A major theme in the history of life is the formation of beneficial associations between symbiotic microbes and plants and animals (1). The pervasiveness of these associations in every ecosystem illustrates how the acquisition of a microbe’s physiological capacity confers substantial fitness benefits to hosts (1). However, dependence on mutualistic microbes becomes a liability if antagonistic microbes attack or out-compete beneficial ones (2). Therefore, mechanisms to preserve beneficial microbes must be a widespread, although poorly understood, component of host-microbe mutualisms. Here, we show that a beetle uses a bacterium to protect its fungal food source from a competitor fungus.
Southern pine beetles (SPB, Dendroctonus frontalis) engage in a beneficial symbiosis with the fungus Entomocorticium sp. A (EsA), which provides nourishment for their developing larvae. Adult SPBs carry EsA in a specialized storage compartment called a mycangium (Fig. 1A); excavate ovipositional galleries within the inner bark and phloem of host pine trees; and inoculate these galleries with EsA (3, 4). The success of the SPB-EsA mutualism is challenged by an antagonistic fungus, Ophiostoma minus (Om), which can out-compete EsA and thereby disrupt SPB larval development (3, 4). Our results indicate that successful maintenance of the SPB-EsA mutualism is likely mediated by an actinomycetous bacterium that produces antibiotics which selectively inhibit Om.
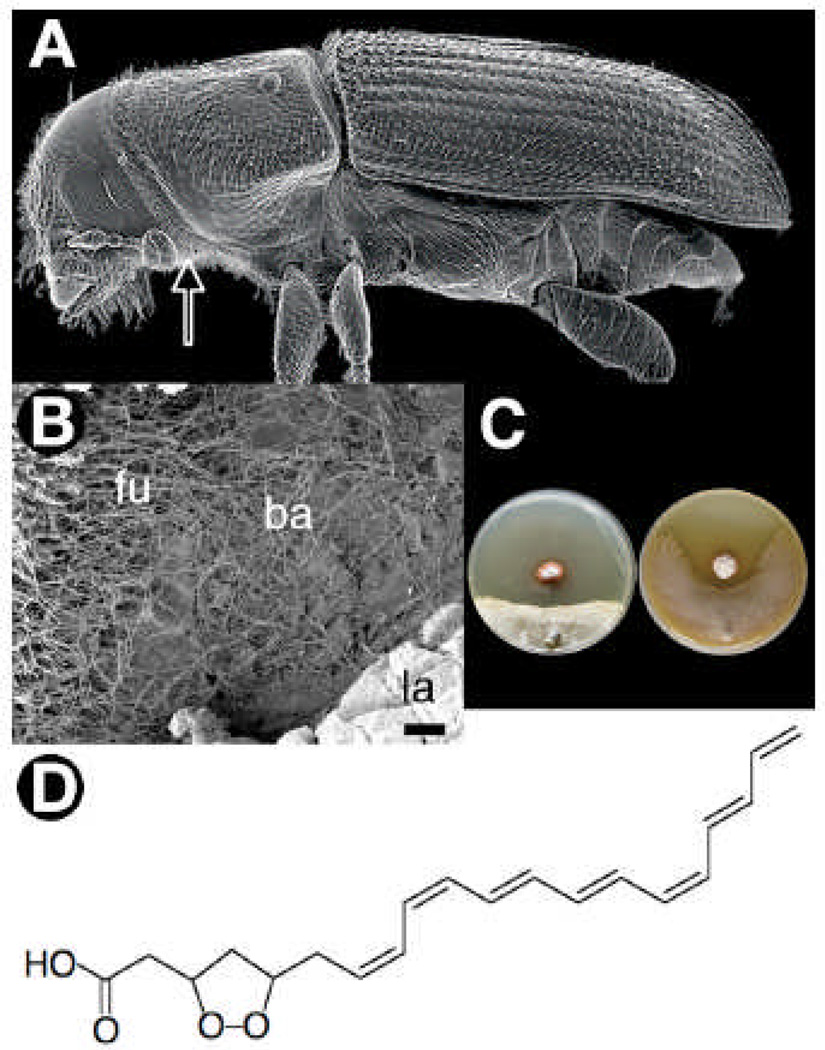
Fig. 1.
(A) SEM micrograph of adult SPB showing the relative placement of the mycangia (arrow), which is used to transport EsA. (B) SEM micrograph from the SPB gallery showing the relative placement of the actinomycetous bacterium (ba), fungus (fu), and beetle larva (la). (C) Representative examples of pairwise bioassay challenges illustrating inhibition of the fungal antagonist Om (left), by a SPB symbiotic actinomycete (strain SPB074). In contrast the SPB’s fungal mutualist EsA is relatively resistant (right) (see SOM for more details). (D) The structure of mycangimycin containing a seven-conjugated double bond chain and a five-membered endoperoxide ring.
The presence of previously unknown actinomycetes within the SPB-EsA mutualisms was established by scanning electron microscopy (SEM) and enrichment culture isolations. SEM revealed unexpected and profuse growth of actinomycetes within the galleries of SPBs, as well as inside the mycangia (Fig. 1B, Fig S1A). Isolations from 110 beetle individuals yielded 846 colony forming units (CFUs) of actinomycetes, including at least one CFU from each of 92 individuals. Out of 164 actinomycete CFUs selected to be transferred to pure culture, 99 isolates had a red morphotype while 65 isolates had a white morphotype. DNA sequence analyses confirmed the visual morphotype distinction, and within each of the two morphotypes there was complete 16S rDNA sequence identity. The two morphotypes form a monophyletic clade closely related toStreptomyces thermosacchari. We also successfully isolated the same red morphotype from 5 of 10 mycangia sampled.
We explored the potential role of the actinomycetes in mediating the SPB fungal community using symbiont pairing bioassays and chemical analyses. The bioassays, which crossed all possible combinations of the two actinomycete morphotypes with EsA and Om, revealed that isolates of the red morphotype produce a diffusible activity that inhibits the beetle’s antagonistic fungus, Om, while only slightly affecting the beneficial fungus, EsA (Fig. 1C, Fig S1B–C). Extensive chemical and spectral analyses on strains of the red morphotype revealed the antifungal molecule responsible for selective inhibition to be a polyene peroxide, which we named ‘mycangimycin’. Mycangimycin (C20H24O4), which has not been previously reported, is a linear 20 carbon carboxylic acid with an endoperoxide linking C-3 and C-5 to form a 1,2-dioxolane and a conjugated cis, cis, trans, trans, cis, trans-heptaene spanning C-7 to C-20 (Fig. 1D). Liquid culture antifungal assays using purified mycangimycin showed Om to be almost 20-times more susceptible (minimal inhibitory concentration (MIC) = 1.0 µM) than EsA (MIC = 19.0 µM) (fig. S1D). The discovery of an actinomycete that is localized in the mycangium and galleries, which produces an antibiotic that selectively suppresses the antagonistic fungus Om, indicates that SPBs engage in an additional mutualism with bacteria to regulate the EsA-Om fungal community. Since other bark-beetle species also depend on successfully maintaining beneficial fungi, tripartite beetle-fungus-bacterium mutualisms may be widespread.
Our discovery parallels earlier work on fungus-farming ants, which use actinomycetes to help protect their fungal gardens from pathogens (5). Taken together, these findings suggest that the use of antibiotic-producing actinomycetes may be a common method to help maintain beneficial microbes. Indeed, considering the importance of pathogens as a driving force in the evolution of all hosts, the benefit of such associations may extend to helping protect plants and animals from pathogens to which they themselves are susceptible (6, 7). If, as seems likely, these associations are widespread, targeting them could be an effective strategy for locating novel biologically active natural products.
Supplementary Material
References
- 1.Moran NA. Curr. Biol. 2006;16:R866. doi: 10.1016/j.cub.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 2.Currie CR, Mueller UG, Malloch D. PNAS. 1999;96:7998. doi: 10.1073/pnas.96.14.7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klepzig KD, Wilkens RT. AEM. 1997;63:612. doi: 10.1128/aem.63.2.621-627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hofstetter RW, et al. Oecologia. 2006;147:679. doi: 10.1007/s00442-005-0312-0. [DOI] [PubMed] [Google Scholar]
- 5.Currie CR, Scott JA, Summerbell RC, Malloch D. Nature. 1999;398:701. [Google Scholar]
- 6.Kaltenpoth M, Gottler W, Herzner G, Strohm E. Curr. Biol. 2005;15:475. doi: 10.1016/j.cub.2004.12.084. [DOI] [PubMed] [Google Scholar]
- 7.Coombs JT, Franco CMM. AEM. 2003;69:5603. doi: 10.1128/AEM.69.9.5603-5608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.We thank A. Adams, S. Adams, C. Booth, E. Caldera, J. Ensign, C. Hsu, Y. Kang, A. Lawrence, W. Monroe, P. Jeffreys, M. Palmisano, A. Pinto, M. Poulsen, K. Raffa, D. Stone and E. Vallery for assistance. Supported provided by the USDA (C.R.C., K.D.K., M.C.Y.), NIH (J.C.), and NSF (C.R.C.).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.