Abstract

Encapsulation of a biological molecule or a molecular complex in a vesicle provides a means of biofriendly immobilization for single molecule studies and further enables new types of analysis if the vesicles are permeable. We previously reported on using DMPC (dimyristoylphosphatidylcholine) vesicles for realizing porous bioreactors. Here, we describe a different strategy for making porous vesicles using a bacterial pore-forming toxin, α-hemolysin. Using RNA folding as a test case, we demonstrate that protein-based pores can allow exchange of magnesium ions through the vesicle wall while keeping the RNA molecule inside. Flow measurements indicate that the encapsulated RNA molecules rapidly respond to the change in the outside buffer condition. The approach was further tested by coencapsulating a helicase protein and its single-stranded DNA track. The DNA translocation activity of E. coli Rep helicase inside vesicles was fueled by ATP provided outside the vesicle, and a dramatically higher number of translocation cycles could be observed due to the minuscule vesicle volume that facilitates rapid rebinding after dissociation. These pores are known to be stable over a wide range of experimental conditions, especially at various temperatures, which is not possible with the previous method using DMPC vesicles. Moreover, engineered mutants of the utilized toxin can potentially be exploited in the future applications.
Introduction
Single molecule fluorescence techniques are revolutionizing biological inquiries both in vitro and in vivo.1−4 In order to observe single molecule reactions for an extended period, molecular movements via diffusion have to be slower than the time scale of the method used to track their movements. Although it is possible to track particles in one dimension, for example cytoskeleton motor proteins moving on actin filaments(5) or microtubules,(6) or in two dimensions, as in the case of membrane protein diffusion,(7) there are many other examples where three-dimensional diffusion would preclude an extended observation time window. Therefore, several methodologies have been developed to keep the molecules of interest under observation for prolonged periods. Immobilization via biotin−streptavidin anchors on bare(8) or polymer-grafted surfaces(9) and confinement within agarose or polyacrylamide gels(10) are very useful strategies, yet care should be taken to ensure minimal perturbation to the activity of biological molecules. A recent report(11) described an elegant technology for the confinement of individual molecules in free solution by means of anti-Brownian electrokinetic trapping (ABEL). ABEL, however, does not allow changing the solution condition while keeping the molecule trapped. An alternative approach entraps a molecule within a small unilamellar vesicle (SUV) which is then tethered on surfaces.12−14 Vesicle encapsulation has been used to study protein folding,(15) RNA folding,(12) DNA folding,(8) DNA−protein interaction,(16) protein−protein interaction,(14) and protein conformational changes.(17) In principle, vesicle encapsulation should provide a more biofriendly environment,(18) which may help avoid artifacts sometimes caused by direct surface tethering of proteins,17,19 because the immediate environment is composed of lipid molecules rather than an artificial solid substrate such as a glass slide. Vesicle encapsulation also obviates the need to modify biological molecules for surface tethering (e.g., biotinylation), offering ease and flexibility to the researchers as was utilized to determine the effect on FRET of dipole orientations of the fluorophores stacked at DNA duplex ends.(20) However, the inability to exchange buffer across the vesicle wall was noted as the major drawback(9) because buffer exchange is commonly practiced either for examining molecules under various conditions, for example by varying concentration of certain chemical agents between measurements,(8) or for triggering certain reactions on demand.(21) Rendering the vesicles porous is therefore essential for fully realizing the potential of the encapsulation approach. This is already achieved for bulk assays,22−24 but their adaptation for single molecule experiments has been limited.
Porous vesicles should also be useful if the measurement condition is not suitable for vesicle formation. In such circumstances, vesicles can be formed under conditions favorable for encapsulation, and the solution condition can then be changed to the desired imaging conditions. Furthermore, because porous vesicles allow buffer exchange without washing out the encapsulated molecules, it would be possible to monitor the interaction between the same pair of biomolecules under various conditions. Finally, the presence of pores prevents the vesicle-to-vesicle variation of intravesicular conditions, for example ion concentrations, emerging from stochastic encapsulation and vesicle size distribution (further discussed in Supporting Information), which may be important for studying systems that display strong heterogeneity.(12) In this regard, pores ensure that the intravesicular condition is in equilibrium with the bulk solution, hence remains identical between vesicles. Clearly, introducing pores on vesicles should greatly expand the venues for single molecule vesicle encapsulation assays.
In order to fully harness the capabilities of vesicle encapsulation and overcome the impermeability barrier, we recently exploited the thermotropic properties of lipid membranes(16) where the membrane permeability of solutes reaches a maximum around membrane’s melting temperature (Tm). We showed previously that the vesicles made of the phospholipids DMPC (dimyristoylphosphatidylcholine) (Tm = 23 °C) can readily allow the passage of K+ (12) and ATP and its analogues(16) at room temperature. Nevertheless, the pores rely on transient formation of lipid packing defects, and their presence and properties may be affected by many factors. An obvious limitation is that the working temperature range is narrow with the membrane being porous only within 3 °C of Tm,(25) making it laborious to carry out measurements at various temperatures. For steady-state measurements, one would need to incubate the sample with the desired buffer at the Tm of the lipid membrane before taking data at a new temperature. Moreover, data acquisition at temperatures different from Tm would be problematic if the encapsulated reaction consumes substrates (e.g., ATP), as their consumption and product accumulation will alter the dynamics. Finally, nonequilibrium kinetic experiments, for example via flow, at various temperatures would require several different lipid compositions with Tm values that match those of the temperatures to be used. We therefore utilized a bacterial toxin, α-hemolysin (aHL), to induce stable pores on SUVs, which had been previously used for bulk measurements in an attempt to build an artificial cell.(22) We proposed using aHL in an earlier report,(16) and the actual implementation is the subject of this article.
aHL is a bacterial toxin secreted by Staphylococcus aureus as water-soluble monomers which incorporate into lipid membranes to form a heptametric pore.(26) There are several reasons for our choice of aHL: (i) Owing to spontaneous incorporation of aHL into the membrane, such pores are easier to make than many other membrane proteins which require difficult reconstitution procedures. (ii) Once formed, aHL pores are stable in a wide variety of buffer conditions, pH, and temperature. (iii) The aHL pore with a narrowest opening of 1.8 nm is not selective, allowing passive passage of molecules up to 3kD both ways.(27) (iv) Finally, available engineered mutants of aHL may be further utilized to achieve unique experimental assays.
Here, we carried out the proof-of-concept experiments using smFRET (single molecule fluorescence resonance energy transfer)28,29 on a home-built total internal reflection fluorescence setup (Figure 1) using three different model systems. First, the hairpin ribozyme whose single molecule behavior under various conditions is well-characterized(30) served as a molecular reporter for the intravesicular buffer condition, in this case magnesium ion concentration. Second, the RNA four-way junction was also used as a well-characterized test system.(31) As the third model system we encapsulated E. coli Rep helicase together with its DNA substrate in order to demonstrate the passage of ATP through aHL pores and to showcase the ability to study transient protein−DNA interactions for a prolonged period.
Figure 1.
A diagram summarizing the experimental scheme (not to scale). (Left) A home-built prism-type total internal reflection microscope with two-color fluorescence detection. (Right) The encapsulation assay. A supported lipid bilayer (a single membrane layer is depicted as one gray stripe) is first formed on a quartz coverslide (yellow) as a cushion. SUVs are then immobilized on the lipid cushion through biotin (brown dots)−streptavidin or neutravidin (blue) linker. The aHL pores on the SUV are shown also in blue. Encapsulated inside SUVs is the biomolecule (orange), depicted in folded and unfolded conformations at two and eight o’clock positions, respectively. The molecule is labeled with a donor (green) and an acceptor (red) which serve as FRET probes. The star shape represents the brighter fluorophore. Black circles represent the chemical agent (e.g., Mg2+ or ATP) relevant to the reaction under study.
Materials and Methods
FRET Measurements
All experiments were carried out using a prism-based two-color total internal reflection fluorescence microscope capable of single molecule FRET analysis(28) at room temperature and with 100 ms exposure time unless otherwise stated. A 532 nm laser was used for donor excitation in FRET measurements. A 633 nm laser was used to excite the acceptor directly to confirm colocalization of a donor-labeled Rep helicase and an acceptor-labeled DNA. FRET efficiency was calculated as the acceptor intensity divided by the total intensity of donor and acceptor fluorescence averaged over 1 s. Data from hundreds of molecules were pooled to obtain FRET histograms. Noise in the measurements could yield FRET values below 0 and above 1 due to negative values after background subtraction for the acceptor and donor intensities, respectively. The leftmost peak in FRET histograms represents donor only population, hence was ignored in data interpretation.
Ribozyme Encapsulation
The encapsulation of the hairpin ribozyme in vesicles, immobilization of SUV on a supported lipid bilayer, RNaseA and nonspecific immobilization controls, imaging, data acquisition, and data analysis were performed as described previously.(12) SUVs encapsulating the ribozyme were formed in a 0.5 mM Mg2+ RNA buffer (10 mM Tris-HCl pH 7.4, 50 mM NaCl, 0.4% (w/w) glucose, and specified amounts of MgCl2) and were then incubated with the surface for specific attachment. The hairpin ribozyme molecules used in these studies did not contain a biotin moiety, thus removal of unencapsulated RNA via gel filtration prior to vesicle tethering was not needed. RNA concentrations during encapsulation were chosen to minimize the probability of encapsulating multiple molecules. Lipid films for the encapsulation were made of EggPC with 1% DPPE−biotin.
Purification and Characterization of aHL
Our initial attempts with the commercial aHL (Calbiochem) were not successful in forming pores despite our efforts that explored a wide variety of experimental conditions. As this failure is likely due to the low concentration of commercially available aHL (the practical maximum concentration we could use was ∼1 μM in monomer), we produced aHL in-house. We overexpressed hexahistidine-tagged aHL from a plasmid in Escherichia coli and purified the protein using a Ni-NTA column, and the protein activity was confirmed by the hemolysis assay (Supporting Information Figure 2). Recombinant aHL concentration was estimated to be ∼36 μM by quantitative Western blotting. For long-term storage, aHL was divided into aliquots, flash frozen, and kept at −80 °C.
aHL Incorporation on SUV
RNA-encapsulating SUVs were diluted (200−500 pM final SUV concentration) in buffer A (10 mM Tris pH 8.0, 200 mM NaCl, 0.1 mg/mL BSA) containing high concentrations (∼36 μM monomer) of purified recombinant aHL. Incubation was carried out at 37 °C for at least 30 min. Various lower concentrations of aHL resulted in the poration of various fractions of SUVs, but 36 μM yielded reproducible results with nearly 100% yield of pore formation as judged by the fraction of molecules that fold when the extravesicular buffer was exchanged from no Mg2+ to 1 or 10 mM Mg2+ condition. Pore formation does not happen when vesicles were incubated with aHL subsequent to immobilization, most likely because most of the aHL was captured by the supported lipid bilayer.
Rep Helicase Encapsulation
Partial duplex DNA (400 nM, double-stranded DNA of 18 base pairs with a 3′ (dT)80 and a Cy5 attached to the junction) and a 400 nM concentration of Rep molecules(32) labeled with a donor fluorophore (Cy3) mixed in a total volume of 200 μL of buffer R (20 mM Tris pH 8.0, 500 mM NaCl) were used to hydrate 2.5 mg of lipid films. In order to achieve, on the average, one molecule of each kind (i.e., one protein vs one DNA) in a 100 nm diameter vesicle, a concentration of 400 nM was chosen. Buffer R contains 500 mM to ensure that the labeled Rep does not aggregate, as when high concentrations of dye-labeled Rep monomers are dissolved under low salt conditions. Upon forming the multilamellar vesicles, 100 or 200 nm (concentrations were scaled accordingly) diameter vesicles were formed via extrusion. Skipping the freeze−thaw cycles was found to be essential in keeping the proteins active (Supporting Information). Finally, unencapsulated Rep molecules were separated from the vesicles by a Ni-NTA column via the histidine tag on the protein. While buffer R had to be used during encapsulation to avoid clumping of Rep, it had to be exchanged with the imaging buffer(4) (10 mM Tris-HCl, pH 7.6, 1 mM ATP, 12 mM MgCl2, 15 mM NaCl, 10% (v/v) glycerol, 0.4% (w/w) glucose, anti-blinking agents such as 2-mercaptoethanol or Trolox, and oxygen scavenging system utilizing catalase and glucose oxidase) during data acquisition because high salt concentration in buffer R does not allow Rep to bind to the DNA. Further details on Rep helicase encapsulation can be found in Supporting Information.
Results and Discussion
Hairpin Ribozyme Experiments
In the presence of high (≥1 mM) Mg2+ concentrations, the ribozyme folds into a compact structure (Figure 2, right panel) bringing the two arms carrying the donor (Cy3) and the acceptor (Cy5) closer and displays high FRET efficiency (∼0.8). The ribozyme is unfolded in the absence of Mg2+ ions and shows low FRET (∼0.2). At the intermediate Mg2+ concentration (0.5 mM), ribozyme spontaneously fluctuates between folded and unfolded conformations, and the FRET signal switches between the low and high values, resulting in two distinct peaks in the FRET histogram.
Figure 2.
(Right) A cartoon summarizing the hairpin ribozyme behavior, where the ribozyme is shown in unfolded (top) and folded (bottom) conformations. Donor and acceptor dyes are depicted in light and dark gray, respectively. (Left) FRET efficiency histograms of single encapsulated hairpin ribozymes. Mg2+ concentrations in the imaging buffer are displayed on each histogram. Not all of the assayed concentrations are shown. (Top) Ribozyme encapsulated in 0.5 mM Mg2+, measured with no Mg2+ outside SUV in the absence of aHL. Regardless of the outside imaging conditions, encapsulated ribozyme exhibited spontaneous fluctuations between folded and unfolded states,(12) which is a typical behavior for 0.5 mM Mg2+. The resultant histogram is hence a superposition of Gaussian distributions peaked around high and low FRET values, in addition to a peak at zero FRET due to the donor-only species. (Middle) Ribozyme encapsulated in 0.5 mM Mg2+, measured with no Mg2+ outside with aHL incorporated. Ribozyme remains stably unfolded. (Bottom) Ribozyme encapsulated in 0.5 mM Mg2+, measured with 0.5 mM Mg2+ subsequent to the measurement described in the middle panel. Ribozyme responds to the change in the outside buffer conditions thanks to the pores.
EggPC vesicles encapsulating the hairpin ribozyme construct were immobilized to the supported lipid bilayer(33) formed on a quartz slide via biotin−streptavidin interaction.(12) For all measurements, nonspecific binding of RNA and the SUVs to the surface was negligible. The fluorescence spots did not disappear after treating the surface with RNaseA, demonstrating that the RNA molecules are protected from digestion through encapsulation.(12) FRET histograms of the encapsulated molecules for all tested conditions (0, 0.5, 1, and 10 mM Mg2+) were indistinguishable from that of the Mg2+ concentration used during encapsulation (0.5 mM) (Figure 2, top left panel), demonstrating a successful encapsulation that yields a well-defined and isolated intravesicular environment.(12)
In contrast, upon treating the vesicles with aHL (Materials and Methods), the encapsulated RNA molecules changed their FRET histograms in response to the changes in the outside buffer condition (Figure 2, middle and bottom left panels). In the absence of Mg2+, RNA molecules were almost completely unfolded (low FRET population only). Under 0.5 mM Mg2+, RNA molecules displayed spontaneous switching between folded and unfolded conformations (coexistence of low and high FRET). At 1 mM or 10 mM Mg2+, RNA molecules were almost completely folded (high FRET only). All conditions were tested multiple times in different sequences, and buffer exchange was fully reversible. We conclude that the aHL pore formation works reliably and allows us to change the intravesicular salt concentration in a reversible fashion, keeping the RNA encapsulated.
Flow Measurements
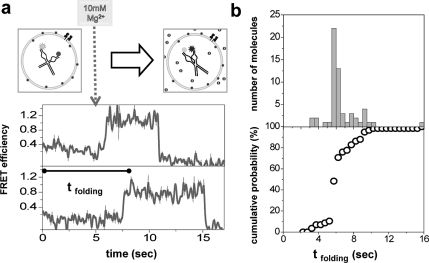
We used a flow delivery system to test how fast the buffer exchange through the pores takes place. We changed the solution condition (from no Mg2+ to 0.5 mM, 1 mM, or 10 mM Mg2+) typically 5−6 s after starting the data acquisition. The initial low FRET signal in the absence of Mg2+ rapidly increased upon flow of Mg2+. Although the flow data is shown only for 10 mM Mg2+ (Figure 3a), all conditions gave qualitatively similar behavior. Folding happened within the first few seconds for the majority of molecules: Only ∼10% of the molecules remained unfolded 3 s after injection of Mg2+ (Figure 3b). We therefore conclude that the number of pores per SUV was high enough (Supporting Information) for changing the buffer condition on a time scale relevant for the hairpin ribozyme folding which typically requires about 1 s to fold at Mg2+ concentrations above 0.5 mM.30,34
Figure 3.
(a) At t = 0, ribozymes encapsulated in porous SUVs are subject to no Mg2+, hence stay unfolded (low FRET). Upon injection of 10 mM Mg2+ into the sample chamber (between fifth and sixth seconds after data acquisition, marked with a dashed arrow), ribozymes rapidly fold (transition to high FRET). The duration for the FRET value to reach ∼0.8 from t = 0 is defined as time of folding (tfolding). The top and the bottom graphs belong to different SUVs (hence different ribozymes) from the same run (thin, black line: raw data; thick, gray line: B-spline smoothed data). (b) Histogram for times of folding (top) and corresponding percentile cumulative folding probability vs time (bottom) obtained from individual traces.
RNA Four-Way Junction Experiments
The RNA four-way junction, which is derived from the hairpin ribozyme by fully basepairing the loop structures, rapidly fluctuates between multiple conformations in the presence of Mg2+. Such fluctuations become slow enough to be measured within our time resolution at room temperature only at high concentrations of Mg2+ (≥50 mM).(30) Previously, we tried to encapsulate the junction in SUVs in order to ensure that the measurements were not altered by the interactions with the surface.(12) However, all of our attempts to encapsulate the junction at 50 mM Mg2+ failed because SUVs did not form at such high Mg2+ concentrations. Having developed the aHL approach, we tackled this problem once more where junction-encapsulating SUVs were first prepared in the absence of Mg2+. Subsequently, pores were formed using aHL, and imaging was carried out in the buffer containing 50 mM Mg2+. The dynamic behavior previously observed for the surface-tethered junction could then be reproduced(30) (Supporting Information Figure 1). aHL can therefore be utilized in such cases for which the measurement conditions are not suitable for the formation of SUV.
Rep Helicase Experiments
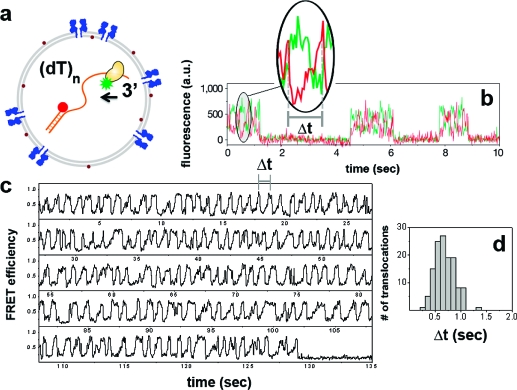
Next, we encapsulated an acceptor (Cy5)-labeled partial duplex DNA with a donor (Cy3)-labeled Rep helicase monomer in SUVs (Materials and Methods and Figure 4a) in order to confirm the ATP passage through aHL pores. A Rep monomer, which cannot unwind duplex DNA by itself,21,35 can translocate on a single-stranded DNA in the 3′ to 5′ direction. When Rep encounters a blockade (such as a double-stranded DNA) on its track, it snaps back to the 3′ end on its substrate. These repetitive translocation events (termed shuttling) continue until the Rep monomer dissociates from DNA.(32) Such measurements were previously carried out on DNA molecules tethered to a quartz surface coated with polymers. For the surface experiments, shuttling takes place a limited number of times (typically less than 10) due to eventual dissociation, and the number of translocation events is thus inadequate for inferring statistically significant information on the shuttling properties from a single Rep helicase (Figure 4b). In contrast, we expected that encapsulation would enable long-term monitoring of shuttling for a single pair of Rep and DNA, as the molecules would quickly rebind within the minuscule SUV volume.
Figure 4.
(a) Rep helicase labeled with donor, and DNA labeled with acceptor at the junction depicted as coencapsulated within a porous SUV. Partial duplex DNA is an 18bp double strand carrying a (dT)80 tail. (b) Rep shuttling data obtained from surface-tethered DNA (modified from ref (32)) with a same length tail. Inset shows a single translocation event where the gradual increase in the acceptor signal (with accompanying decrease in the donor signal) reflects translocation of Rep toward the junction, whereas the abrupt drop marks the snapping back to the 3′ end. The translocation time (Δt) is defined as the period between consecutive snapping events. Individual cycles of shuttling separated in time are due to different Rep molecules from solution binding on the same surface-attached DNA and exhibiting limited repetitive translocations until dissociation. (c) FRET trace of Rep shuttling on a (dT)80 DNA within 100 nm diameter porous SUVs. In marked contrast to the surface experiments, a single Rep−DNA pair shows over 140 translocation events until acceptor photobleaching near 130 s). Time resolution was 30 ms. (d) Δt histograms can be built from a single Rep−DNA pair. Similar statistics for surface measurements (b) can only be obtained by merging data from translocations events from many shuttling cycles exhibited by different Rep molecules.
Upon surface immobilization of SUVs, and subsequent ATP injection, shuttling events were observed within SUVs (Figure 4c), indicating the ATP passage through aHL. The translocation activity was observed only when aHL was incorporated and ATP was present. Number of repetitive translocation events that can be observed from an individual Rep was substantially higher in the SUVs compared to the surface assays as anticipated. When encapsulated, a single Rep−DNA pair exhibited shuttling essentially until photobleaching of the fluorophores. Such enhancement for the number of shuttling events makes it possible to observe more than 100 cycles of translocation from an individual Rep−DNA pair and to obtain the kinetic parameters reliably free of complications arising from single molecule heterogeneity (Figure 4d).
In conclusion, in order to fully harness the capabilities of the vesicle encapsulation, we described here a generalizable methodology for making robustly porous vesicles. aHL pores utilized in this study allowed rapid passage of Mg2+ as well as ATP through membranes of phospholipids SUVs. Compared to the thermothropically rendered defects, aHL pores are well-defined and stable at various temperatures. In the future, engineered aHL mutants of which opening/closing can be controlled via different chemical or optical means27,36,37 may be exploited to design novel experimental platforms.
Acknowledgments
aHL plasmid (p17Sf1A-H5) was obtained from Stephen Cheley. The hairpin ribozyme constructs were gifts from David Lilley and Tim Wilson. Original plasmids for Rep helicase expression were obtained from Timothy Lohman. We also thank Hagan Bayley, Yuji Ishitsuka, Cathy Murphy, Jerome Mathe, and Amit Meller for useful discussions and experimental help. Funding was provided by grants from the National Institutes of Health (GM 065367, GM 074526) and the National Science Foundation (PHY 0645550, PHY 082613).
Supporting Information Available
Vesicle-to-vesicle variation of effective reagent concentration in the absence of pores, more details about Rep helicase encapsulation, and the number of aHL pores required for rapid buffer exchange. This information is available free of charge via the Internet at http://pubs.acs.org/.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Joo C.; McKinney S. A.; Nakamura M.; Rasnik I.; Myong S.; Ha T. Cell 2006, 126, 515–27. [DOI] [PubMed] [Google Scholar]
- Joo C.; Balci H.; Ishitsuka Y.; Buranachai C.; Ha T. Annu. Rev. Biochem. 2008, 77, 51–76. [DOI] [PubMed] [Google Scholar]
- Moerner W. E. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 12596–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X. S.; Choi P. J.; Li G. W.; Lee N. K.; Lia G. Annu. Rev. Biophys. 2008, 37, 417–44. [DOI] [PubMed] [Google Scholar]
- Yildiz A.; Forkey J. N.; McKinney S. A.; Ha T.; Goldman Y. E.; Selvin P. R. Science 2003, 300, 2061–5. [DOI] [PubMed] [Google Scholar]
- Yildiz A.; Tomishige M.; Vale R. D.; Selvin P. R. Science 2004, 303, 676–8. [DOI] [PubMed] [Google Scholar]
- Lommerse P. H.; Snaar-Jagalska B. E.; Spaink H. P.; Schmidt T. J. Cell Sci. 2005, 118, 1799–809. [DOI] [PubMed] [Google Scholar]
- Lee J. Y.; Okumus B.; Kim D. S.; Ha T. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 18938–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amirgoulova E. V.; Groll J.; Heyes C. D.; Ameringer T.; Rocker C.; Moller M.; Nienhaus G. U. ChemPhysChem 2004, 5, 552–555. [DOI] [PubMed] [Google Scholar]
- Dickson R. M.; Norris D. J.; Tzeng Y. L.; Moerner W. E. Science 1996, 274, 966–9. [DOI] [PubMed] [Google Scholar]
- Cohen A. E.; Moerner W. E. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 4362–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumus B.; Wilson T. J.; Lilley D. M.; Ha T. Biophys. J. 2004, 87, 2798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukobza E.; Sonnenfeld A.; Haran G. J. Phys. Chem. B 2001, 105, 12165–12170. [Google Scholar]
- Benitez J. J.; Keller A. M.; Ochieng P.; Yatsunyk L. A.; Huffman D. L.; Rosenzweig A. C.; Chen P. J. Am. Chem. Soc. 2008, 130, 2446–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades E.; Gussakovsky E.; Haran G. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 3197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse I.; Okumus B.; Joo C.; Ha T. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 12646–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickler M.; Hessling M.; Ratzke C.; Buchner J.; Hugel T. Nat. Struct. Mol. Biol. 2009, 16, 281–6. [DOI] [PubMed] [Google Scholar]
- Chiu D. T.; Wilson C. F.; Ryttsen F.; Stromberg A.; Farre C.; Karlsson A.; Nordholm S.; Gaggar A.; Modi B. P.; Moscho A.; Garza-Lopez R. A.; Orwar O.; Zare R. N. Science 1999, 283, 1892–5. [DOI] [PubMed] [Google Scholar]
- Talaga D. S.; Lau W. L.; Roder H.; Tang J.; Jia Y.; DeGrado W. F.; Hochstrasser R. M. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 13021–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal A.; Arslan S.; Okumus B.; Wilson T. J.; Giraud G.; Norman D. G.; Ha T.; Lilley D. M. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 11176–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T.; Rasnik I.; Cheng W.; Babcock H. P.; Gauss G. H.; Lohman T. M.; Chu S. Nature 2002, 419, 638–41. [DOI] [PubMed] [Google Scholar]
- Noireaux V.; Libchaber A. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 17669–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterhalter M.; Hilty C.; Bezrukov S. M.; Nardin C.; Meier W.; Fournier D. Talanta 2001, 55, 965–71. [DOI] [PubMed] [Google Scholar]
- Meier W.; Nardin C.; Winterhalter M. Angew. Chem., Int. Ed. 2000, 39, 4599–4602. [PubMed] [Google Scholar]
- Bolinger P. Y.; Stamou D.; Vogel H. J. Am. Chem. Soc. 2004, 126, 8594–5. [DOI] [PubMed] [Google Scholar]
- Bhakdi S.; Bayley H.; Valeva A.; Walev I.; Walker B.; Kehoe M.; Palmer M. Arch. Microbiol. 1996, 165, 73–9. [DOI] [PubMed] [Google Scholar]
- Chang C. Y.; Niblack B.; Walker B.; Bayley H. Chem. Biol. 1995, 2, 391–400. [DOI] [PubMed] [Google Scholar]
- Roy R.; Hohng S.; Ha T. Nat. Methods 2008, 5, 507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T.; Enderle T.; Ogletree D. F.; Chemla D. S.; Selvin P. R.; Weiss S. Proc. Natl. Acad. Sci. U.S.A. 1996, 93, 6264–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan E.; Wilson T. J.; Nahas M. K.; Clegg R. M.; Lilley D. M.; Ha T. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 9308–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohng S.; Wilson T. J.; Tan E.; Clegg R. M.; Lilley D. M.; Ha T. J. Mol. Biol. 2004, 336, 69–79. [DOI] [PubMed] [Google Scholar]
- Myong S.; Rasnik I.; Joo C.; Lohman T. M.; Ha T. Nature 2005, 437, 1321–5. [DOI] [PubMed] [Google Scholar]
- Chan Y. H.; Boxer S. G. Curr. Opin. Chem. Biol. 2007, 11, 581–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X.; Kim H.; Pereira M. J.; Babcock H. P.; Walter N. G.; Chu S. Science 2002, 296, 1473–6. [DOI] [PubMed] [Google Scholar]
- Cheng W.; Hsieh J.; Brendza K. M.; Lohman T. M. J. Mol. Biol. 2001, 310, 327–50. [DOI] [PubMed] [Google Scholar]
- Bayley H. Sci. Am. 1997, 277, 62–7. [DOI] [PubMed] [Google Scholar]
- Russo M. J.; Bayley H.; Toner M. Nat. Biotechnol. 1997, 15, 278–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.