC, and rsmY/rsmZ genes in other microorganisms and may represent upstream activation sites (UASs), recognized by the phosphorylated forms of the GacA/UvrY/LetA family of regulatory proteins (Kulkarni et al., 2006). Indeed, in L. pneumophila, LetA bound to fragments containing these sequences (Sahr et al., 2009), and in P. fluorescens CHA0, a deletion of this sequence caused a marked reduction in the expression of rsmZ and completely abolished the control by GacA (Heeb et al., 2002).
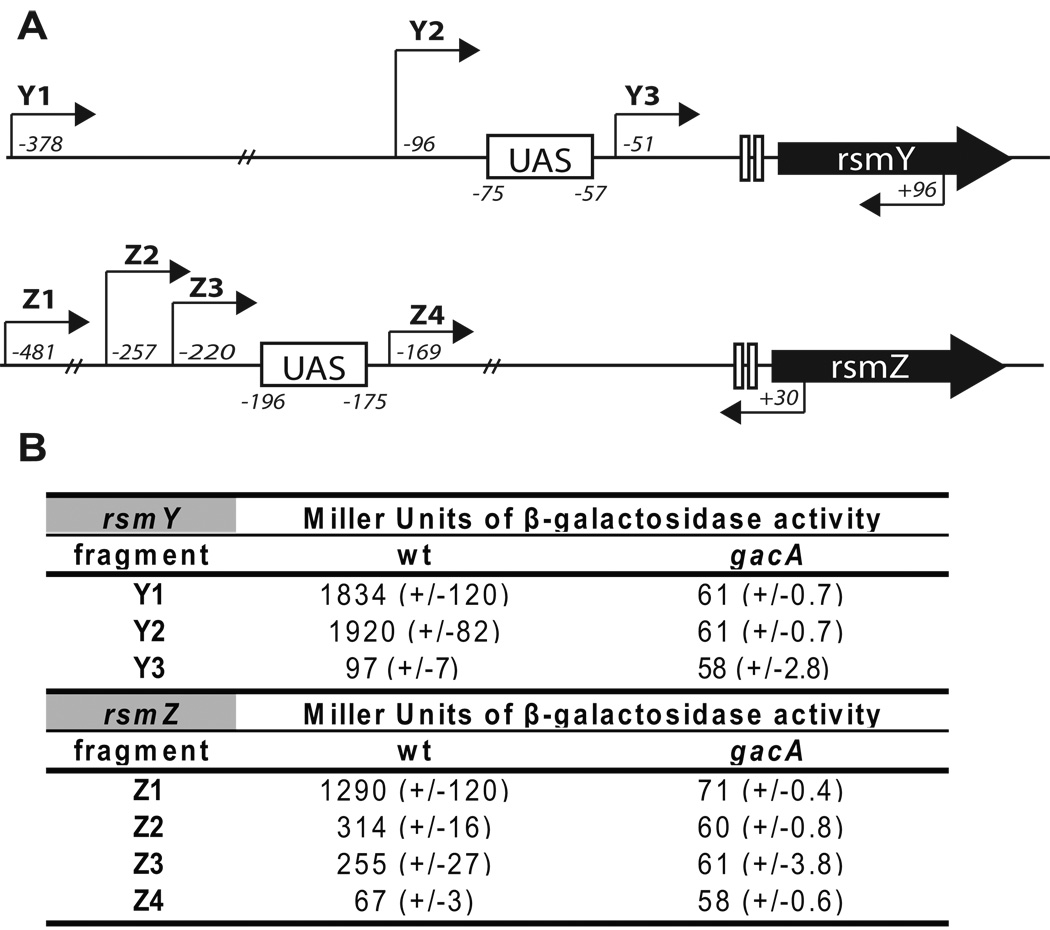
We constructed transcriptional lacZ fusions using truncated fragments of the upstream regulatory regions of rsmY and rsmZ. For both rsmY and rsmZ, activities of those reporter fusions that lacked the conserved UAS site were reduced to the levels detected in the gacA mutant (Figure 5). For rsmY, fusion to a fragment extending 21 bps upstream of the UAS was as active and GacA-dependent, as the fusion ending 303 bps upstream of the UAS, suggesting that it contained all the elements necessary for the full expression of rsmY. In the case of rsmZ, fusions to fragments extending as far as 61 bps upstream of the UAS showed only partial activity when compared to the fragment that ended 285 bps upstream of the UAS, suggesting that the sequences considerably further upstream of the conserved UAS element play a role in the activation of rsmZ expression (Figure 5). Overall, these results confirm that the predicted UAS is required for the GacA-dependent expression of the rsmY and rsmZ genes and these regulatory sequences most likely represent the binding sites for phosphorylated GacA.
Figure 5. Location of the putative GacA regulatory sites upstream of the rsmY and rsmZ genes.
A. Arrows indicate the location of primers used to amplify the sequences that were fused to a lacZ reporter. Italicized numbers indicate the position of the beginning (top arrows) and the end (bottom arrows) nucleotide of each fragment relative to the transcription start site of rsmY or rsmZ.
B. Activities of a lacZ reporter fused to various fragments upstream of the rsmY and rsmZ genes as indicated in A. The activities were measured in P. aeruginosa wild type and gacA mutant strains and are shown as Miller Units of β-galactosidase activity (with standard deviation shown in parentheses). Cultures were grown for 7 hours at 37°C before the assay.
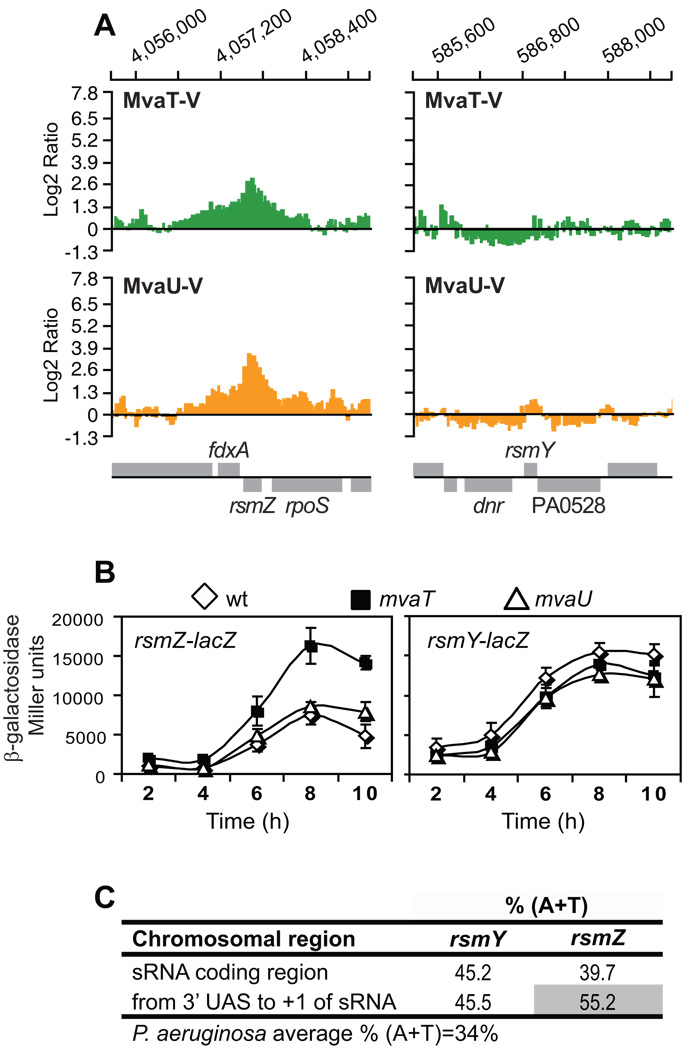
The differences in length and structure of the sequences upstream of the rsmY an rsmZ genes that are required for the expression of the two sRNAs, and the overall lower levels of expression of rsmZ compared to rsmY (Brencic and Lory, 2009), suggest that factors other than GacA may play a role in the regulation of the expression of each of the two sRNAs. We have previously used ChIP-on-chip analyses to identify interactions of MvaT and MvaU, two members of the H-NS family of global regulatory proteins, with regions on the P. aeruginosa chromosome (Castang et al., 2008). Among hundreds of chromosomal fragments, bound to MvaT, MvaU, or both, immunoprecipitation with either one of these proteins enriched for fragments containing the rsmZ gene. In contrast, we detected no enrichment of probes near the rsmY gene (Figure 6A). In order to test the influence of MvaT and MvaU on rsmZ expression, we introduced rsmY and rsmZ transcriptional lacZ fusions into mvaT and mvaU mutants of P. aeruginosa. In the mvaT mutant, expression of the rsmZ-lacZ fusion was increased about two fold throughout the course of growth, while the expression of the rsmY-lacZ construct was unaffected by the absence of MvaT (Figure 6B). The lack of any effect of the mvaU deletion on rsmZ expression is consistent with the observations made for the roles of MvaT and MvaU in expression of other target genes. The two proteins are functionally redundant, and a deletion of mvaU results in an increase in the amount of MvaT in the cell. It has been proposed that this compensatory effect could be sufficient to mask any role MvaU might play in the repression of target genes (Vallet-Gely et al., 2005).
Figure 6. Role of MvaT and MvaU in the regulation of rsmY and rsmZ.
A. VSV-G-tagged MvaT (MvaT-V) and MvaU (MvaU-V) associate with intergenic and coding sequences of rsmZ (left panel) but not of rsmY (right panel). Genes corresponding to the enriched regions are represented below the charts above the horizontal line if their orientation is left to right, and below the horizontal line if their orientation is right to left.
B. Effect of mvaT and mvaU deletions on the expression of rsmZ (left panel) and rsmY (right panel), as measured by Miller assays using rsmZ-lacZ and rsmY-lacZ reporter fusions.
C. The region between 3' of the UAS site and transcription start site (+1) of rsmZ is significantly enriched for A and T compared to the overall composition of various P. aeruginosa genomes and compared to the rsmZ coding sequence.
MvaT and MvaU are known to preferentially bind AT-rich regions of the chromosome (Castang et al., 2008). We examined the A+T composition of the rsmY and rsmZ coding regions and of the regions between the UAS element and the transcription start site of each sRNA (Figure 6C). The 174-bp region between the GacA binding site and the rsmZ transcriptional start site is highly enriched for A and T (55%) compared to the overall composition of various P. aeruginosa genomes, with an average % (A+T) content of 34%, as well as compared to the A+T-composition of the rsmZ coding sequence (39%) (Figure 6C). Therefore, it is likely that this region represents one or more MvaT or MvaU binding sites. Both the rsmY coding sequence and the rsmY upstream region are also enriched for A and T (45%), although the enrichment is not as striking and the A+T compositions of two regions are indistinguishable (Figure 6C).
DISCUSSION
RsmY and RsmZ-like small regulatory RNAs (sRNAs) act by antagonizing the activities of RNA-binding translational regulators of the CsrA/RsmA family (Babitzke and Romeo, 2007; Lapouge et al., 2008; Lucchetti-Miganeh et al., 2008). RsmA/CsrA proteins bind to structured segments at the 5' ends of target mRNAs, and prevent or facilitate translational initiation, which is reversed following sequestration of RsmA by the sRNAs. The transcription of genes encoding the Csr/Rsm sRNAs in all γ-proteobacteria is controlled by a two-component regulatory system, consisting of a sensor-kinase and response regulator, BarA and UvrY, BarA and SirS, LetS and LetA, GacS and GacA in E. coli, Salmonella, Legionella and Pseudomonas species, respectively. In P. aeruginosa the RsmY/Z/A post-transcriptional regulatory system controls the expression of over 500 genes (Brencic and Lory, 2009; Burrowes et al., 2006) and the transcription of RsmY and RsmZ depends on the GacS/GacA pathway, which is antagonized by the RetS sensor kinase (Goodman et al., 2009). We have shown that the expression of two selected reciprocally controlled phenotypes (type III secretion and biofilm formation) is coordinated such that a mutant in retS phenocopies a mutation in rsmA, and the phenotypes resulting from a gacA mutation are identical to those observed in strains where the genes for rsmY and rsmZ have been deleted. We also compared transcript levels in the retS mutant and retS gacS double mutant to those in wild type cells. For each gene, we found that changes in transcript levels caused by the retS mutation depended on the presence of GacS. On the other hand, comparison of mRNA levels in a gacA mutant and rsmYZ double mutant showed that transcript concentrations for all genes were almost indistinguishable between the two strains. Combined, these results provided evidence that the components of the RsmA, RsmY and RsmZ post-translational regulatory system may be the primary, if not the sole mediators of the signals transduced by the RetS/GacS/GacA pathway. To test this hypothesis further, we carried out ChIP-on-chip analysis to determine the genome-wide distribution of GacA binding sites. We detected specific GacA association only to regions adjacent to the rsmY and rsmZ genes, and we showed that this association is positively influenced by a retS mutation. We therefore conclude that phosphorylation of GacA by GacS results in transcription of only two genes, and the broad effect on mRNA levels influenced by GacS/GacA (and therefore RetS) is due to the activities of two sRNAs RsmY and RsmZ on the translational regulator RsmA. It is important to note, however, that not all of the genes that are affected by the retS, gacA or rsmYZ mutations are expected to be directly regulated by RsmA via mRNA binding. In fact, the effect of the RetS/GacS/A/RsmY/Z/A on many of these genes, including the T3S genes, is likely to be a consequence of RsmA-mediated regulation of transcriptional regulators of those genes (Brencic and Lory, 2009).
The two-component systems that regulate transcription of the Csr/Rsm family of sRNAs control a variety of cellular functions in different microorganisms. It is conceivable that in most instances the regulatory effect will be achieved by control of transcription of the sRNA genes with the subsequent post-transcriptional effect of these regulatory RNAs on CsrA/RsmA translational regulators bound to their mRNA targets. However, unlike what we have observed for the GacS/A system in P. aeruginosa, in other bacterial species, direct binding of the response regulator to promoters other than those for the sRNAs has been demonstrated. In Salmonella enterica ser. Typhimurium, SirA directly controls the expression of csrB and csrC, but also the expression of the HilA and HilC transcription factors (Teplitski et al., 2003). Furthermore, the Salmonella BarA/SirA system can affect the expression of the operon encoding the type I fimbriae by both direct transcriptional and indirect post-transcriptional mechanisms. The transcription of the fim genes is directly regulated by SirA, and the same regulatory protein also controls the translation and/or stability of the fim gene transcripts through its regulation of csrB and csrC transcription (Teplitski et al., 2006). In Legionella pneumophila, the expression of genes involved in flagellar functions is regulated by the LetS/LetA system independently of RsmY and RsmZ (Sahr et al., 2009). Therefore, utilizing genome-wide technologies such as ChIP-on-chip analyses, a more careful examination of direct interactions of the response regulators of the GacA/UvrY/SirA/LetA family with their regulatory targets could identify more complex regulatory networks in diverse organisms, capable of amplifying or directing the flow of input signals towards specific groups of genes. However, in P. aeruginosa the GacA regulatory activity is limited to only two promoters.
Given the membrane localization and a prominent periplasmic domain of the two-component sensor kinases, it is expected that the GacS/GacA/RsmY/Z-type regulatory pathways are controlled by external signals that regulate the autokinase activity of the sensor-kinase. However, in certain instances, additional regulatory elements were shown to influence the expression of the Csr/Rsm small RNAs. In Erwinia caratovora, the FlhDC complex, the master regulator of flagella production, positively regulates the expression of the response regulator GacA and represses the transcription of HexA, a LysR-like protein shown to control the expression of the sRNA gene rsmB (Cui et al., 2008; Mukherjee et al., 2000). In E. coli, the CsrD protein that contains GGDEF and EAL domains regulates the levels of the small RNAs CsrB and CsrC by controlling their degradation rates (Suzuki et al., 2006). In P. aeruginosa, the RNA chaperone Hfq binds and stabilizes RsmY but not RsmZ and therefore influences RsmY levels post-transcriptionally (Sonnleitner et al., 2006).
The transcription rates of the rsmY and rsmZ genes in P. aeruginosa are clearly distinct; rsmY is expressed at approximately two-fold higher levels than rsmZ. We have shown that a considerably longer region upstream of the conserved UAS element is required for the expression of rsmZ compared to the region that is required for the expression of rsmY. Furthermore, there is a distinct difference in the location of the conserved UAS element relative to the transcription start sites of the two small RNAs. The UAS site is located 57 upstream of rsmY and 174 bps upstream of rsmZ (Table S4). This pattern appears to be conserved among other organisms phylogenetically related to P. aeruginosa, including Pseudomonas putida, Pseudomonas fluorescens, and Pseudomonas syringae (Table S4). The location of the UAS relative to the rsmY and rsmZ genes in L. pneumophila is also similar to those in the Pseudomonaceae, while this pattern is not conserved in E. coli, Salmonella typhimurium, Vibrio cholerae or Yersinia pestis, where the distances between the UAS sequences and the genes encoding sRNAs CsrB, CsrC ranges between 150 to 211 bp. The differences in length and structure of the sequences upstream of the rsmY an rsmZ genes that are required for the expression of the two sRNAs, and the overall difference in the levels of expression of rsmZ and rsmY (Brencic and Lory, 2009) suggest that additional factors may be contributing to the control of the expression of each of the two sRNAs.
We have shown that a region of DNA adjacent to rsmZ, but not rsmY, associates with MvaT and MvaU, two histone-like nucleoid structuring proteins related to H-NS (Castang et al., 2008), and that MvaT repressed transcription of rsmZ while it had no effect on the expression of rsmY. Similar to H-NS binding to sequences with a strong bias towards adenine and thymine deoxyribonucleotides (A and T), MvaT and MvaU preferentially associate with regions of the chromosome where the % (A+T) is relatively high (Castang et al., 2008). We found that the 174-bp region between the GacA binding site and the rsmZ transcriptional start site is highly enriched for A and T (55%) compared to the overall composition of various P. aeruginosa genomes, with an average % A+T of 34% (Mathee et al., 2008; Stover et al., 2000). Therefore, it is likely that this region contains one or more MvaT or MvaU binding sites, whose occupancy results in transcriptional repression. Since repression of gene expression by H-NS-like proteins is often counteracted by the activities of specific transcriptional regulators, it is conceivable that GacA performs this anti-silencing function, or that displacement of bound MvaT or MvaU is carried out by an as yet unidentified transcription factor. The differences in regulation of cellular concentrations of RsmY and RsmZ may provide a clue to understanding the benefits of having two seemingly redundant RNAs. In principle, this arrangement may allow a more efficient and precise regulatory response via a gene dosage effect by providing more possibilities for integrating various signals resulting in the regulation of the overall levels of the sRNAs.
Recent recognition of sRNAs as key intermediates in stress and nutritional responses in bacteria highlighted the importance of post-transcriptional control mechanisms for successful environmental adaptation. In several microorganisms, up to a hundred different sRNAs have been identified (Altuvia, 2007) and given the likelihood of each regulating more than a single mRNA, these elements can account for the regulation of expression of a substantial fraction of the transcriptome. Indeed, the global impact of the RetS/Gac/Rsm signal transduction pathway acting through rsmY and rsmZ may greatly exceed the range of many prokaryotic signal transduction pathways that act through transcriptional regulators.
EXPERIMENTAL PROCEDURES
Plasmids, Strains and Culture Conditions
All strains used in this study were derived from P. aeruginosa strain PAK (D. Bradley) or PAO1 (A. Rietsch). Primer sequences and a detailed description of cloning procedures used for construction of lacZ fusions, deletion mutants and other strains used in the study are provided in Supporting experimental procedures. P. aeruginosa strains were maintained in Luria-Bertani (LB) broth with antibiotics as required (150 µg/ml carbenicillin, 75 µg/ml gentamicin, 25 µg/ml Irgasan, 50 µg/ml tetracycline). Growth incubations were performed at 37°C at 300 rpm shaking in baffled flasks for transcriptional profiling or in culture tubes for β-galactosidase assays and chromatin immunoprecipitation analyses.
Transcriptional Profiling
For the retS/gacS experiment, the cultures were grown and processed exactly as described by Goodman and colleagues (Goodman et al., 2004), such that direct comparisons between the gacS retS transcriptome and the previously published retS and wt transcriptomes were possible. For the gacA/rsmYZ experiment, duplicate cultures were grown overnight in LB. These cultures were inoculated at an optical density at 600 nm (OD600) of 0.01 into LB and grown for 7 hours (OD600 ~ 6.0). RNA was extracted, reverse transcribed, fragmented, and labeled as described (Wolfgang et al., 2003) with one exception; at the time of harvesting, 20% of the culture volume of stop solution (95% ethanol, 5% phenol, pH 4.7) was added. The processed samples were hybridized to Affymetrix GeneChip P. aeruginosa Microarrays (Affymetrix), and the chips were washed and scanned according to the provided protocol. The 5,678 probe sets specific to strain PAK were filtered for statistically significant differences (Student's t test p value ≤ 0.05), signal above noise level (Present call in at least two samples), and a minimum 2-fold change using commercially available software (GeneSpring).
β-galactosidase Assays
All measurements of β-galactosidase activity were carried out in duplicates or triplicates starting with overnight cultures. Each experiment was performed at least twice. Overnight cultures were grown in LB with antibiotics as appropriate. Cultures were diluted 1:100 into LB, and grown for 4 or 7 h before being assayed. For the time course of rsmY and rsmZ expression, cultures were sampled at each time point and assayed immediately. The amounts of β-galactosidase activity were quantified as described (Sambrook et al., 1989). In experiments involving the type III secretion genes, assay cultures were grown in calcium-depleted conditions which were achieved by addition of nitrilotriacetic acid and CaCl2, each to a final concentration of 10 mM.
Biofilm Assay
In a modification of the biofilm ring assay (O'Toole and Kolter, 1998), overnight cultures were diluted to OD600 =0.0025 in LB and triplicate 1-ml aliquots were dispensed into glass tubes. Following static incubation in a humidified 37°C chamber, the media was removed and tubes were washed gently with distilled water.
Chromatin Immunoprecipitation (ChIP)
5-ml volumes of LB broth were inoculated at a starting OD600 of ~0.03, and grown at 37 °C with aeration, to an OD600 of ~1.4. Crosslinking and ChIP were then performed as described previously (Castang et al., 2008).
ChIP-on-Chip
Triplicate cultures of PAO1 ΔretS GacA-V, each grown in eight×5-ml volumes of LB as described above, were used for chromatin immunoprecipitation. Approximately 250 ng of ChIP and input DNA from each replicate was subjected to one round of amplification using the BioPrime Array CGH Genomic Labeling System kit (Invitrogen). Resulting sample DNA was labeled and hybridized to custom P. aeruginosa PAO1 high-density oligonucleotide arrays, by Roche NimbleGen, Inc., as previously described (Castang et al., 2008).
ChIP-on-Chip Data Analysis
To control for any nonspecific enrichment, the average log2 values from three mock VSV-G ChIP-on-chip experiments were subtracted from each array prior to data analysis (Castang et al., 2008). Data from each array were smoothed using a sliding window average, with a window size of 6 probes and a step size of 3 probes. Following smoothing, a fixed percentile cut-off method was used to determine “peaks” of GacA-V enrichment (Castang et al., 2008). The threshold level used for analysis was set at 99.9% and only peaks that were isolated in all three arrays were considered to be significant. Such a stringent cut-off was used as we were unable to validate enrichment in regions that were isolated using lower threshold levels (data not shown).
ChIP Quantitative PCR
qPCR was performed using iTaq SYBR green with ROX (BioRad) and an Applied Biosystems StepOnePlus detection system. ChIP fold enrichment values were calculated as described (Castang et al., 2008), and represent the relative abundance of a sequence of interest compared to a negative control region. All ChIP fold enrichment values represent the average of two biological replicates.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Keith Turner (Children’s Hospital, Boston) for writing the Perl script used for ChIP-on-chip data smoothing. We also thank Andy Goodman (Washington University, St. Louis) for helpful discussions and valuable insights during the initial stages of this study. This work was supported by National Institutes of Health Grants AI069007 and AI057754 (to S.LD.), and AI021451 (to SL).
REFERENCES
- Altuvia S. Identification of bacterial small non-coding RNAs: experimental approaches. Curr Opin Microbiol. 2007;10:257–261. doi: 10.1016/j.mib.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Baker CS, Eory LA, Yakhnin H, Mercante J, Romeo T, Babitzke P. CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J Bacteriol. 2007;189:5472–5481. doi: 10.1128/JB.00529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brencic A, Lory S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol. 2009;72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrowes E, Baysse C, Adams C, O'Gara F. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology. 2006;152:405–418. doi: 10.1099/mic.0.28324-0. [DOI] [PubMed] [Google Scholar]
- Castang S, McManus HR, Turner KH, Dove SL. H-NS family members function coordinately in an opportunistic pathogen. Proc Natl Acad Sci U S A. 2008;105:18947–18952. doi: 10.1073/pnas.0808215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Chatterjee A, Hasegawa H, Dixit V, Leigh N, Chatterjee AK. ExpR, a LuxR homolog of Erwinia carotovora subsp. carotovora, activates transcription of rsmA, which specifies a global regulatory RNA-binding protein. J Bacteriol. 2005;187:4792–4803. doi: 10.1128/JB.187.14.4792-4803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Chatterjee A, Yang H, Chatterjee AK. Regulatory network controlling extracellular proteins in Erwinia carotovora subsp. carotovora: FlhDC, the master regulator of flagellar genes, activates rsmB regulatory RNA production by affecting gacA and hexA (lrhA) expression. J Bacteriol. 2008;190:4610–4623. doi: 10.1128/JB.01828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey AK, Baker CS, Romeo T, Babitzke P. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA. 2005;11:1579–1587. doi: 10.1261/rna.2990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune DR, Suyemoto M, Altier C. Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar Typhimurium. Infect Immun. 2006;74:331–339. doi: 10.1128/IAI.74.1.331-339.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Mack TR, Stock AM. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem Sci. 2007;32:225–234. doi: 10.1016/j.tibs.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009;23:249–259. doi: 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudapaty S, Suzuki K, Wang X, Babitzke P, Romeo T. Regulatory interactions of Csr components: the RNA binding protein CsrA activates csrB transcription in Escherichia coli. J Bacteriol. 2001;183:6017–6027. doi: 10.1128/JB.183.20.6017-6027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeb S, Blumer C, Haas D. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J Bacteriol. 2002;184:1046–1056. doi: 10.1128/jb.184.4.1046-1056.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay E, Dubuis C, Haas D. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc Natl Acad Sci U S A. 2005;102:17136–17141. doi: 10.1073/pnas.0505673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay E, Humair B, Denervaud V, Riedel K, Spahr S, Eberl L, Valverde C, Haas D. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J Bacteriol. 2006;188:6026–6033. doi: 10.1128/JB.00409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni PR, Cui X, Williams JW, Stevens AM, Kulkarni RV. Prediction of CsrA-regulating small RNAs in bacteria and their experimental verification in Vibrio fischeri. Nucleic Acids Res. 2006;34:3361–3369. doi: 10.1093/nar/gkl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapouge K, Schubert M, Allain FH, Haas D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol. 2008;67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Cui Y, Mukherjee A, Chatterjee AK. Characterization of a novel RNA regulator of Erwinia carotovora ssp. carotovora that controls production of extracellular enzymes and secondary metabolites. Mol Microbiol. 1998;29:219–234. doi: 10.1046/j.1365-2958.1998.00924.x. [DOI] [PubMed] [Google Scholar]
- Lucchetti-Miganeh C, Burrowes E, Baysse C, Ermel G. The post-transcriptional regulator CsrA plays a central role in the adaptation of bacterial pathogens to different stages of infection in animal hosts. Microbiology. 2008;154:16–29. doi: 10.1099/mic.0.2007/012286-0. [DOI] [PubMed] [Google Scholar]
- Mathee K, Narasimhan G, Valdes C, Qiu X, Matewish JM, Koehrsen M, Rokas A, Yandava CN, Engels R, Zeng E, Olavarietta R, Doud M, Smith RS, Montgomery P, White JR, Godfrey PA, Kodira C, Birren B, Galagan JE, Lory S. Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci U S A. 2008;105:3100–3105. doi: 10.1073/pnas.0711982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNealy TL, Forsbach-Birk V, Shi C, Marre R. The Hfq homolog in Legionella pneumophila demonstrates regulation by LetA and RpoS and interacts with the global regulator CsrA. J Bacteriol. 2005;187:1527–1532. doi: 10.1128/JB.187.4.1527-1532.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Cui Y, Ma W, Liu Y, Ishihama A, Eisenstark A, Chatterjee AK. RpoS (sigma-S) controls expression of rsmA, a global regulator of secondary metabolites, harpin, and extracellular proteins in Erwinia carotovora. J Bacteriol. 1998;180:3629–3634. doi: 10.1128/jb.180.14.3629-3634.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Cui Y, Ma W, Liu Y, Chatterjee AK. HexA of Erwinia carotovora ssp. carotovora strain Ecc71 negatively regulates production of RpoS and rsmB RNA, a global regulator of extracellular proteins, plant virulence and the quorum-sensing signal, N-(3-oxohexanoyl)-L-homoserine lactone. Environ Microbiol. 2000;2:203–215. doi: 10.1046/j.1462-2920.2000.00093.x. [DOI] [PubMed] [Google Scholar]
- O'Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- Pessi G, Williams F, Hindle Z, Heurlier K, Holden MT, Camara M, Haas D, Williams P. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J Bacteriol. 2001;183:6676–6683. doi: 10.1128/JB.183.22.6676-6683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahr T, Brüggemann H, Jules M, Lomma M, Albert-Weissenberger C, Cazalet C, Buchrieser C. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol Microbiol. 2009;72:641–762. doi: 10.1111/j.1365-2958.2009.06677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sonnleitner E, Schuster M, Sorger-Domenigg T, Greenberg EP, Blasi U. Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol Microbiol. 2006;59:1542–1558. doi: 10.1111/j.1365-2958.2006.05032.x. [DOI] [PubMed] [Google Scholar]
- Sorger-Domenigg T, Sonnleitner E, Kaberdin VR, Blasi U. Distinct and overlapping binding sites of Pseudomonas aeruginosa Hfq and RsmA proteins on the non-coding RNA RsmY. Biochem Biophys Res Commun. 2007;352:769–773. doi: 10.1016/j.bbrc.2006.11.084. [DOI] [PubMed] [Google Scholar]
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Babitzke P, Kushner SR, Romeo T. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 2006;20:2605–2617. doi: 10.1101/gad.1461606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplitski M, Goodier RI, Ahmer BM. Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J Bacteriol. 2003;185:7257–7265. doi: 10.1128/JB.185.24.7257-7265.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplitski M, Al-Agely A, Ahmer BM. Contribution of the SirA regulon to biofilm formation in Salmonella enterica serovar Typhimurium. Microbiology. 2006;152:3411–3424. doi: 10.1099/mic.0.29118-0. [DOI] [PubMed] [Google Scholar]
- Vallet-Gely I, Donovan KE, Fang R, Joung JK, Dove SL. Repression of phase-variable cup gene expression by H-NS-like proteins in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2005;102:11082–11087. doi: 10.1073/pnas.0502663102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde C, Haas D. Small RNAs controlled by two-component systems. Adv Exp Med Biol. 2008;631:54–79. doi: 10.1007/978-0-387-78885-2_5. [DOI] [PubMed] [Google Scholar]
- Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci U S A. 2006;103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I, Baker CS, Georgellis D, Babitzke P, Romeo T. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol. 2003;48:657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- Wolfgang MC, Lee VT, Gilmore ME, Lory S. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev Cell. 2003;4:253–263. doi: 10.1016/s1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.