Abstract
Rationale
COPD exacerbations reduce quality of life and increase mortality. Genetic variation may explain the substantial variability seen in exacerbation frequency among COPD subjects with similar lung function. We analyzed whether polymorphisms in five candidate genes previously associated with COPD susceptibility also demonstrate association with COPD exacerbations.
Methods
Eighty-eight single nucleotide polymorphisms in microsomal epoxide hydrolase (EPHX1), transforming growth factor beta 1 (TGFB1), SERPINE2, glutathione S-transferase pi (GSTP1), and surfactant protein B (SFTPB) were genotyped in 389 non-Hispanic white participants in the National Emphysema Treatment Trial. Exacerbations were defined as COPD-related emergency room visits or hospitalizations using Centers for Medicare and Medicaid Services claims data.
Measurements and Main Results
216 subjects (56%) experienced one or more exacerbations during the study period. An SFTPB promoter polymorphism, rs3024791, was associated with COPD exacerbations (p=0.008). Logistic regression models confirmed the association with rs3024791 (p = 0.007). Poisson regression models demonstrated association of multiple SFTPB SNPs with exacerbation rates: rs2118177 (p = 0.006), rs2304566 (p = 0.002), rs1130866 (p = 0.04), and rs3024791 (p = 0.002). Polymorphisms in EPHX1, GSTP1, TGFB1, and SERPINE2 did not demonstrate association with COPD exacerbations.
Conclusions
Variants in SFTPB are associated with COPD susceptibility and COPD exacerbation frequency.
Keywords: association analysis, COPD, exacerbations, genetics, surfactant protein B, single nucleotide polymorphisms
Introduction
The Global Initiative for Chronic Obstructive Pulmonary Disease (GOLD) defines COPD exacerbations as acute events within the natural course of the disease that exceed daily variation in baseline dyspnea, cough, and/or sputum that may merit a change in regular medication.[1] COPD exacerbations are associated with a range of outcomes, from slightly worsening respiratory symptoms to death. Frequent COPD exacerbations may hasten decline in lung function[2, 3] and increase mortality.[4] COPD exacerbations have a substantial personal [5, 6] and public impact [7, 8] including reduction in quality of life, lost wages, and increased health care costs. Exacerbations resulting in hospitalization are responsible for the greatest proportion of healthcare costs related to COPD.[8–10] Efforts to reduce or prevent COPD exacerbations, particularly those associated with hospitalizations, would dramatically improve the health status of COPD subjects and diminish the economic burden imposed by the disease.
Genetic determinants of COPD exacerbations have not been clearly identified [11, 12] despite the observation that COPD exacerbations are more frequent in individuals with prior exacerbations. Variation in exacerbation frequency could relate to different COPD subtypes or to differential susceptibility to infections. We hypothesized that genetic variants associated with COPD susceptibility may influence the occurrence and frequency of COPD exacerbations. To test this hypothesis, we analyzed eighty-eight polymorphisms in five genes previously associated with COPD susceptibility in at least two studies, for association with COPD exacerbations.[13, 14]
Materials and Methods
Study Participants
389 non-Hispanic white subjects enrolled in the NETT Genetics Ancillary Study were included in this analysis. The NETT was a multi-center, randomized, clinical trial comparing conventional medical therapy for severe emphysema to lung volume reduction surgery (LVRS).[15–17] Briefly, eligibility criteria for participation in the NETT were as follows: 1) FEV1 ≤ 45% predicted, 2) evidence of hyperinflation manifested by an elevated total lung capacity and residual volume on pulmonary function testing, 3) bilateral emphysema on high resolution computed tomographic scanning of the chest, and 4) successful completion of pre-determined performance goals during pulmonary rehabilitation. Active cigarette smoking was an exclusion criterion.[16] As the purpose of our study was to determine the role of genetic influences on the susceptibility to or the frequency of COPD exacerbations, nine individuals who were randomized to LVRS but did not undergo surgery were considered medically treated in our analyses.
Data collection
Information regarding COPD-related emergency room visits or hospitalizations was available for an eight-year study period. Enrollment into the NETT began in January 1998. Exacerbation event data was collected for all participants beginning one year prior to randomization from claims records of the Centers for Medicare and Medicaid Services (CMS). Visit records with a principal ICD9-CM diagnosis code of 491, 492, 493, or 496 were eligible for inclusion. Emergency room visits and hospitalizations on the same calendar date were considered as one event. Independent emergency room visits and hospitalizations separated by a period of 14 or more days were included as unique events. The BODE index [18], a composite multidimensional score, was modified to include the post-pulmonary rehabilitation FEV1 values, 6-minute walk test, body mass index (BMI), and the University of California San Diego Pulmonary Rehabilitation Shortness-of-Breath Questionnaire.[19] The co-morbidity score is derived from the Charlson co-morbidity index that has been modified for use with administrative data.
SNP Selection and Genotyping
Detailed procedural descriptions of the genotyping and SNP selection for EPHX1, GSTP1, SFTPB [13, 20], SERPINE2 [14], and TGFB1 [21] have been previously published. Briefly, genotype data for European Americans (CEU) from the SeattleSNPs Program for Genomic Applications (http://pga.mbt.washington.edu/) and the International Hap Map Project [22] were used to select linkage disequilibrium-tagging SNPs for all five genes. Pairwise LD-tagging was achieved with Tagger (http://www.broad.mit.edu/mpg/tagger/) for SNPs with a minimum minor allele frequency of 0.1 and an r2 of 0.9. Additional SNPs in SERPINE2 were obtained from DNA sequencing of all exon and exon-intron boundaries.[14] Three platforms were used for genotyping: allele specific hybridization (Illumina Golden Gate Assay, San Diego, CA), the 5′ to 3′ exonuclease assay (TaqMan, Applied Biosystems, Foster City, CA), or with unlabeled minisequencing reactions and mass spectrometry (Sequenom, San Diego, CA). All polymorphisms analyzed in the five candidate genes are listed in the online data supplement. (Table S1 in the Online Data Supplement) The linkage disequilibrium (LD) map of the five SNPs in SFTPB was created in Haploview.[23]
Statistical Analysis
Quantitative data with a normal distribution are presented as means ± standard deviations. Variables with a non-normal distribution are presented as medians ± interquartile ranges. Polymorphisms with significant Cochran-Armitage tests for trend (p ≤ 0.05),[24] a genotype-based test for association (SAS/Genetics; Cary, NC), were entered into multivariable logistic regression models. The logistic regression models for probability of any exacerbation analyzed linear trends with additive genetic coding. The models were adjusted for the following potential confounders: age, FEV1 percent predicted, pack-years of smoking, gender (1 = male, 0 = female), surgery status (1 = underwent LVRS, 0 = received medical therapy), and surgery-by-genotype interaction. There were no significant surgery-by-genotype interactions and the interaction terms were not included in the final models. CMS claims data were available for the period from 1997–2004. In order to analyze exacerbation rates, account for correlated data in the form of multiple exacerbations in a single individual, and allow for a differential effect of medical therapy versus lung volume reduction surgery on exacerbation frequency, Poisson rate regression models were constructed.[25] The Poisson rate models, accounting for varying observation times, were constructed under an additive mode of inheritance and were adjusted for age, FEV1 percent predicted, pack-years of smoking, and surgery. The multi-state Poisson models assumed no difference between the treatment groups prior to randomization. To determine if surgical treatment influenced the frequency of exacerbations, exacerbation counts were compared in the surgical patients after LVRS and after randomization in medical patients. There were no significant surgery-by-genotype interactions in the Poisson models, and the interaction terms were removed from the final models. Haplotype analysis was performed on haplotypes ≥ 5% frequency using the haplo.stats [26] package in R (version 2.5.1). The sliding window haplotype analysis assessed adjacent 2-loci, 3-loci, or 4-loci combinations. Empirical p-values for significant global haplotype and sliding window analyses were obtained through permutation of the p-values with 1000 simulations. The haplotype analysis was adjusted for age, gender, FEV1 percent predicted, pack-years of smoking, and treatment. All SFTPB and TGFB1 SNPs were in Hardy-Weinberg equilibrium. Two GSTP1 SNPs, one EPHX1 SNP, and one SERPINE2 SNP were out of Hardy-Weinberg equilibrium and were removed from further analysis.
Results
The demographic and clinical characteristics of the NETT participants are displayed in Table 1. Over the period of study, COPD exacerbations defined as COPD-related emergency department visits or hospitalizations occurred in 56% of subjects. Detailed characterizations of the epidemiologic predictors of exacerbations in NETT participants have been reported elsewhere.[27, 28]
Table 1.
Demographic Characteristics of 389 NETT Subjects*
| Age (years) | 67 ± 6 |
| Pack-years | 66 ± 30 |
| FEV1 (% predicted, post BD) | 28 ± 7 |
| Modified BODE (median ± IQR) | 5 ± 3 |
| Gender (% male) | 64% |
| LVRS | 53% |
| Exacerbations (% with > 1) ‡ | 56% |
| Bronchodilator therapy | 99% |
| Inhaled corticosteroid therapy | 71% |
| Inhaled anticholinergic therapy | 82% |
| Current systemic corticosteroid therapy | 24% |
Values are + standard deviation unless otherwise listed
Exacerbations ranged from 0–29 events per subject
post BD = after bronchodilator administration
IQR = interquartile range
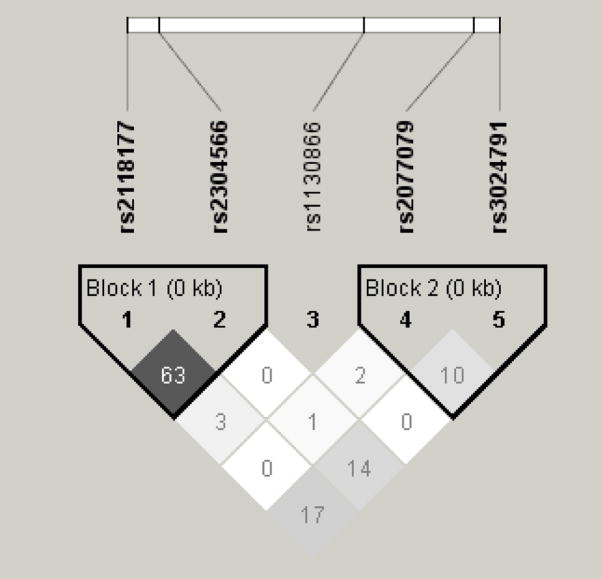
Cochran-Armitage trend tests for SNPs in EPHX1, TGFB1, GSTP1, and SERPINE2 were not significant. Those genes were not analyzed further. The SFTPB genotype frequencies for study participants are displayed in Table 2. Pairwise linkage disequilibrium was determined for the five SFTPB SNPs by r2 (Figure 1). There was generally low LD between the SFTPB SNPs. The highest r2 was 0.6 between rs211877 and rs2304566.
Table 2.
Genotype Frequencies for SFTPB SNPs in 389 non-Hispanic White COPD Subjects*
| SNP | Genotype | Count | Frequency |
|---|---|---|---|
| rs2118177 | C/C | 55 | 0.14 |
| T/C | 159 | 0.42 | |
| T/T | 166 | 0.44 | |
| rs2304566 | G/G | 28 | 0.07 |
| A/G | 139 | 0.37 | |
| A/A | 214 | 0.56 | |
| rs1130866 | T/T | 113 | 0.29 |
| T/C | 189 | 0.49 | |
| C/C | 85 | 0.22 | |
| rs2077079 | C/C | 52 | 0.14 |
| A/C | 176 | 0.47 | |
| A/A | 143 | 0.39 | |
| rs3024791 | G/G | 276 | 0.73 |
| G/A | 94 | 0.25 | |
| A/A | 9 | 0.02 |
Genotype counts do not sum to 389 due to missing genotypes: 9 missing genotypes for rs2118177, 8 missing genotypes for rs2304566, 2 missing genotypes for rs1130866, 8 missing genotypes for rs2077079, 10 missing genotypes for rs3024791
Figure 1.
Linkage Disequilibrium Map for Five SFTPB SNPs in 389 NETT Subjects
rs211877 - rs2304566 r2 0.6
rs2118177 - rs1130866 r2 0.03
rs211877 - rs2077079 r2 0.0
rs211877 - rs3024791 r2 0.2
rs2304566 - rs1130866 r2 0.01
rs2304566 - rs2077079 r2 0.01
rs2304566 - rs3024791 r2 0.2
rs1130866 - rs2077079 r2 0.02
rs1130866 - rs3024791 r2 0.0
rs2077079 - rs3024791 r2 0.1
One SNP in surfactant protein B, rs3024791, a promoter polymorphism, was significantly associated with probability of any exacerbation in the unadjusted Cochran-Armitage trend test (p=0.008). Cochran-Armitage trend tests were non-significant for the remaining SFTPB SNPs. (Table 3.)
Table 3.
Association Analysis of SFTPB SNPs for Probability of Any COPD Exacerbation
| SNP | Allele Frequency Non-exacerbators | Allele Frequency Exacerbators | p-value Allele Test | p-value Armitage Trend Test |
|---|---|---|---|---|
| rs2118177 | 0.39 | 0.33 | 0.11 | 0.13 |
| rs2304566 | 0.29 | 0.23 | 0.10 | 0.10 |
| rs1130866 | 0.50 | 0.44 | 0.10 | 0.10 |
| rs2077079 | 0.35 | 0.40 | 0.20 | 0.20 |
| rs3024791 | 0.19 | 0.12 | 0.007 | 0.008 |
After adjustment for potentially relevant clinical confounders such as age, gender, FEV1, smoking history, medical versus surgical treatment, and follow-up time, having more copies of the minor allele of rs3024791, OR 0.6 [95% CL 0.4, 0.9], p = 0.007 was associated with a reduction in the odds of experiencing COPD exacerbations, independent of the treatment effect.(Table 4.)
Table 4.
| SNP | Unadjusted Odds [CL] | p-value | Adjusted Odds [CL] | p-value |
|---|---|---|---|---|
| rs2118177 | 0.8 [0.6 – 1.1] | 0.1 | 0.8 [0.6 – 1.1] | 0.2 |
| rs2304566 | 0.8 [0.6 – 1.1] | 0.1 | 0.8 [0.5 – 1.1] | 0.1 |
| rs1130866 | 0.8 [0.6 – 1.1] | 0.1 | 0.8 [0.6 – 1.1] | 0.1 |
| rs2077079 | 1.2 [0.9 – 1.7[ | 0.2 | 1.2 [0.9 – 1.6] | 0.3 |
| rs3024791 | 0.6 [0.4 – 0.9] | 0.008 | 0.6 [0.4 – 0.9] | 0.007 |
Adjusted for age, gender, pack-years of smoking, FEV1 % predicted, treatment
216 NETT subjects with at least one exacerbations
Number of COPD exacerbations during this period = 575
Enrollment into the NETT occurred between January 1998 and July 2002. Enrollment during the latter years of the study resulted in only 29 months for CMS data to accrue post-randomization. As a sensitivity analysis to account for variable follow-up time, an analysis of claims data was limited to 3.3 years (1 year prior to randomization and 2.3 years after randomization) in the logistic regression models. Seven individuals had < 2.3 years of follow-up time. All seven individuals died after randomization. Their data were not excluded from this analysis. Controlling for relevant covariates in an additive model, the T allele of rs3024791 was significantly associated with a reduction in the odds of COPD exacerbations, OR 0.6 (95% CI 0.4–0.9), p = 0.02.
Of the 389 study subjects in the 8-year period of study, 216 participants (56%) experienced 575 exacerbations resulting in an overall exacerbation rate of 0.29 exacerbation events per person-year. The majority of the cohort experienced ≤ 4 total events (median = 1, interquartile range = 2). Stratified by medical versus surgical treatment, 183 subjects (47%) were treated non-surgically and 206 subjects (53%) underwent LVRS. In the medical group, 110 subjects experienced 307 exacerbations resulting in 0.32 events per person year in the medical arm. In the surgically treated group, 106 subjects experienced 268 exacerbations resulting in an exacerbation frequency of 0.25 events per person-year in the surgical arm. Delayed surgical therapy could artificially affect the exacerbation frequency by reducing the number of exacerbations experienced during the post-operative period. Most individuals, however, underwent LVRS soon after randomization. The median time to LVRS in this exacerbation cohort was 11 days (interquartile range = 6 days). LVRS was performed in these subjects within 20 days (90th percentile).
In the Poisson analyses (Table 5), four of five polymorphisms in SFTPB were significantly associated with COPD exacerbation counts.
Table 5.
Poisson Regression of COPD Exacerbation Rates
| Estimate | SE | 95% CI | p-value | |
|---|---|---|---|---|
| rs2118177 | −0.30 | 0.12 | [−0.5, −0.07] | 0.01 |
| Age | −0.03 | 0.01 | [−0.05, −0.005] | 0.02 |
| FEV1 % predicted | −0.03 | 0.01 | [−0.05, −0.01] | 0.01 |
| Treatment Group | −0.55 | 0.19 | [−0.9, −0.18] | 0.004 |
| Pack-years | 0.003 | 0.003 | [−0.004, 0.01] | 0.4 |
| Gender | −0.14 | 0.19 | [−0.5, 0.2] | 0.5 |
| rs2304566 | −0.37 | 0.12 | [−0.6, −0.1] | 0.002 |
| Age | −0.03 | 0.01 | [−0.05, −0.004] | 0.02 |
| FEV1 % predicted | −0.03 | 0.01 | [−0.05, −0.008] | 0.007 |
| Treatment Group | −0.52 | 0.19 | [−0.9, −0.2] | 0.005 |
| Pack-years | 0.003 | 0.003 | [−0.004, 0.01] | 0.4 |
| Gender | −0.17 | 0.18 | [−0.5, 0.19] | 0.4 |
| rs1130866 | −0.23 | 0.1 | [−0.42, −0.04] | 0.02 |
| Age | −0.03 | 0.01 | [−0.05, −0.005] | 0.02 |
| FEV1 % predicted | −0.03 | 0.01 | [−0.05, −0.007] | 0.01 |
| Treatment Group | −0.50 | 0.2 | [−0.9, −0.1] | 0.008 |
| Pack-years | 0.003 | 0.003 | [−0.004, 0.01] | 0.4 |
| Gender | −0.12 | 0.19 | [−0.5, 0.3] | 0.5 |
| rs3024791 | −0.61 | 0.18 | [−1.0, −0.3] | 0.0007 |
| Age | −0.03 | 0.01 | [−0.05, −0.005] | 0.02 |
| FEV1 % predicted | −0.03 | 0.01 | [−0.05, −0.008] | 0.009 |
| Treatment Group | −0.53 | 0.19 | [−0.9, −0.2] | 0.006 |
| Pack-years | 0.003 | 0.003 | [−0.004, 0.01] | 0.4 |
| Gender | −0.10 | 0.19 | [−0.5, 0.2] | 0.5 |
| rs2077079 | 0.01 | 0.12 | [−0.2, 0.2] | 0.9 |
| Age | −0.03 | 0.01 | [−0.05, −0.004] | 0.02 |
| FEV1 % predicted | −0.03 | 0.01 | [−0.05, −0.005] | 0.02 |
| Treatment Group | −0.50 | 0.19 | [−0.9, −0.1] | 0.01 |
| Pack-years | 0.002 | 0.003 | [−0.004, 0.01] | 0.5 |
| Gender | −0.1 | 0.19 | [−0.5, 0.3] | 0.6 |
Treatment Group (1 = LVRS, 0 = medical)
Gender (1 = male, 0 = female)
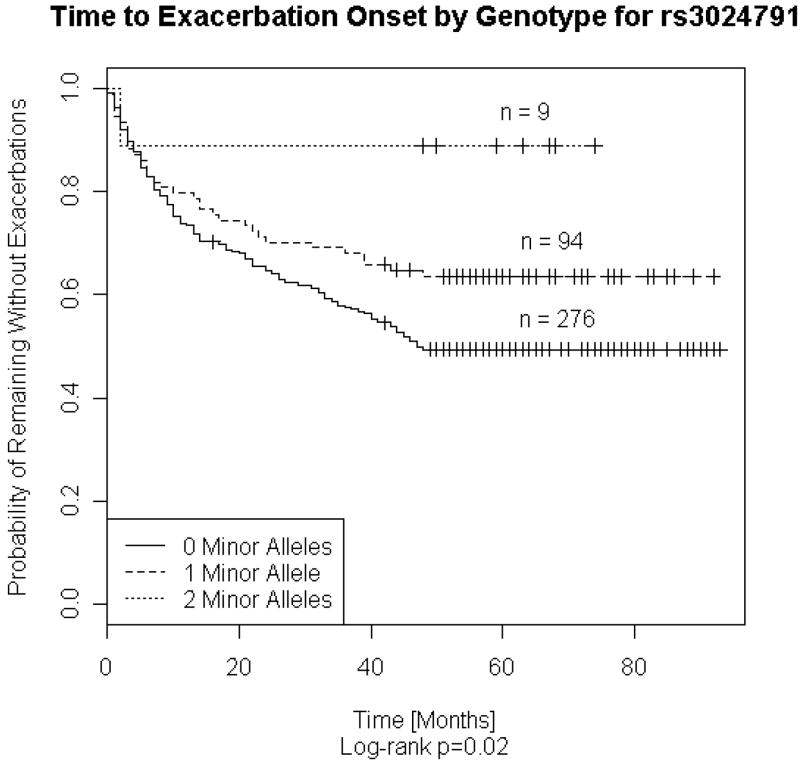
The models were constructed with additive genetic coding and adjusted for age, gender, lung function, pack-years, and treatment. Inheritance of one or more copies of minor alleles of the following variants: rs2118177, rs2304566, rs1130866, and rs3024791, was associated with a reduction in exacerbation counts. (Illustrated with genotype survival curves for rs3024791, depicting time to onset of exacerbations, in Figure 2) The findings were robust with models constructed with exacerbations separated by thirty days, truncating follow-up at four years, or using the entire CMS data collected over eight years.
Figure 2.
Time to COPD Exacerbation Onset for 379 NETT Subjects Stratified SFTPB rs3024791 Genotype
Log rank test was used to determine whether two or more survival curves are identical. (p = 0.02)
There were10 missing genotypes for rs3024791.
For the binary exacerbations phenotype, exacerbations ≥ 1 vs. zero exacerbations, haplotype analysis was performed on SFTPB haplotypes of 5% or greater frequency. The global haplotype score p-value for all five SNPs, based on 1000 simulations, was p = 0.03. The C-G-C-A-A haplotype (rs2118177-rs2304566-rs1130866-rs2077079-rs3024791) had the strongest significance (p = 0.009) and a frequency of 6%. Sliding window haplotype analysis was performed with empirical p-values determined by simulation. The strongest association was observed for the two SNP haplotype (rs2077079-rs3024791), p = 0.006; however haplotypes with three loci (rs1130866-rs2077079-rs3024791), p = 0.01, and four loci (rs2304566-rs1130866-rs2077079-rs3024791), p = 0.01, were also significant. The adjusted global haplotype score for all SNPs included the following covariates: age, gender, lung function, pack-years, and treatment received. After 1000 simulations, the adjusted global haplotype p-value was p = 0.03.
Discussion
The adverse consequences of COPD exacerbations are varied; alteration in quality of life, increased health care costs and utilization, accelerated lung function decline, and increased mortality. We have demonstrated, in individuals with severe chronic airflow limitation, that polymorphisms in surfactant protein B are associated with susceptibility to COPD exacerbations that are substantial enough to warrant emergency department care or hospitalizations. We have also demonstrated that polymorphisms in surfactant protein B are associated with exacerbation frequency. There are few studies of the genetic determinants of COPD exacerbations. Yang and colleagues analyzed the relationship of a polymorphism in mannose binding lectin (MBL2) to COPD exacerbations, COPD susceptibility, and mannose binding lectin levels. In this 2-year retrospective study, the MBL2 codon 54 B allele was associated with an increased frequency of hospital admissions [OR 4.9 (95% CI 1.7 – 14.4), p = 0.011] for COPD exacerbations and reduced mannose binding lectin levels, p < 0.001.[11] No association was demonstrated between the polymorphisms and COPD susceptibility. Takabatake et al performed a study of Japanese male COPD subjects for association of four SNPs in three chemokine genes (CCL11, CCL5, and CCL1) with COPD exacerbations. From the analysis of two years of retrospective data, frequent COPD exacerbations were associated with a CCL1 SNP (rs2282691) under a dominant model of inheritance. The same SNP associated with mortality in a 30-month prospective evaluation of exacerbations [Cox proportional hazards OR 5.93 (95 % CI 1.28–27.48), p = 0.023].[12]
Pulmonary alveolar surfactant is composed of phospholipids and four surfactant proteins that reduce surface tension and prevent alveolar collapse at low lung volumes. Surfactant protein B promotes adhesion and spreading of surfactant phospholipids and stabilizes the phospholipid monolayer.[29, 30] Polymorphisms in SFTPB have been associated with COPD susceptibility in a Mexican population[31] and in a German study.[32] Guo et al found association between the Thr131Ile polymorphism (1580 C/T) and COPD susceptibility (OR 3.7 [95% CI, 1.2–12.1], p = 0.03). Seifart et al studied variants in intron 4 (insertions or deletions) in chronic bronchitis or COPD subjects and controls. The only significant difference between the groups was in a subset analysis; variants in intron 4 were more frequent in the COPD subjects with respiratory failure compared to the population-based controls (OR 4.9 [95% CI, 1.8–13.6], p = 0.003) or the matched controls (OR 3.55 [95% CI, 1.4–9.4], p =0.017). Hersh and colleagues found significant association of the SFTPB Thr131Ile variant (rs1130866) with COPD susceptibility in a case-control study of 304 NETT participants and 441 smoking control subjects only in the presence of a gene-by-smoking interaction term, p = 0.01.[13] In the Boston Early-Onset COPD Study, this SNP was significant in individuals with moderate-to-severe airflow obstruction, p = 0.03. A short tandem repeat (D2S388) in SFTPB has also been demonstrated to be associated with the six-minute walk distance, a test for functional impairment, in subjects with severe COPD.[19] Variants in SFTPB have also been reported to be associated with ARDS [33, 34] and community-acquired pneumonia.[35]
Alterations in the hydrophobic surfactant proteins, B and C, are associated with chronic lung disease and acute respiratory distress syndromes that may be due to deficiency of the bioactive surfactant peptides and/or the intracellular accrual of harmful proteins.[36] In addition to the biophysical properties of alveolar stabilization, improving pulmonary compliance, removal of particulate matter, and enhancing mucociliary clearance, surfactant and its components have immunologic properties.[37] Whether the significant polymorphisms in our study have qualitative or quantitative impact on surfactant protein B levels or function is not currently known. The Thr131Ile non-synonymous SNP is located within a functional glycosylation recognition sequence; it has been postulated that alteration of glycosylation could affect the tertiary structure and processing of surfactant protein B.[35] In a study of hospitalized patients (n= 53) with pulmonary and non-pulmonary diseases (28% COPD), a surfactant protein B promoter polymorphism, rs3024791, reduced transcriptional activity in transfected cells.[38] This SNP was potentially felt to be a genetic cause for the individual variability in surfactant protein B mRNA levels. It was not felt to be sufficient to cause surfactant protein B deficiency but could contribute to a loss of function in the presence of multiple mutations or in the presence of a predisposition for lung disease in combination with other environmental factors. In our study, rs3024791 was significantly associated with COPD exacerbations and exacerbation frequency.
A unique strength of this analysis is that we demonstrate association of surfactant protein B with COPD exacerbation occurrence and frequency; however, our study has limitations. Our study was based on medical claims data, but we did not review actual hospital records to verify diagnosis coding. Our study found association with exacerbations significant enough to warrant emergency care or hospitalization in a medically insured cohort, but we do not have data on less serious exacerbations that only resulted in escalation of medical therapy such as new prescriptions of antibiotics and/or corticosteroids. We acknowledge that health care utilization varies dependent upon access and availability. Additionally, mild exacerbations could be clinically significant and result in emergency room visits of hospitalization for individuals with severe COPD; all individuals in this study were GOPD stage III or greater. In the absence of biomarkers that objectively identify COPD exacerbations, we arbitrarily chose a threshold of 14 days to differentiate new from relapsing exacerbations. However, we found no difference in our results using a more stringent threshold of thirty days (data not shown). Our study was not designed to determine the effect of the polymorphisms on surfactant protein B levels or function. Though our findings are a significant addition to the increasing body of evidence for the importance of SFTPB in COPD pathogenesis, our study results have not yet been replicated. Additionally, our most significant observations for rs2024791 would withstand the scrutiny of multiple comparisons with five genes but would not meet the more stringent threshold of a Bonferroni correction for 88 SNPs. Lastly, we did not formally test for population stratification in our ethnically homogeneous cohort.
We provide additional evidence for the importance of polymorphisms in surfactant protein B in COPD pathophysiology. We demonstrate association of COPD exacerbations and a potentially functional polymorphism in surfactant protein B. Additional research is needed to precisely determine the mechanisms by which these variants influence susceptibility to exacerbations. Further determining the genetic influences on COPD exacerbations may ultimately identify individuals at risk and foster the development of new therapeutic targets and treatment strategies.
Supplementary Material
Acknowledgments
The authors wish to thank Janne Abullarade, Soma Datta, and Barbara J. Klanderman, Ph.D. for their expert assistance.
Financial Disclosure: This study was funded with the following grants: HL007427, HL075478, HL71393, and HL082541 from the National Institutes of Health (NIH). NIH – K08, HL072918, funded DLD. NIH – K08, HL080242, funded CPH. The National Emphysema Treatment Trial (NETT) was supported by contracts with the National Heart, Lung, and Blood Institute (N01HR76101, N01HR76102, N01HR76103, N01HR76104, N01HR76105, N01HR76106, N01HR76107, N01HR76108, N01HR76109, N01HR76110, N01HR76111, N01HR76112, N01HR76113, N01HR76114, N01HR76115, N01HR76116, N01HR76118, and N01HR76119) the Centers for Medicare and Medicaid Services, and the Agency for Healthcare Research and Quality.
References
- 1.Rabe KF, Hurd S, Anzueto A, Barnes PJ, Buist SA, Calverley P, Fukuchi Y, Jenkins C, Rodriguez-Roisin R, van Weel C, Zielinski J. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007;176(6):532–555. doi: 10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- 2.Donaldson GC, Seemungal TA, Bhowmik A, Wedzicha JA. Relationship between exacerbation frequency and lung function decline in chronic obstructive pulmonary disease. Thorax. 2002;57(10):847–852. doi: 10.1136/thorax.57.10.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silverman EK. Exacerbations in chronic obstructive pulmonary disease: do they contribute to disease progression? Proceedings of the American Thoracic Society. 2007;4(8):586–590. doi: 10.1513/pats.200706-068TH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soler-Cataluna JJ, Martinez-Garcia MA, Roman Sanchez P, Salcedo E, Navarro M, Ochando R. Severe acute exacerbations and mortality in patients with chronic obstructive pulmonary disease. Thorax. 2005;60(11):925–931. doi: 10.1136/thx.2005.040527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seemungal TA, Donaldson GC, Paul EA, Bestall JC, Jeffries DJ, Wedzicha JA. Effect of exacerbation on quality of life in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 1998;157(5 Pt 1):1418–1422. doi: 10.1164/ajrccm.157.5.9709032. [DOI] [PubMed] [Google Scholar]
- 6.Donaldson GC, Wilkinson TM, Hurst JR, Perera WR, Wedzicha JA. Exacerbations and time spent outdoors in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2005;171(5):446–452. doi: 10.1164/rccm.200408-1054OC. [DOI] [PubMed] [Google Scholar]
- 7.Hilleman DE, Dewan N, Malesker M, Friedman M. Pharmacoeconomic evaluation of COPD. Chest. 2000;118(5):1278–1285. doi: 10.1378/chest.118.5.1278. [DOI] [PubMed] [Google Scholar]
- 8.Halpern MT, Stanford RH, Borker R. The burden of COPD in the U.S.A. : results from the Confronting COPD survey. Respiratory Medicine. 2003;97 (Suppl C):S81–89. doi: 10.1016/s0954-6111(03)80028-8. [DOI] [PubMed] [Google Scholar]
- 9.Miravitlles M, Murio C, Guerrero T, Gisbert R. Pharmacoeconomic evaluation of acute exacerbations of chronic bronchitis and COPD. Chest. 2002;121(5):1449–1455. doi: 10.1378/chest.121.5.1449. [DOI] [PubMed] [Google Scholar]
- 10.Celli BR, Barnes PJ. Exacerbations of chronic obstructive pulmonary disease. Eur Respir J. 2007;29(6):1224–1238. doi: 10.1183/09031936.00109906. [DOI] [PubMed] [Google Scholar]
- 11.Yang IA, Seeney SL, Wolter JM, Anders EM, McCormack JG, Tunnicliffe AM, Rabnott GC, Shaw JG, Dent AG, Kim ST, Zimmerman PV, Fong KM. Mannose-binding lectin gene polymorphism predicts hospital admissions for COPD infections. Genes and Immunity. 2003;4(4):269–274. doi: 10.1038/sj.gene.6363961. [DOI] [PubMed] [Google Scholar]
- 12.Takabatake N, Shibata Y, Abe S, Wada T, Machiya J, Igarashi A, Tokairin Y, Ji G, Sato H, Sata M, Takeishi Y, Emi M, Muramatsu M, Kubota I. A single nucleotide polymorphism in the CCL1 gene predicts acute exacerbations in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;174(8):875–885. doi: 10.1164/rccm.200603-443OC. [DOI] [PubMed] [Google Scholar]
- 13.Hersh CP, Demeo DL, Lange C, Litonjua AA, Reilly JJ, Kwiatkowski D, Laird N, Sylvia JS, Sparrow D, Speizer FE, Weiss ST, Silverman EK. Attempted replication of reported chronic obstructive pulmonary disease candidate gene associations. American Journal of Respiratory cell and Molecular Biology. 2005;33(1):71–78. doi: 10.1165/rcmb.2005-0073OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeMeo D, Mariani T, Lange C, Lake S, Litonjua A, Celedon J, Reilly J, Chapman HA, Sparrow D, Spira A, Beane J, Pinto-Plata V, Speizer FE, Shapiro S, Weiss ST, Silverman EK. The SERPINE2 gene is associated with chronic obstructive pulmonary disease. Proceedings of the American Thoracic Society. 2006;3(6):502. doi: 10.1513/pats.200603-070MS. [DOI] [PubMed] [Google Scholar]
- 15.Rationale and design of the National Emphysema Treatment Trial (NETT) A prospective randomized trial of lung volume reduction surgery. J Thorac Cardiovasc Surg. 1999;118(3):518–528. doi: 10.1016/s0022-5223(99)70191-1. [DOI] [PubMed] [Google Scholar]
- 16.Patients at high risk of death after lung-volume-reduction surgery. The New England Journal of Medicine. 2001;345(15):1075–1083. doi: 10.1056/NEJMoa11798. [DOI] [PubMed] [Google Scholar]
- 17.Fishman A, Martinez F, Naunheim K, Piantadosi S, Wise R, Ries A, Weinmann G, Wood DE. A randomized trial comparing lung-volume-reduction surgery with medical therapy for severe emphysema. The New England Journal of Medicine. 2003;348(21):2059–2073. doi: 10.1056/NEJMoa030287. [DOI] [PubMed] [Google Scholar]
- 18.Celli BR, Cote CG, Marin JM, Casanova C, Montes de Oca M, Mendez RA, Pinto Plata V, Cabral HJ. The body-mass index, airflow obstruction, dyspnea, and exercise capacity index in chronic obstructive pulmonary disease. The New England Journal of Medicine. 2004;350(10):1005–1012. doi: 10.1056/NEJMoa021322. [DOI] [PubMed] [Google Scholar]
- 19.Hersh CP, Demeo DL, Lazarus R, Celedon JC, Raby BA, Benditt JO, Criner G, Make B, Martinez FJ, Scanlon PD, Sciurba FC, Utz JP, Reilly JJ, Silverman EK. Genetic association analysis of functional impairment in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;173(9):977–984. doi: 10.1164/rccm.200509-1452OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hersh CP, Demeo DL, Reilly JJ, Silverman EK. Xenobiotic metabolizing enzyme gene polymorphisms predict response to lung volume reduction surgery. Respir Res. 2007;8(1):59. doi: 10.1186/1465-9921-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Celedon JC, Lange C, Raby BA, Litonjua AA, Palmer LJ, DeMeo DL, Reilly JJ, Kwiatkowski DJ, Chapman HA, Laird N, Sylvia JS, Hernandez M, Speizer FE, Weiss ST, Silverman EK. The transforming growth factor-beta1 (TGFB1) gene is associated with chronic obstructive pulmonary disease (COPD) Human Molecular Genetics. 2004;13(15):1649–1656. doi: 10.1093/hmg/ddh171. [DOI] [PubMed] [Google Scholar]
- 22.A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics (Oxford, England) 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 24.Freidlin B, Zheng G, Li Z, Gastwirth JL. Trend tests for case-control studies of genetic markers: power, sample size and robustness. Human Heredity. 2002;53(3):146–152. doi: 10.1159/000064976. [DOI] [PubMed] [Google Scholar]
- 25.Suissa S. Statistical treatment of exacerbations in therapeutic trials of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;173(8):842–846. doi: 10.1164/rccm.200508-1338PP. [DOI] [PubMed] [Google Scholar]
- 26.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. American Journal of Human Genetics. 2002;70(2):425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan VS, Ramsey SD, Make BJ, Martinez FJ. Physiologic variables and functional status independently predict COPD hospitalizations and emergency department visits in patients with severe COPD. Copd. 2007;4(1):29–39. doi: 10.1080/15412550601169430. [DOI] [PubMed] [Google Scholar]
- 28.Washko GR, Fan VS, Ramsey SD, Mohsenifar Z, Martinez F, Make BJ, Sciurba FC, Criner GJ, Minai O, Decamp MM, Reilly JJ. The effect of lung volume reduction surgery on chronic obstructive pulmonary disease exacerbations. Am J Respir Crit Care Med. 2008;177(2):164–169. doi: 10.1164/rccm.200708-1194OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griese M. Pulmonary surfactant in health and human lung diseases: state of the art. Eur Respir J. 1999;13(6):1455–1476. doi: 10.1183/09031936.99.13614779. [DOI] [PubMed] [Google Scholar]
- 30.Steagall WK, Lin JP, Moss J. The C/A(−18) polymorphism in the surfactant protein B gene influences transcription and protein levels of surfactant protein B. American Journal of Physiology. 2007;292(2):L448–453. doi: 10.1152/ajplung.00307.2006. [DOI] [PubMed] [Google Scholar]
- 31.Guo X, Lin HM, Lin Z, Montano M, Sansores R, Wang G, DiAngelo S, Pardo A, Selman M, Floros J. Surfactant protein gene A, B, and D marker alleles in chronic obstructive pulmonary disease of a Mexican population. Eur Respir J. 2001;18(3):482–490. doi: 10.1183/09031936.01.00043401. [DOI] [PubMed] [Google Scholar]
- 32.Seifart C, Plagens A, Brodje D, Muller B, von Wichert P, Floros J. Surfactant protein B intron 4 variation in German patients with COPD and acute respiratory failure. Disease Markers. 2002;18(3):129–136. doi: 10.1155/2002/194075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong MN, Wei Z, Xu LL, Miller DP, Thompson BT, Christiani DC. Polymorphism in the surfactant protein-B gene, gender, and the risk of direct pulmonary injury and ARDS. Chest. 2004;125(1):203–211. doi: 10.1378/chest.125.1.203. [DOI] [PubMed] [Google Scholar]
- 34.Lin Z, Pearson C, Chinchilli V, Pietschmann SM, Luo J, Pison U, Floros J. Polymorphisms of human SP-A, SP-B, and SP-D genes: association of SP-B Thr131Ile with ARDS. Clinical Genetics. 2000;58(3):181–191. doi: 10.1034/j.1399-0004.2000.580305.x. [DOI] [PubMed] [Google Scholar]
- 35.Quasney MW, Waterer GW, Dahmer MK, Kron GK, Zhang Q, Kessler LA, Wunderink RG. Association between surfactant protein B + 1580 polymorphism and the risk of respiratory failure in adults with community-acquired pneumonia. Critical Care Medicine. 2004;32(5):1115–1119. doi: 10.1097/01.ccm.0000124872.55243.5a. [DOI] [PubMed] [Google Scholar]
- 36.Whitsett JA, Weaver TE. Hydrophobic surfactant proteins in lung function and disease. The New England Journal of Medicine. 2002;347(26):2141–2148. doi: 10.1056/NEJMra022387. [DOI] [PubMed] [Google Scholar]
- 37.van Iwaarden JF, Claassen E, Jeurissen SH, Haagsman HP, Kraal G. Alveolar macrophages, surfactant lipids, and surfactant protein B regulate the induction of immune responses via the airways. American Journal of Respiratory Cell and Molecular Biology. 2001;24(4):452–458. doi: 10.1165/ajrcmb.24.4.4239. [DOI] [PubMed] [Google Scholar]
- 38.Thomas KH, Meyn P, Suttorp N. Single nucleotide polymorphism in 5'-flanking region reduces transcription of surfactant protein B gene in H441 cells. American Journal of Physiology. 2006;291(3):L386–390. doi: 10.1152/ajplung.00193.2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.