Abstract
Virtual screening of small molecules against a protein target often identifies the correct pose, but the ranking in terms of binding energy remains a difficult problem, resulting in unacceptable numbers of false positives and negatives. To investigate this problem, the performance of three docking programs, FRED, QXP/FLO, and GLIDE, along with their five different scoring functions, was evaluated with the engineered cavity in cyctochrome c peroxidase (CCP). This small cavity is negatively charged and completely buried from solvent. A test set of 60 molecules, experimentally identified as 43 “binders” and 17 “non-binders”, were tested with the CCP binding site. The docking methods’ performance is quantified by the ROC curve and their reproduction of crystal poses. The effects from generation of different ligand tautomers and inclusion of water molecule in the cavity are also discussed.
INTRODUCTION
Since the emergence of an increasing number of 3D biological structures, molecular docking has become a major computational method for discovering and designing new ligands for biological targets.1–7 Since the pioneering work of Kuntz et al.,8,9 many docking programs have been developed, based on different physicochemical approximations. 10,11 In each program, a pose generator needs to be combined with one or more fast-scoring functions. Many studies have been carried out to evaluate their performance.2,13–25
Docking programs are designed to reproduce a ligand’s pose seen in the crystal structure. They are usually parametrized with a training set, containing a large number of diverse protein–ligand crystal structures, and then evaluated with one or more test sets. The optimal parameters are chosen to give overall best results. Usually two criteria are considered for evaluation of the docking performance: reproduction of the crystal structure of protein–ligand complex structures26–37 and ranking of ligands in the databases according to binding energies.38–45
Even though some docking tools and studies have been tailored to work with particular kinds of protein–ligand systems,46,47 most commercial docking programs focus on generality and might not produce accurate results when dealing with unusual systems. Scoring functions, particularly empirical scoring functions, have common weaknesses. For instance, larger ligands tend to receive better scores. Docking programs also have a tendency to position ligands to match the shape of the protein surface. Therefore, ligands with large groups projecting into the solvent in the crystallographic structure are not handled well. Asymmetric ligands with symmetric features (“pseudosymmetrical” molecules) also are difficult to dock. Docking programs sometimes position them at a 180° angle to the crystal binding mode. These “near-native” poses might be ranked better than the crystal poses; thus the docking programs would discard the correct poses and pick the “near-native” poses with poor rmsd values.30,48
Since each docking program has its own algorithms and parameters, it is advisable to use caution in different situations, depending on the size, hydrophobicity, and solvation accessibility of the binding pocket. However, discovering the best docking performance for a specific case is not straightforward. Shoichet et al. recently published studies on model charged and hydrophobic binding pockets with the program DOCK3.5.54.49–51 This paper expands on their research of the charged model binding site in cyctochrome c peroxidase (CCP). The study focuses on investigatiion of the docking performance of different docking programs. Three docking programs have been studied: FRED,52 QXP/FLO,12,53 and GLIDE.54
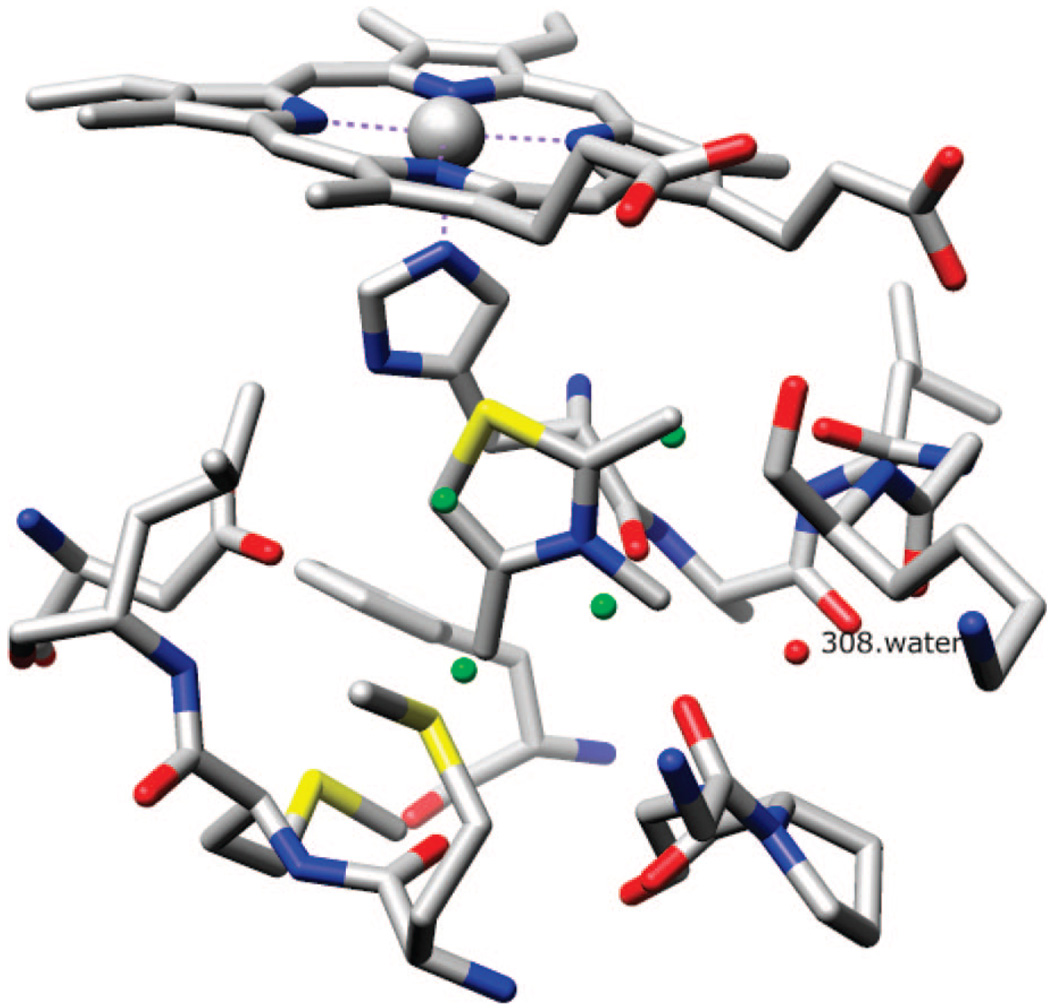
A model binding pocket needs to be small and simple enough to study. An example is buried nonpolar cavity of T4 lysozyme created by mutations.55,56 The model in this study is an engineered pocket in cyctochrome c peroxidase (CCP) with a substitution (Trp191 → Gly).57 The CCPW191G pocket is completely buried but negatively charged. Several exposed backbone carbonyl groups and Asp235, ligated by five water molecules and a potassium ion, contribute to the negative charge (Figure 1). This binding pocket has been studied extensively, with 43 known ligands58 and 17 compounds that do not bind to this pocket.59 Most binders are small heterocycles with a single positive charge. For 35 known binders, X-ray crystal structures of the complex have been determined. In addition to the study by Shoichet, Brooks et al. also used this model pocket for their λ-dynamics approach.60,61 Olson and colleagues tested AutoDock3.0’s docking ability on this pocket with 33 compounds (16 binders and 17 nonbinders).62 Olson’s study included clustering of the stochastic docking results, and the adjusted score was the average predicted binding energy augmented with a penalty equal to the number of clusters of 100 docking trials in units of kilocalories per mole. The method was able to separate binders and nonbinders with the adjusted score.
Figure 1.
Cavity of CCP W191G, with bound ligand 2,3,4-trimethyl-1,3-thiazole (PDB code 1AC4). Water molecule 308 (red) is conserved in all structures. Four water molecules (green) are superimposed from unbound CCPW191G (PDB code 1CMU). (This figure was made using Chimera.76)
FRED (fast rigid exhaustive docking), developed by OpenEye Scientific Software, Inc. is a fast docking program based on multiconformer docking instead of the more widely used incremental construction.21 The ligands’ low-energy conformations are pre-generated by OMEGA, part of the OpenEye suite.63,64 FRED rigidly fits these conformers into a pre-defined binding site and ranks the poses by scoring functions. Among the eight scoring functions implemented in FRED, two functions, Chemgauss3 and Zapbind, 52 include desolvation terms in their scores and are therefore chosen for this study. The Chemgauss3 scoring function entails these interactions: steric, hydrogen bond, metal–ligand, ligand, and protein desolvation. Each interaction is described by a base function that is smoothed by convolution with a Gaussian function.65 The Zapbind scoring function combines a surface area contact term and an electrostatic interaction term calculated using the Poisson–Boltzmann (PB) solvent approximation.66 The surface area term is calculated using a Gaussian-based method, while the PB energy is calculated using ZAP. Zapbind is very sensitive to the presence of even minor atom clashes, and therefore, the ligand coordinates require a force-field refinement. Zapbind is the most computationally expensive scoring function among all the ones integrated in FRED. All other functions typically can examine and score all possible poses of a ligand in the binding pocket within seconds.
QXP/FLO uses Monte Carlo perturbation with energy minimization to perform full conformational searches for flexible molecules.12 Between the initial perturbation and energy minimization, an additional fast search step produces approximate low-energy structures.
GLIDE (grid-based ligand docking from energetics) applies a series of filters to search for plausible locations of the ligands in the binding pocket before docking to reduce the number of pose candidates. A grid is used to represent the shape and properties of the receptor by different sets of fields. Ligand conformations need to be exhaustively enumerated in the ligand torsion angle space. After initial screening to locate promising ligand poses, ligand geometries are minimized in the binding pocket using a standard molecular mechanics (OPLS-AA force field67) and a distance-dependent dielectric model. The top candidates are then refined by a Monte Carlo procedure to examine nearby torsional minima. To rank the best docked poses, a model energy function combining empirical and force-field terms are used.68,69 In addition to the standard precision (SP) scoring function, GLIDE also offers a new scoring function of extra precision (XP) that incorporates novel terms and has been shown to improve the selection of actual binding poses.70 Both scoring functions are investigated in this study.
All three programs are widely used in molecular docking, but a detailed evaluation for docking into charged pockets has not been reported so far. Our study investigates their performance on CCPW191G binding pocket with different docking protocols by quantitative comparison of binder/nonbinder separation, and docked poses’ RMSDs. We also evaluate the importance of including HOH308 in binding pocket during docking.
METHODS
The CCPW191G receptor structure was downloaded from the PDB Web site71 (PDB code 1AC4). All water molecules and the potassium ion in the pocket were removed, except for water molecule 308 (HOH308). SMILES strings of 43 binders and 17 nonbinders were downloaded from Prof. Shoichet’s research Web site.58,59 MOE72 was used to convert SMILES into 3D structures, add hydrogens, calculate AM1BCC charges,73 and obtain minimized geometries. Visual inspection was carried out to ensure that the 3D structures were correct. Unless mentioned otherwise, default parameters were used for structure preparation and docking.
DOCKING PROTOCOLS
FRED
FRED52 requires a set of low-energy conformations for each ligand. The conformers were generated by OMEGA74 and stored in a single binary file. Because of the small size of the ligands, the rms threshold between different conformers of OMEGA was set to 0.1Å, instead of the default value of 0.8 Å. A total of 102 conformers were generated for 60 molecules.
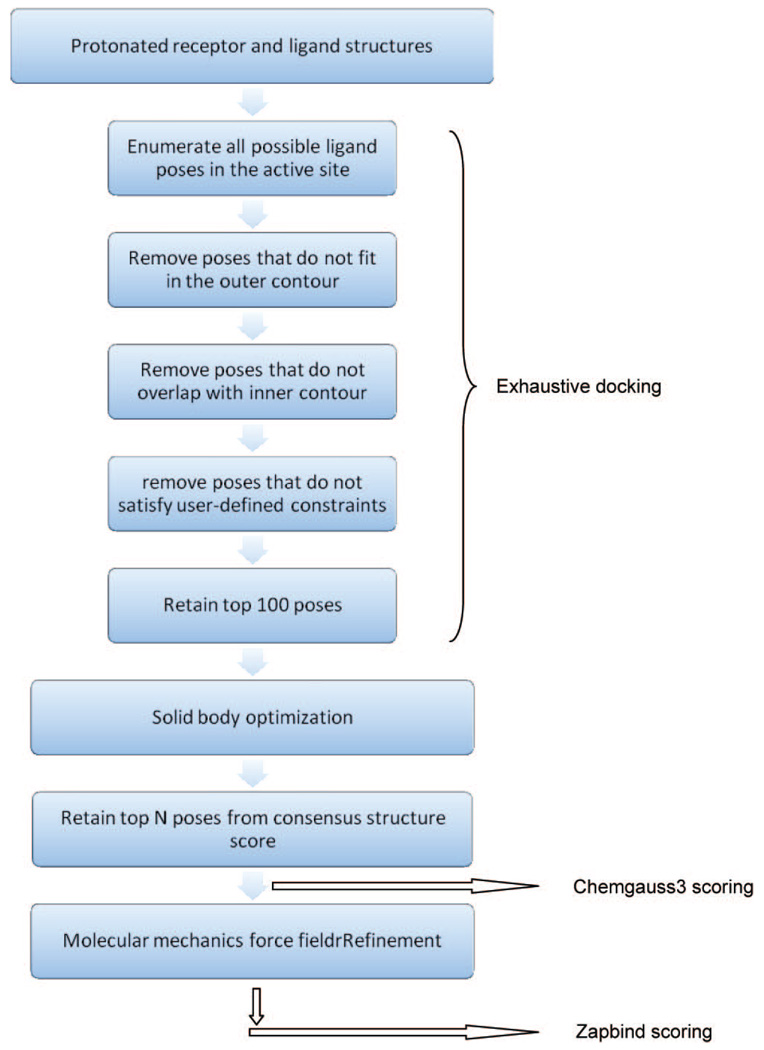
FRED docking consists of 4 steps: exhaustive docking, optimization, consensus structure, and the optional force field refinement. During exhaustive docking, a pose ensemble is generated by rigidly rotating and translating each conformer within the active site. The active site is defined by a box of 5 Å (default) extension in all directions from the bound ligand. Two complementary volumes, named the inner and the outer contour, are generated by building two isocontours of the same shape potential grid at different contour levels. The generated pose has to overlap with the inner contour and not exceed the outer contour. All surviving poses are scored with a scoring function (default = Chemgauss3), and the top 100 (default) poses are passed to optimization. In optimization, a systematic solid body optimization is done by rigidly rotating and translating the poses at half the step size used in exhaustive docking. Chemgauss3 (default) is used in this step to score the poses during optimization. The poses then go to consensus structure, in which the poses with the top consensus scores (default = PLP, Chemgauss3, and OEChemscore) are retained, and all other poses are discarded. The user-defined number of top poses is written out with their scores (except for Zapbind scoring function). For Zapbind scoring, a Merck molecular mechanics force field refinement needs to be performed. The refinement consists of full coordinate optimization of all ligand atoms. It is required for Zapbind, which is extremely sensitive to small atom–atom clashes. The flowchart illustrates the FRED docking protocol (Chart 1).
Chart 1.
Flowchart of the Docking Methodology of FRED
In our study, the receptor structure was protonated using Molprobity,75 which also examines potential misinterpretations of the crystallographers that involve “iso-steric” χ side-chain terminal flips of asparagine, glutamine, and histidine. The heme cofactor was protonated with MOE. Hydrogen atoms were added to HOH308 manually. Visual inspection was done, and the bonds and protonation states were carefully corrected. AM1BCC charges were added to the receptor using Chimera76 and to HOH308 and all heme atoms using MOE. FRED_receptor77 was used to prepare the receptor structure file. A box around the bound ligand molecule 2,3,4-trimethylthiazole from the crystal structure was created by a 5 Å extension in all directions to define the binding site. The box volume was 1913 Å3; the inner and outer contours are 43 and 243 Å3, respectively. No other constraints were applied in the setup.
Default parameters were used in docking. Docked ligands were ranked by the Chemgauss3 scoring function without Merck molecular mechanics force field refinement. The docked poses then underwent the refinement and were ranked by the Zapbind scoring function. In the rmsd result, please note that Chemgauss3 rmsd refers to the docked pose before the refinement; Zapbind rmsd applies to the poses after the refinement.
QXP/FLO
The receptor, the heme cofactor, and HOH308 were protonated the same manner as for the FRED docking preparation (receptor by Molprobity, heme by MOE, HOH308 manually). We did not use QXP/FLO’s protonation and hydrogen optimization tools. Instead, we removed nonpolar hydrogen atoms on the receptor and ligands using QXP/FLO. The protonated 3D structure was loaded into QXP/FLO,53 and the working set was defined as 7 Å from the bound ligand 2,3,4-trimethylthiazole. All nonpolar hydrogen atoms on the receptor, and the ligands were removed.
Docking was carried out using the MCDOCK conformational searching/energy minimization procedure of QXP. The receptor was held rigid. (The binding site marker atoms are listed in the Supporting Information) The ligands were subjected to 100 cycles of Monte Carlo conformational searching and energy minimization. For each small molecule, the 10 lowest-energy conformations were saved.
GLIDE
The receptor structure from the PDB was prepared with the “Protein Preparation Wizard” of Maestro.78 The heme cofactor and the iron ion’s charges and connectivity were carefully inspected. The receptor’s structure was protonated, charged, and refined using Maestro. The receptor grid for future docking was also generated. All ligand structures were prepared with Ligprep.79 All tautomers and different protonation states were generated.
The refined receptor with and without HOH308 and protonation-enumerated ligand structures were loaded into Maestro. Two docking runs were carried out, with SP or XP scoring functions. Ten poses per ligand were saved for each docking run. No other changes were applied to the default docking setting.
ROC Curves
To quantitatively compare all docking methods, the docking result was analyzed using the ROC (receiver operating characteristic) curve,80 which describes a docking method’s ability to avoid false positives and false negatives. Since the true positives and the true negatives are known in this study, the ROC curve is particularly suitable. The effectiveness of a docking method can be quantified by the area under the ROC curve (AUC). A theoretically perfect performance has an AUC value of 1.0; while a random selection performance presents with a 0.5 AUC value.
rmsd Calculations
The rmsd values have been calculated over heavy atoms of the docked pose and the crystal pose of the same ligand when applicable. To avoid overestimation of the rmsd values, symmetry operators have been included in the calculation routine to the interchange equivalent atoms in the symmetrical molecules. The “best pose” is defined as the docked pose that is the nearest to the experimental binding mode, whereas the “top pose” is defined as the docked pose that is ranked first.
RESULTS AND DISCUSSION
Binder/Nonbinder Separation
All docking results are analyzed in terms of ROC (receiver operating characteristics) curves, and the AUC values are shown in Table 1. The actual scores and rankings are listed in the Supporting Information.
Table 1.
AUC Values of All Docking Tests
| AUC value | ||
|---|---|---|
| FRED | no HOH308, Chemgauss3 | 0.63 |
| no HOH308, Zapbind | 0.88 | |
| with HOH308, Chemgauss3 | 0.61 | |
| with HOH308, Zapbind | 0.83 | |
| QXP/FLO | no HOH308 | 0.92 |
| with HOH308 | 0.89 | |
| GLIDE | no HOH308, SP scoring | 0.66 |
| no HOH308, XP scoring | 0.59 | |
| with HOH308, SP scoring | 0.67 | |
| with HOH308, XP scoring | 0.61 |
FRED
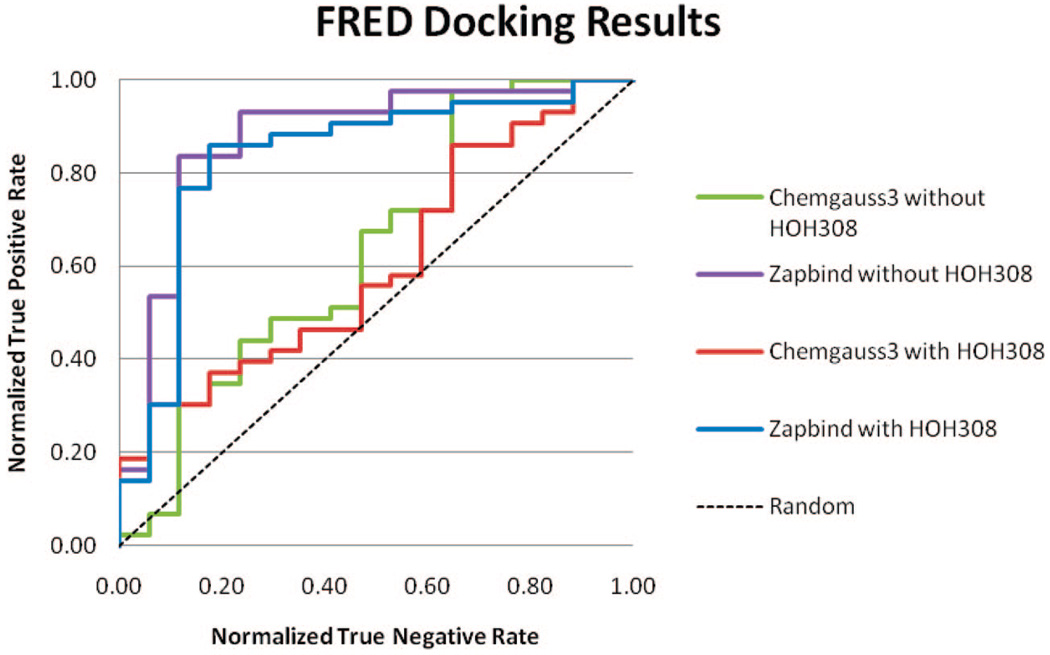
Results of all four docking tests (with Chemgauss3 or Zapbind scoring; HOH308 present or absent in the binding pocket) are shown in Figure 2. The Chemgauss3 scoring produced less satisfying results and did not separate binder from nonbinder ligands well. The ROC curve even fell below random selection in the middle. The average AUC (with or without HOH308) of Chemgauss3 scoring is 0.62. Most ligands in our test set are small and hydrophilic, and the CCPW191G cavity is deeply buried. Kellenberger reported that FRED usually fails for these type of ligands.30 The ChemScore scoring function was used in their study.
Figure 2.
ROC curve of FRED docking results.
However, Zapbind scoring produced much better results than Chemgauss3, with an average AUC of 0.855. The Zapbind score has two components: the “Zap” and “Area” terms. After reviewing both components of all Zapbind scores, we observed that the Area term does not distinguish much among all binders and nonbinders. The Area term accounts for buried area contribution to binding, and it is calculated using a Gaussian-based method. The “Zap” term, which is calculated using the Poisson–Boltzmann solvent approximation, is responsible for separating binders from nonbinders.
The inclusion of HOH308 in binding pocket did not improve docking result. For both scoring functions, the inclusion of HOH308 actually made the AUC value slightly worse.
Two nonbinders cannot be ranked acceptably by either scoring functions in FRED: methylammonium and dimethylammonium. Both compounds are acyclic, unlike all other ligands. For Chemgauss3, the scoring function requires at least three connected heavy atoms to generate atom types. Therefore, both compounds are too small to be properly assigned atom types. Chemgauss3 could not calculate energy contributions except for steric interactions, thus both compounds received artificially low scores. For Zapbind scoring, both compounds have unusually favorable contributions from the Poisson-Boltzmann solvent approximations, which results in higher ranks than all other nonbinders.
In addition, consensus ranking was also tried with FRED, in which the poses returned from exhaustive docking were scored by both Chemgauss3 and Zapbind scoring functions, and the consensus “score” is the sum of that pose’s rank in both scoring functions’ lists. However, consensus scoring cannot outperform the best scoring function.81 The consensus rank shows that it did not separate binders and nonbinders as well as Zapbind, but it did outperform Chemgauss3.
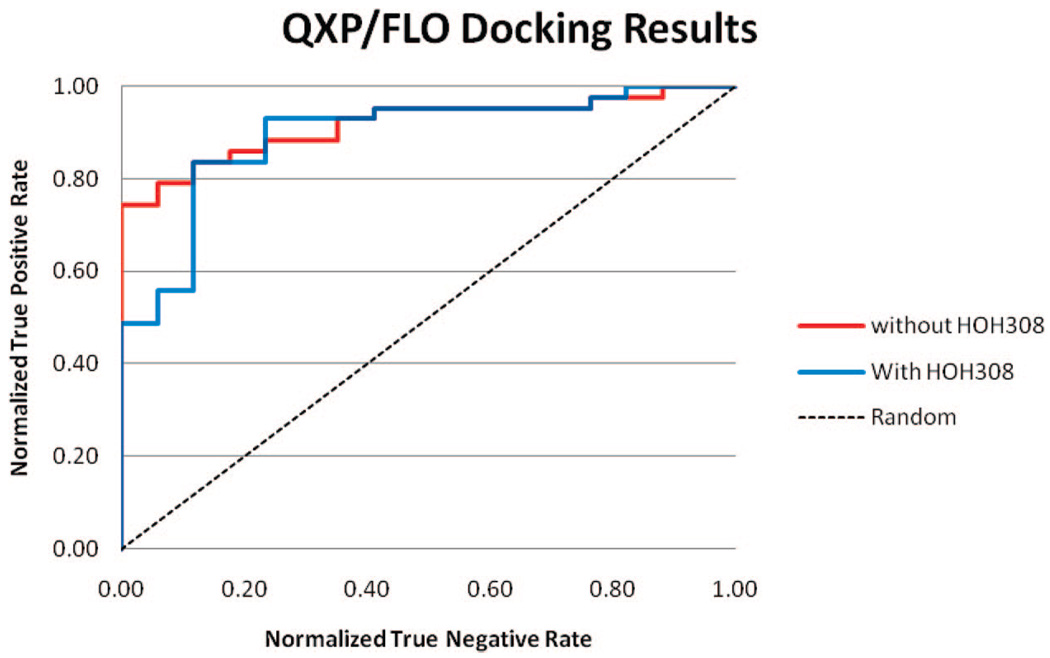
QXP/FLO
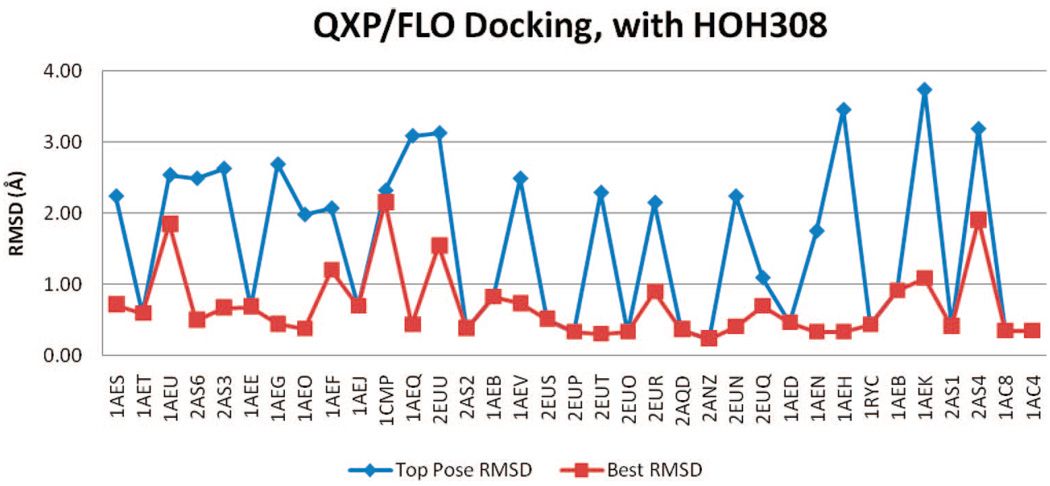
QXP/FLO did a very good job in separating binders and nonbinders, with or without HOH308 in binding pocket (Figure 3). The average AUC value was 0.905. The inclusion of HOH308 again did not result in any improvement. In fact, it reduced the AUC value slightly. From the ROC curves, it can be seen that docking without HOH308 was able to identify 74% of the binders before any non-binders; while for docking with HOH308, that number dropped to less than 50%.
Figure 3.
ROC curve of QXP/FLO docking results.
The two outliers observed in FRED docking, methylammonium, and dimethylammonium, still ranked top two among all nonbinders in QXP/FLO docking. Both methods reached limitations when docking unusually small molecules, even though they worked well for other molecules.
It should also be noted that lowest-ranked molecule, isoniazide had positive scores. According to its FLO energy terms, isoniazide has a strongly positive VDW energy contribution (more than 40.2 kJ/mol). It should also be noted that isoniazide has the highest molecular weight among all binders and nonbinders. Therefore, QXP/FLO was not able to dock isoniazide without clashing into the protein because of its size, which resulted in the abnormally high VDW energy contributions.
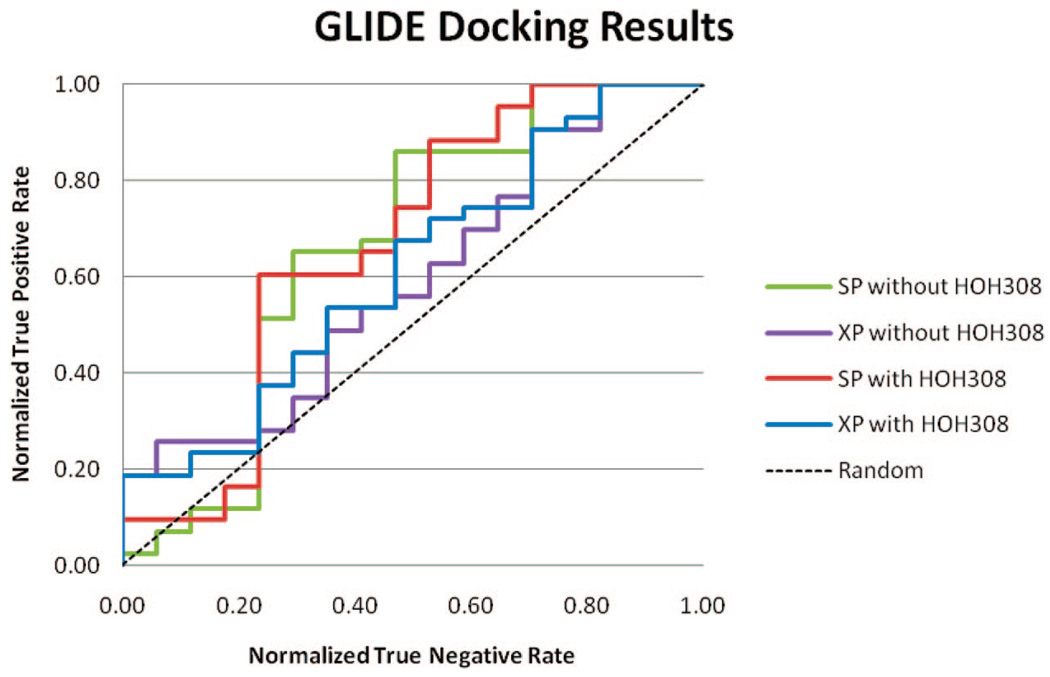
GLIDE
The ROC curves of all GLIDE docking results are shown in Figure 4. AUC values of all four docking trials are lower than 0.70, lower than FRED with Zapbind scoring and QXP/FLO. XP (extra precision) scoring surprisingly performed worse than SP (standard precision) from the AUC values. The inclusion of HOH308 did not significantly affect the result.
Figure 4.
ROC curve of GLIDE docking results.
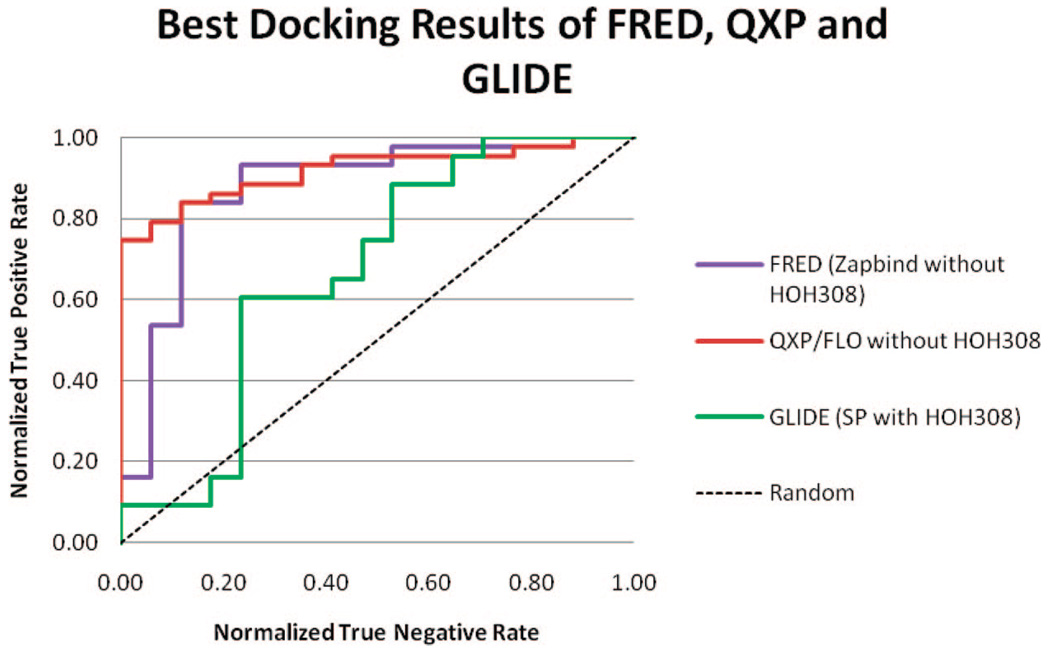
Figure 5 shows the ROC curves of the best result from FRED, QXP/FLO, and GLIDE. QXP/FLO outperformed the other two methods by ranking more than 74% of the binders higher than any nonbinders.
Figure 5.
ROC curve of best results from FRED, QXP, and GLIDE docking.
RMSDs of the Predicted Poses
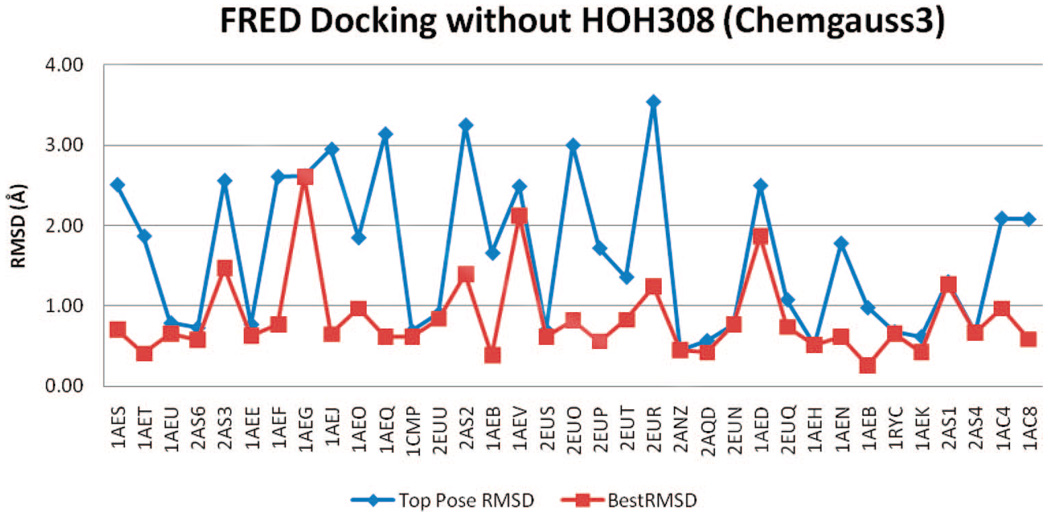
Among the 43 binder ligands, 35 have crystal structures for comparison. All rmsd results are illustrated in Figure 6–Figure 15. The ligands are listed in the order of their molecular weights. The successfully docked ligand is defined as having TP-rmsd or BP-rmsd less than 1 Å. Table 2 shows the number of successfully docked binders for all docking runs.
Figure 6.
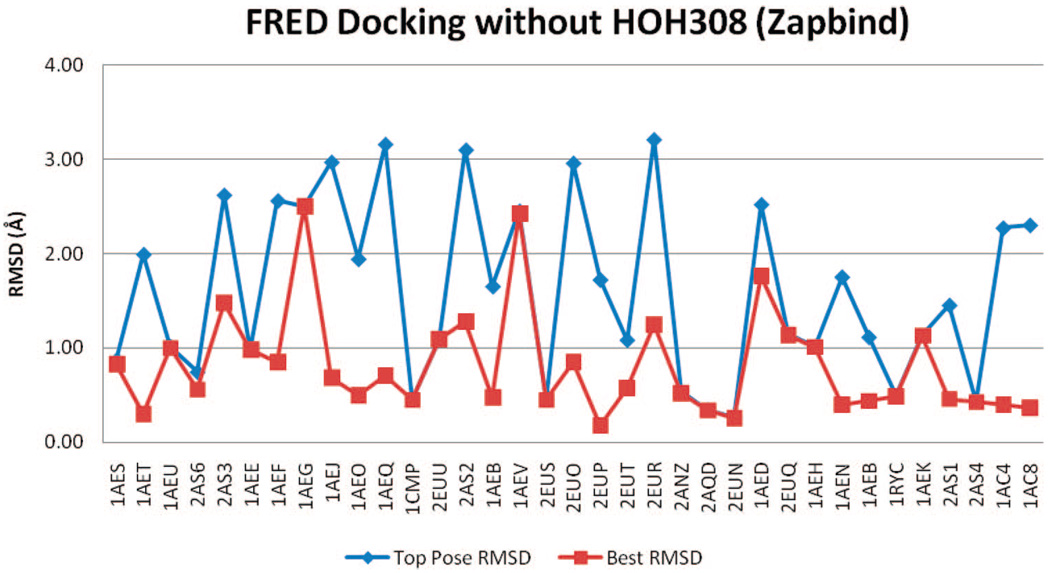
rmsd results using FRED with Chemgauss3 scoring and HOH308 removed.
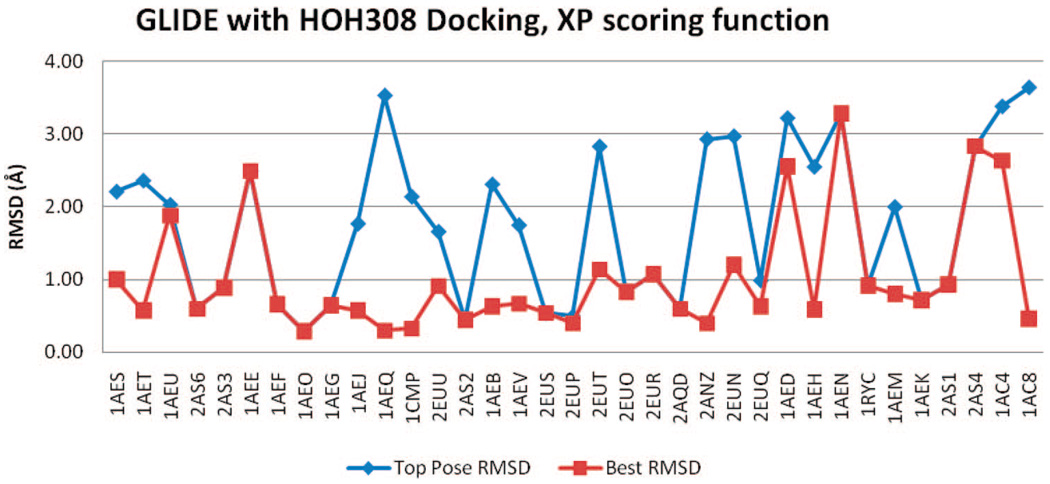
Figure 15.
rmsd results using GLIDE (XP) with HOH308.
Table 2.
Statistical Summary of RMSDs on 35 Binders with Crystal Structures Docked by Different Programs
| rmsd statistics (Å) | no. of ligands that have rmsd < 1 Å among top 10 poses | ||||||
|---|---|---|---|---|---|---|---|
| TP-rmsd | BP-rmsd | ||||||
| mean | SDa | mean | SDa | TP-rmsd | BP-rmsd | ||
| FRED | no HOH308, Chemgauss3 | 1.65 | 0.95 | 0.85 | 0.51 | 14 | 28 |
| no HOH308, Zapbind | 1.61 | 0.94 | 0.82 | 0.56 | 10 | 25 | |
| with HOH308, Chemgauss3 | 1.93 | 1.09 | 0.92 | 0.57 | 12 | 25 | |
| with HOH308, Zapbind | 1.82 | 1.09 | 0.91 | 0.60 | 10 | 23 | |
| QXP/FLO | no HOH308 | 1.27 | 1.10 | 0.52 | 0.28 | 21 | 34 |
| with HOH308 | 1.58 | 1.12 | 0.69 | 0.49 | 16 | 29 | |
| GLIDE | no HOH308, SP scoring | 2.04 | 1.08 | 0.71 | 0.42 | 8 | 29 |
| no HOH308, XP scoring | 1.99 | 1.74 | 0.95 | 0.73 | 11 | 26 | |
| with HOH308, SP scoring | 2.44 | 0.89 | 0.70 | 0.41 | 4 | 29 | |
| with HOH308, XP scoring | 1.79 | 1.06 | 1.01 | 0.79 | 14 | 26 | |
SD = standard deviation.
FRED
Figure 6 and Figure 7 show RMSDs of docked poses using FRED, with Chemgauss3 and Zapbind scoring, in the docking test, while HOH308 was removed from binding pocket. It shows RMSDs of both the top pose (pose with lowest energy) and the best pose (pose with lowest rmsd to the crystal structure among the top 10 poses).
Figure 7.
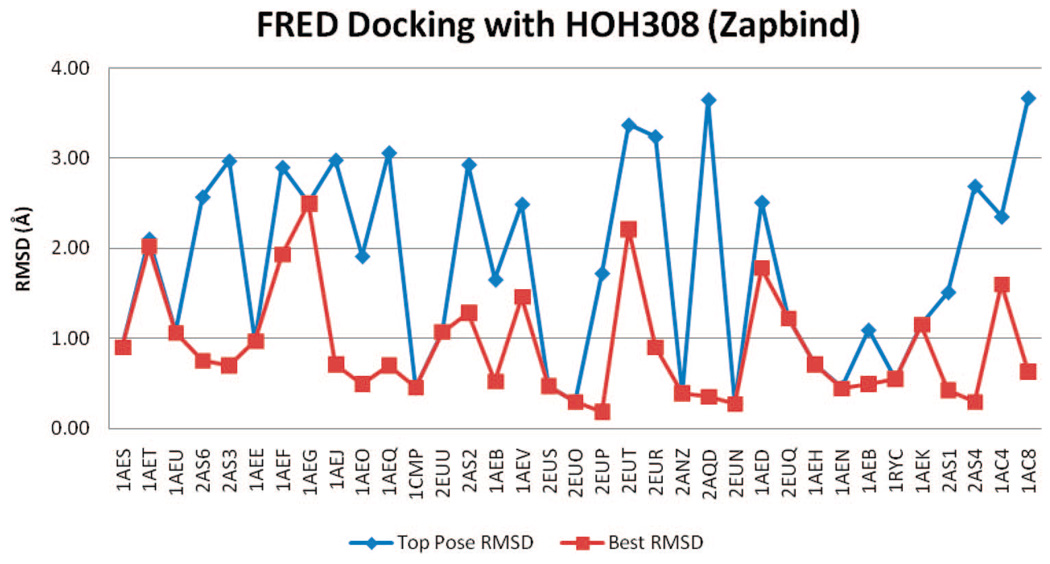
rmsd results using FRED with Zapbind scoring and HOH308 removed.
According to the FRED developers, refinement, which is required for Zapbind scoring, is a “double-edged sword”. While it removes atomic clashes in ligands, it tends to move a ligand to a poorer geometry relative to the crystal pose. The reason is that crystal poses are often not at a force-field minimum and, therefore, cannot be located by force field refinement.
FRED performed reasonably well at reproducing the binders’ crystal poses. Before refinement (Chemgauss3), 14 of 35 binders had TP-rmsd values of less than 1 Å; 28 binders had BP-RMSDs better than 1 Å. The refinement made rmsd results were slightly worse. After refinement (Zapbind), 10 binder ligands had TP-rmsd values better than 1 Å, and 25 had BP-rmsd values better than 1 Å (Table 2).
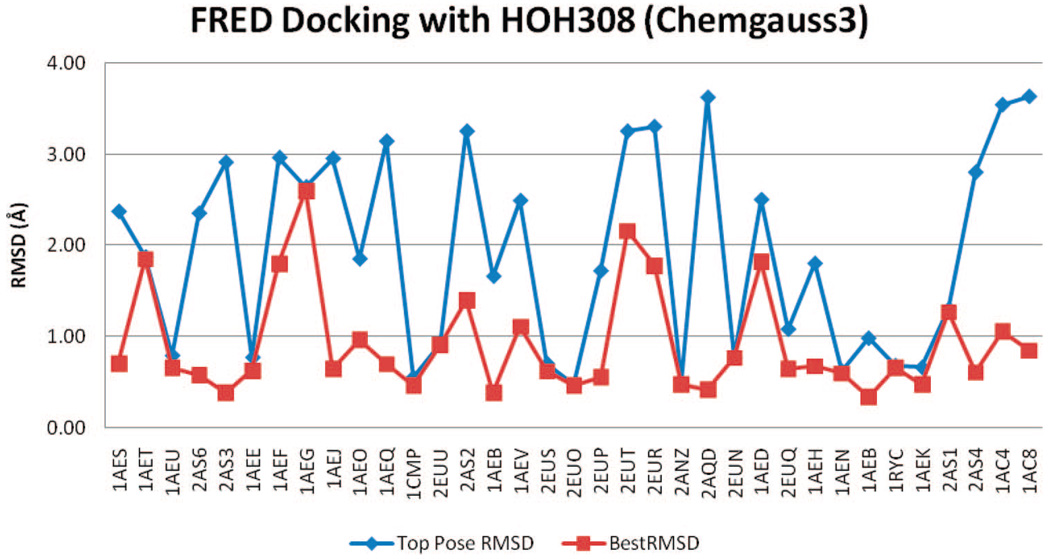
Figure 8 and Figure 9 show the rmsd values of docked poses using FRED, Chemgauss3, and Zapbind scoring, in the docking test with HOH308 in binding pocket. Compared to the results without HOH308 in pocket, both the TP-rmsd and BP-rmsd values are slightly worse. Before refinement, 12 ligands have TP-rmsd values better than 1 Å; 25 ligands have BP-rmsd values better than 1 Å, and 10 binder ligands have TP-rmsd values better than 1 Å; 23 have BP-rmsd values better than 1 Å (Table 2).
Figure 8.
rmsd results using FRED with Chemgauss3 scoring and with HOH308 in binding pocket.
Figure 9.
rmsd results using FRED and Zapbind scoring with HOH308 in binding pocket.
Docking without HOH308 gives better means and standard deviations of the TP-rmsd and BP-rmsd values than docking with HOH308. The best mean TP-rmsd (1.61 Å) and BP-rmsd (0.82 Å) of FRED docking are from Zapbind scoring without HOH308, consistent with the ROC plots.
QXP/FLO
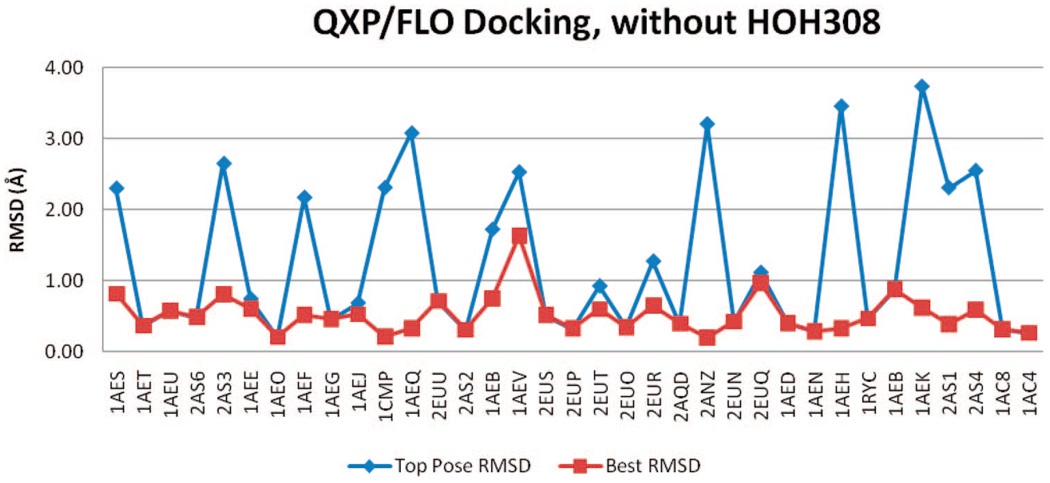
QXP/FLO outperformed the other two methods in reproducing the binders’ crystal poses. Between the two tests, the docking without HOH308 appeared to be superior: 21 binders have TP-rmsd values better than 1 Å, and 34 have BP-rmsd values better than 1 Å (Table 2 and Figure 10 and Figure 11). In other words, QXP/FLO can reproduce almost all binders’ crystal poses with less than 1 Å rmsd, among its top 10 docked poses. The only ligand that has a BP-rmsd worse than 1 Å is 2-aminothiazole (PDB code 1AEV). The mean TP-rmsd (1.27 Å) and BP-rmsd (0.52 Å) values are better than all other docking results.
Figure 10.
rmsd results using QXP/FLO with HOH308 removed.
Figure 11.
rmsd results using QXP/FLO with HOH308 kept in pocket.
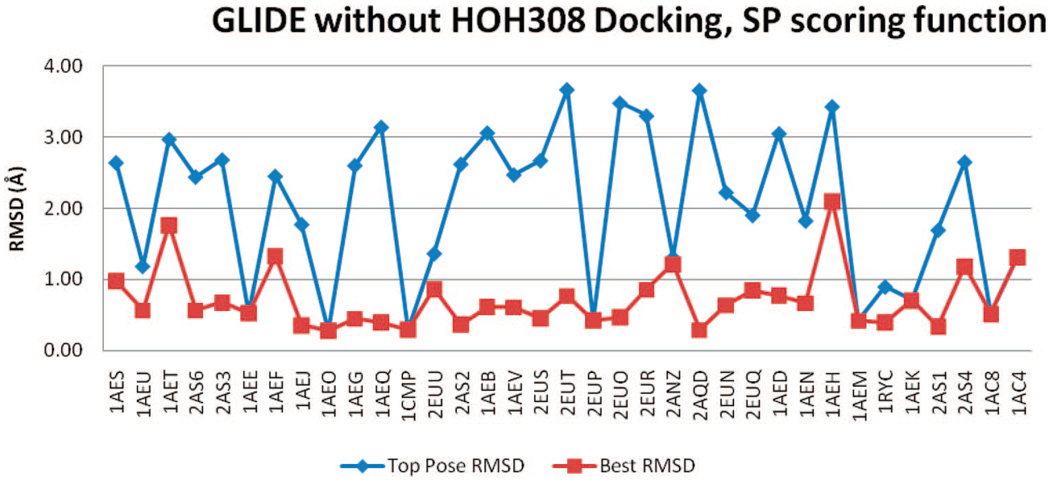
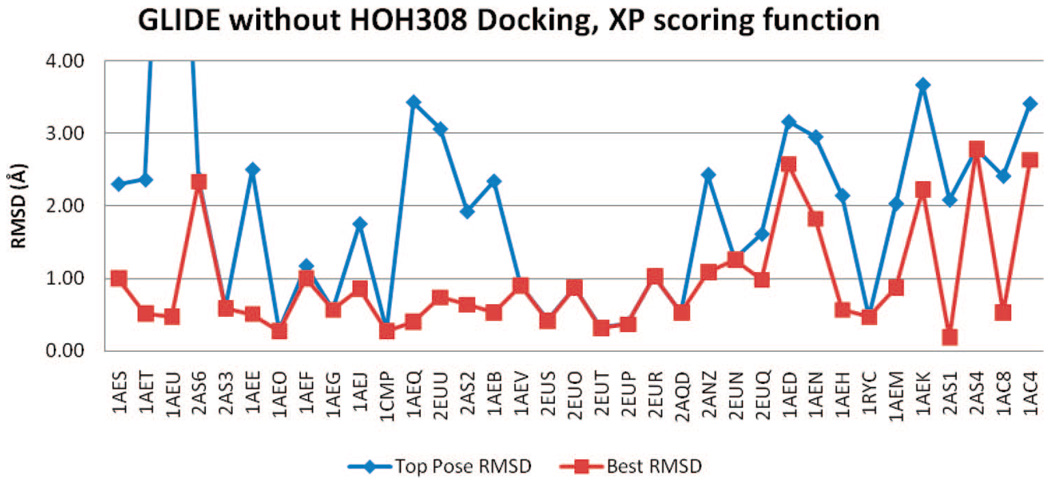
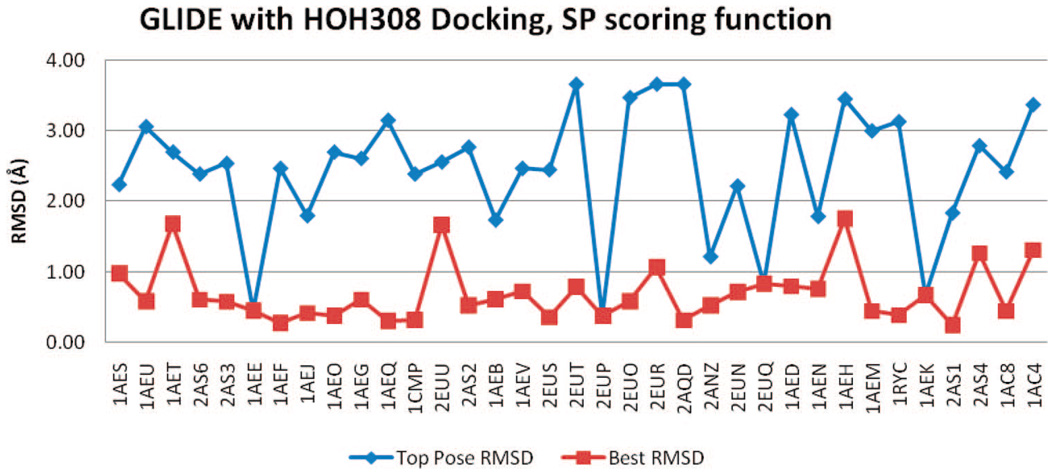
GLIDE
The GLIDE rmsd results of all four docking trials, with or without HOH308, SP, or XP scoring functions, are included in Figure 12–Figure 15. Regardless of the inclusion of HOH308, SP scoring has slightly better BP-rmsd values than XP scoring, but more no. 1 poses in XP scoring are closer to crystal poses than those in SP scoring (Table 2). The overall rmsd results of GLIDE are worse than those of QXP/FLO; they are also slightly worse than those of FRED.
Figure 12.
rmsd results using GLIDE (SP) without HOH308.
One ligand, 2-methylimidazole (PDB code 1AEU), had an erroneously high TP-rmsd value (10 Å) with GLIDE, in XP scoring without HOH308. During docking, GLIDE incorrectly placed the ligand in a pocket at the other side of the heme cofactor, resulting in the bad rmsd. Two of the top 10 poses (nos. 1 and 4) were put in this incorrect pocket, and they were in contact with PRO145, forming one hydrogen bond with the carbonyl oxygen. Similarly, GLIDE also docked another nonbinder ligand, tetrazole, into this pocket. In this case, all 10 poses were positioned in this PRO145 pocket. None of the other binders or nonbinders were docked in this pocket.
For GLIDE, a box with the default diameter of 10 Å is set up in the active site. The diameter midpoint of each docked ligand is required to remain within this box. To avoid erroneous ligand positioning, a smaller diameter of 8 Å was tested in docking with XP scoring and no HOH308. No “out of area” docking was discovered; the mean TP-rmsd improved to 1.61 Å, compared to 1.99 Å using the default diameter. However, no improvements were seen in the mean BP-rmsd and the AUC value. The same smaller diameter was also tested with other docking conditions (SP with or without HOH308, XP with HOH308), no significant improvements were shown in RMSDs and AUC values. In some cases, the result was worse. Therefore, using the smaller midpoint box can avoid the “out of area” docking, but does not seem to affect GLIDE docking results considerably.
Tautomer Generation
The possibility that GLIDE’s unsatisfying result was related to the ligand tautomers was also studied. For comparison, ChemAxon’s Marvin82 was used to confirm whether Ligprep’s tautomer generation was complete.
Ligprep generated 98 different protonation states and tautomers for all 60 binders and nonbinders. Marvin’s Calculator Plugins82 was used on these 98 isomers to see if additional tautomers could be generated. Default parameters were used, except “protect aromaticity” and “protect charge” were unselected. Only two ligands (4-amino-5-imidazole carboxamide and isoniazide) had new tautomers with at least 1% distribution among all tautomers of the same ligand. All other new tautomers showed 0% distribution and thus were considered insignificant. This led to the conclusion that Ligprep’s tautomer enumeration is practically complete and that the docking result was not affected by tautomer generation in this case.
HOH308’s Effect on Docking
The importance of HOH308 in docking had been noted in other studies. In reports by Rosenfeld et al.62 and Brenk et al.,49 HOH308 was kept in the binding pocket during docking. In Rosenfeld’s study, another cavity water HOH401 was also optionally kept in binding pocket when stochastic docking gave ambiguous result, such as 2-aminothazole, 3-aminopyridine, 4-aminopyridine, and imidazole. In two cases (2-aminothazole and 3-aminopyridine), docked poses (with HOH401) became more consistent to the crystal pose than docking without HOH401. In the other two cases (4-aminopyridine and imidazole), docking results could not be improved and disordered ligand binding in the cavity was believed to be the cause.
However, HOH308 did not seem to affect docking results in our study. Of all three methods, docking without HOH308 seemed to produce better results indicated by the ROC curves and rmsd results. The inclusion of HOH308 showed little or no improvements.
Moreover, the decision to keep the cavity water or not cannot be made without comprehensive experimental studies. Brenk et al. discovered that, with extensive crystallography study, HOH308 is likely to take more than one position in the CCPW191G binding pocket according to the ligand binding modes.49 In this study, HOH308 was prohibited from moving during docking. However, the improvement on CCPW181G docking from a movable HOH308 remains intriguing for future studies.
CONCLUSION
CCP W191G is an excellent model for docking exploration. It has a fairly small cavity, which is completely negatively charged and buried from solvent. It also has been studied extensively, resulting in sets of binders with crystal structures and nonbinders. The test of separating binders and nonbinders using three popular docking programs, FRED, QXP/FLO, and GLIDE, showed that, even when studying such a small binding pocket, docking programs have limitations in ranking the ligands. The inclusion of HOH308 in active site showed little effect on docking results in this study. Among all docking runs, QXP/FLO outperformed the other two methods in separating binders and nonbinders and in accurately reproducing crystal poses.
Supplementary Material
Figure 13.
rmsd results using GLIDE (XP) without HOH308.
Figure 14.
rmsd results using GLIDE (SP) with HOH308.
ACKNOWLEDGMENT
We thank Dr. Anthony Nicholls from OpenEye Scientific Software, Inc., and Dr. Colin McMartin for providing us their software. We also wish to acknowledge Prof. Wim G. J. Hol for providing a supportive environment in the Biomolecular Structure Center. Supported by NIH Grant AI067921.
Abbreviations
- TP-rmsd
top pose rmsd
- BP-rmsd
best pose rmsd
- CCP
cyctochrome c peroxidase
- BD
binder
- NB
nonbinder
- VDW
van der Waals
- ROC
receiver operating characteristics
- AUC
area under ROC curve.
Footnotes
Supporting Information Available: Ranking results from all docking methods, the binding site marker atoms during QXP/FLO docking, and all ligands’ chemical names, structures, binding affinities, the crystal structures’ PDB codes, crystallography data, and primary citations. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES AND NOTES
- 1.Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discovery. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 2.Shoichet BK. Virtual screening of chemical libraries. Nature. 2004;432:862–865. doi: 10.1038/nature03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abagyan R, Totrov M. High-throughput docking for lead generation. Curr. Opin. Chem. Biol. 2001;5:375–382. doi: 10.1016/s1367-5931(00)00217-9. [DOI] [PubMed] [Google Scholar]
- 4.Alvarez JC. High-throughput docking as a source of novel drug leads. Curr. Opin. Chem. Biol. 2004;8:365–370. doi: 10.1016/j.cbpa.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Jenkins JL, Shapiro R. Identification of small-molecule inhibitors of human angiogenin and characterization of their binding interactions guided by computational docking. Biochemistry. 2003;42:6674–6687. doi: 10.1021/bi034164e. [DOI] [PubMed] [Google Scholar]
- 6.Vangrevelinghe E, Zimmermann K, Schoepfer J, Portmann R, Fabbro D, Furet P. Discovery of a potent and selective protein kinase CK2 inhibitor by high-throughput docking. J. Med. Chem. 2003;46:2656–2662. doi: 10.1021/jm030827e. [DOI] [PubMed] [Google Scholar]
- 7.Jozwiak K, Ravichandran S, Collins JR, Wainer IW. Interaction of noncompetitive inhibitors with an immobilized α3β4 nicotinic acetylcholine receptor investigated by affinity chromatography, quantitative structure–activity relationship analysis, and molecular docking. J. Med. Chem. 2004;47:4008–4021. doi: 10.1021/jm0400707. [DOI] [PubMed] [Google Scholar]
- 8.Kuntz ID, Blaney JM, Oatley SJ, Langidge R, Ferrin TE. A geometric approach to macromolecule–ligand interactions. J. Mol. Biol. 1982;161:269–288. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- 9.Brooijmans N, Kuntz ID. Molecular recognition and docking algorithms. Annu. Rev. Biophys. Biomol. Struct. 2003;32:335–373. doi: 10.1146/annurev.biophys.32.110601.142532. [DOI] [PubMed] [Google Scholar]
- 10.Halperin I, Ma B, Wolfon H, Nussinov R. Principles of docking: An overview of search algorithms and a guide to scoring functions. Proteins. 2002;47:409–443. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- 11.Taylor RD, Jewsbury PJ, Essex JW. A review of protein–small molecule docking methods. J. Comput.-Aided. Mol. Des. 2002;16:151–166. doi: 10.1023/a:1020155510718. [DOI] [PubMed] [Google Scholar]
- 12.McMartin C, Bohacek RS. QXP: Powerful, rapid computer algorithms for structure-based drug design. J. Comput.-Aided Mol. Des. 1997;11:333–344. doi: 10.1023/a:1007907728892. [DOI] [PubMed] [Google Scholar]
- 13.Totrov M, Abagyan R. Flexible protein–ligand docking by global energy optimization in internal coordinates. Proteins. 1997;1:215–220. doi: 10.1002/(sici)1097-0134(1997)1+<215::aid-prot29>3.3.co;2-i. [DOI] [PubMed] [Google Scholar]
- 14.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 15.Baxter CA, Murray CW, Clark DE, Westhead DR, Eldridge MD. Flexible docking using Tabu search and an empirical estimate of binding affinity. Proteins. 1998;33:367–382. [PubMed] [Google Scholar]
- 16.Morris G, Goodsell D, Halliday R, Huey R, Hart W, Belew R, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 17.Kramer B, Rarey M, Lengaue rT. Evaluation of the FLEXX incremental construction algorithm for protein–ligand docking. Proteins. 1999;37:228–241. doi: 10.1002/(sici)1097-0134(19991101)37:2<228::aid-prot8>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 18.Moustakas DT, Lang PT, Pegg S, Pettersen E, Kuntz ID, Brooijmans N, Rizzo RC. Development and validation of a modular, extensible docking program: DOCK 5. J. Comput-Aided Mol. Des. 2006;20:601–619. doi: 10.1007/s10822-006-9060-4. [DOI] [PubMed] [Google Scholar]
- 19.Diller DJ, Merz KM., Jr High throughput docking for library design and library prioritization. Proteins. 2001;43:113–124. doi: 10.1002/1097-0134(20010501)43:2<113::aid-prot1023>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 20.David L, Luo R, Gilson MK. ligand–receptor docking with the mining minima optimizer. J. Comput.-Aided Mol. Des. 2001;15:157–171. doi: 10.1023/a:1008128723048. [DOI] [PubMed] [Google Scholar]
- 21.McGann MR, Almond HR, Nicholls A, Grant JA, Brown FK. Gaussian docking functions. Biopolymers. 2003;68:76–90. doi: 10.1002/bip.10207. [DOI] [PubMed] [Google Scholar]
- 22.Jain AN. Surflex: Fully automatic flexible molecular docking using a molecular similarity-based search engine. J. Med. Chem. 2003;46:499–511. doi: 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]
- 23.Venkatachalam CM, Jiang X, Oldfield T, Waldman M. Ligandfit: A novel method for the shape-directed rapid docking of ligands to protein active sites. J. Mol. Graphics Modell. 2003;21:289–307. doi: 10.1016/s1093-3263(02)00164-x. [DOI] [PubMed] [Google Scholar]
- 24.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein–ligand docking using GOLD. Proteins. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 25.Taylor RD, Jewsbury PJ, Essex JW. FDS: Flexible ligand and receptor docking with a continuum solvent model and soft-core energy function. J. Comput. Chem. 2003;24:1637–1656. doi: 10.1002/jcc.10295. [DOI] [PubMed] [Google Scholar]
- 26.Stahl M, Rarey M. Detailed analysis of scoring functions for virtual screening. J. Med. Chem. 2001;44(7):1035–1042. doi: 10.1021/jm0003992. [DOI] [PubMed] [Google Scholar]
- 27.Schulz-Gasch T, Stahl M. Binding site characteristics in structure-based virtual screening: Evaluation of current docking tools. J. Mol. Model. 2003;9(1):47–57. doi: 10.1007/s00894-002-0112-y. [DOI] [PubMed] [Google Scholar]
- 28.Wang R, Lu Y, Wang S. Comparative evaluation of 11 scoring functions for molecular docking. J. Med. Chem. 2003;46(12):2287–2303. doi: 10.1021/jm0203783. [DOI] [PubMed] [Google Scholar]
- 29.Perola E, Walters WP, Charifson PS. A detailed comparison of current docking and scoring methods on systems of pharmaceutical relevance. Proteins: Struct., Funct., Bioinf. 2004;56(2):235–249. doi: 10.1002/prot.20088. [DOI] [PubMed] [Google Scholar]
- 30.Kellenberger E, Rodrigo J, Muller P, Rognan D. Comparative evaluation of eight docking tools for docking and virtual screening accuracy. Proteins: Struct., Funct., Bioinf. 2004;57(2):225–242. doi: 10.1002/prot.20149. [DOI] [PubMed] [Google Scholar]
- 31.Kontoyianni M, McClellan LM, Sokol GS. Evaluation of docking performance: Comparative data on docking algorithms. J. Med. Chem. 2004;47(3):558–565. doi: 10.1021/jm0302997. [DOI] [PubMed] [Google Scholar]
- 32.Xing L, Hodgkin E, Liu Q, Sedlock D. Evaluation and application of multiple scoring functions for a virtual screening experiment. J. Comput.-Aided Mol. Des. 2004;18(5):333–344. doi: 10.1023/b:jcam.0000047812.39758.ab. [DOI] [PubMed] [Google Scholar]
- 33.Cummings MD, DesJarlais RL, Gibbs AC, Mohan V, Jaeger EP. Comparison of automated docking programs as virtual screening tools. J. Med. Chem. 2005;48:962–976. doi: 10.1021/jm049798d. [DOI] [PubMed] [Google Scholar]
- 34.Chen H, Lyne PD, Giordanetto F, Lovell T, Li J. On evaluating molecular-docking methods for pose prediction and enrichment factors. J. Chem. Inf. Model. 2006;46(1):401–415. doi: 10.1021/ci0503255. [DOI] [PubMed] [Google Scholar]
- 35.Warren GL, Andrews CW, Capelli A, Clarke B, La Londe J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS. A critical assessment of docking programs and scoring functions. J. Med. Chem. 2006;49(20):5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 36.Gehlhaar DK, Verkhivker GM, Rejto PA, Sherman CJ, Fogel DB, Fogel LJ, Freer ST. Molecular recognition of the inhibitor AG-1343 by HIV-1 protease: Conformationally flexible docking by evolutionary programming. Chem. Biol. 1995;2:317–324. doi: 10.1016/1074-5521(95)90050-0. [DOI] [PubMed] [Google Scholar]
- 37.Welch W, Ruppert J, Jain AN. Hammerhead: Fast, fully automated docking of flexible ligands to protein binding sites. Chem. Biol. 1996;3:449–462. doi: 10.1016/s1074-5521(96)90093-9. [DOI] [PubMed] [Google Scholar]
- 38.Muegge I, Martin YC, Hajduk PJ, Fesik SW. Evaluation of PMF scoring in docking weak ligands to the FK506 binding protein. J. Med. Chem. 1999;42:2498–2503. doi: 10.1021/jm990073x. [DOI] [PubMed] [Google Scholar]
- 39.Bissantz C, Folkers G, Rognan D. Protein-based virtual screening of chemical databases: 1. Evaluation of different docking/scoring combinations. J. Med. Chem. 2000;43:4759–4767. doi: 10.1021/jm001044l. [DOI] [PubMed] [Google Scholar]
- 40.Gohlke H, Hendlich M, Klebe G. Knowledge-based scoring function to predict protein–ligand interactions. J. Mol. Biol. 2000;295:337–356. doi: 10.1006/jmbi.1999.3371. [DOI] [PubMed] [Google Scholar]
- 41.Pearlman DA, Charifson PS. Are free energy calculations useful in practice? A comparison with rapid scoring functions for the p38 MAP kinase protein system. J. Med. Chem. 2001;44:3417–3423. doi: 10.1021/jm0100279. [DOI] [PubMed] [Google Scholar]
- 42.Terp GE, Johansen BN, Christensen IT, Jørgensen FS. A new concept for multidimensional selection of ligand conformations (Multiselect) and multidimensional scoring (Multiscore) of protein–ligand binding affinities. J. Med. Chem. 2001;44:2333–2343. doi: 10.1021/jm001090l. [DOI] [PubMed] [Google Scholar]
- 43.Buzko OV, Bishop AC, Shokat KM. Modified AutoDock for accurate docking of protein kinase inhibitors. J. Comput-Aided Mol. Des. 2002;16:113–127. doi: 10.1023/a:1016366013656. [DOI] [PubMed] [Google Scholar]
- 44.Clark RD, Strizhev A, Leonard JM, Blake JF, Matthew JB. Consensus scoring for ligand/protein interactions. J. Mol. Graphics Model. 2002;20:281–295. doi: 10.1016/s1093-3263(01)00125-5. [DOI] [PubMed] [Google Scholar]
- 45.Gohlke H, Klebe G. Approaches to the description and prediction of the binding affinity of small-molecule ligands to macromolecular receptors. Angew. Chem., Int. Ed. 2002;41:2644–2676. doi: 10.1002/1521-3773(20020802)41:15<2644::AID-ANIE2644>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 46.eHiTS, version 6.2. Toronto ON, Canada: SimBioSys Inc.; 2008. [Google Scholar]
- 47.Laederach A, Reilly PJ. Modeling protein recognition of carbohydrates. Proteins. 2005;60:591–597. doi: 10.1002/prot.20545. [DOI] [PubMed] [Google Scholar]
- 48.Andersson CD, Thysell E, Lindström A, Bylesjö M, Raubacher F, Linusson A. A multivariate approach to investigate docking parameters’ effects on docking performance. J.Chem. Inf. Model. 2007;47:1673–1687. doi: 10.1021/ci6005596. [DOI] [PubMed] [Google Scholar]
- 49.Brenk R, Vetter SW, Boyce SE, Goodin DB, Shoichet BK. Probing molecular docking in a charged model binding site. J. Mol. Biol. 2006;357:1449–1470. doi: 10.1016/j.jmb.2006.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei BQ, Baase WA, Weaver LH, Matthews BW, Shoichet BK. A model binding site for testing scoring functions in molecular docking. J. Mol. Biol. 2002;322:339–355. doi: 10.1016/s0022-2836(02)00777-5. [DOI] [PubMed] [Google Scholar]
- 51.Lorber DM, Shoichet BK. Hierarchical docking of databases of multiple ligand conformations. Curr. Top. Med. Chem. 2005;5:739–749. doi: 10.2174/1568026054637683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.FRED, version 2.2.1. Santa Fe, NM: Openeye Scientific Software Inc.; 2007. [Google Scholar]
- 53.QXP/FLO, version 99.6. Colebrook, CT: ThistleSoft Inc.; 1999. [Google Scholar]
- 54.GLIDE, version 3.5. Portland, OR: Schrodiner Inc.; 2008. [Google Scholar]
- 55.Eriksson AE, Baase WA, Wozniak JA, Matthews BW. A cavity-containing mutant of T4 lysozyme is stabilized by buried benzene. Nature. 1992;355:371–373. doi: 10.1038/355371a0. [DOI] [PubMed] [Google Scholar]
- 56.Morton A, Baase WA, Matthews BW. Energetic origins of specificity of ligand binding in an interior nonpolar cavity of T4 lysozyme. Biochemistry. 1995;34:8564–8575. doi: 10.1021/bi00027a006. [DOI] [PubMed] [Google Scholar]
- 57.Fitzgerald MM, Churchill MJ, McRee DE, Goodin DB. Small molecule binding to an artificially created cavity at the active site of cytochrome c peroxidase. Biochemistry. 1994;33:3807–3818. [PubMed] [Google Scholar]
- 58.Shoichet Laboratory Homepage. Under “Take Aways → CCP binders”. San Francisco, CA: University of California, San Francisco; [accessed Apr 19, 2007]. http://shoichetlab.compbio.ucsf.edu/ [Google Scholar]
- 59.Shoichet Laboratory Homepage. Under “Take Aways → CCP nonbinders”. San Francisco, CA: University of California, San Francisco; [accessed Apr 19, 2007]. http://shoichetlab.compbio.ucsf.edu/ [Google Scholar]
- 60.Banba S, Guo ZY, Brooks CL. Efficient sampling of ligand orientations and conformations in free energy calculations using the λ-dynamics method. J. Phys. Chem., Ser. B. 2000;104:6903–6910. [Google Scholar]
- 61.Banba S, Brooks CL. Free energy screening of small ligands binding to an artificial protein cavity. J. Chem. Phys. 2000;113:3423–3433. [Google Scholar]
- 62.Rosenfeld RJ, Goodsell DS, Musah RA, Morris GM, Goodin DB, Olson AJ. Automated docking of ligands to an artificial active site: Augmenting crystallographic analysis with computer modeling. J. Comput-Aided Mol. Des. 2003;17:525–536. doi: 10.1023/b:jcam.0000004604.87558.02. [DOI] [PubMed] [Google Scholar]
- 63.Boström J. Reproducing the conformations of protein-bound ligands: A critical evaluation of several popular conformational searching tools. J. Comput-Aided Mol. Des. 2001;15:1137–1152. doi: 10.1023/a:1015930826903. [DOI] [PubMed] [Google Scholar]
- 64.Boström J, Greenwood JR, Gottfries J. Assessing the performance of OMEGA with respect to retrieving bioactive conformations. J. Mol. Graphics Modell. 2003;21:449–462. doi: 10.1016/s1093-3263(02)00204-8. [DOI] [PubMed] [Google Scholar]
- 65.ED user manual, version 2.2. Santa Fe, NM: Openeye Scientific Software Inc.; 2007. [Google Scholar]
- 66.Grant JA, Pickup BT, Nicholls A. A smooth permittivity function for Poisson–Boltzmann solvation methods. J. Comput. Chem. 2001;22(6):608–640. [Google Scholar]
- 67.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996;118:11225–11236. [Google Scholar]
- 68.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 69.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 70.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision Glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein–ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 71. [accessed Apr 2, 2007];RCSB Protein Data Bank. http://http://www.rcsb.org.
- 72.MOE, version 200608. Montreal, Quebec, Canada: Chemical Computing Group Inc.; 2006. [Google Scholar]
- 73.Jakalian A, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic charges. AM1-BCC Model:II. Parameterization and validation. J. Comput. Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 74.OMEGA, version 2.2.0. Santa Fe, NM: Openeye Scientific Software Inc.; 2007. [Google Scholar]
- 75. [accessed Jun 14, 2007];Molprobity. http://molprobity.biochem.duke.edu/
- 76.Chimera, version 1.2422. San Francisco, CA: University of California; 2007. http://www.cgl.ucsf.edu/chimera/ [Google Scholar]
- 77.FRED_receptor, version 2.2.1. Santa Fe, NM: Openeye Scientific Software Inc.; 2007. [Google Scholar]
- 78.Maestro, version 7.0. Portland, OR: Schrodiner Inc.; 2008. [Google Scholar]
- 79.LigPrep. Portland, OR: Schrodiner Inc.; 2008 version 16016. [Google Scholar]
- 80.Hand D, Mannila H, Smyth P. Principles of Data Mining. Cambridge, MA: The MIT Press; 2001. [Google Scholar]
- 81.Teramoto R, Fukunishi H. Supervised consensus scoring for docking and virtual screening. J. Chem. Inf. Model. 2007;47:526–534. doi: 10.1021/ci6004993. [DOI] [PubMed] [Google Scholar]
- 82.Marvn, version 5.0.6. Budapest, Hungary: ChemAxon Kft.; 2008. http://www.chemaxon.com. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.