Abstract
Understanding the underlying physics of the binding of small molecule ligands to protein active sites is a key objective of computational chemistry and biology. It is widely believed that displacement of water molecules from the active site by the ligand is a principal (if not the dominant) source of binding free energy. Although continuum theories of hydration are routinely used to describe the contributions of the solvent to the binding affinity of the complex, it is still an unsettled question as to whether or not these continuum solvation theories describe the underlying molecular physics with sufficient accuracy to reliably rank the binding affinities of a set of ligands for a given protein. Here we develop a novel, computationally efficient, descriptor of the contribution of the solvent to the binding free energy of a small molecule and its associated receptor that captures the effects of the ligand displacing the solvent from the protein active site with atomic detail. This descriptor quantitatively predicts (R2=0.81) the binding free energy differences between congeneric ligand pairs for the test system factor Xa, elucidates physical properties of the active site solvent that appear to be missing in most continuum theories of hydration, and identifies several features of the hydration of the factor Xa active site relevant to the structure-activity-relationship of its inhibitors.
Introduction
Understanding the underlying physics of the binding of small molecule ligands to protein active sites is a key objective of computational chemistry and biology. While a wide range of techniques exist for calculating binding free energies, ranging from methods that should be accurate in principle (e.g., free energy perturbation theory) to relatively simple approximations based on empirically derived scoring functions, no completely satisfactory and robust approach has yet been developed. Furthermore, physical insight into the sources of binding affinity is, arguably, as important as computing accurate numbers; as such, insight would be extremely valuable in the design of pharmaceutical candidate molecules.
It is widely believed that displacement of water molecules from the active site by the ligand is a principal (if not the dominant) source of binding free energy. Water molecules solvating protein active sites are often entropically unfavorable due to the orientational and positional constraints imposed by the protein surface, or energetically unfavorable due to the water molecule’s inability to form a full complement of hydrogen bonds when solvating the protein surface. This leads to free energy gains when a ligand that is suitably complementary to the active site displaces these waters into bulk solution, thus providing a relatively more favorable environment. Free energy perturbation methods are capable of computing these free energy gains explicitly (within the accuracy of the force field used in the simulations), but are computationally very expensive. Empirical scoring functions require negligible computational effort for a single ligand, but it has proven very difficult to achieve high accuracy and robustness.
“Standard” empirical scoring functions are dominated by lipophilic atom-atom contact terms that reward the close approach of lipophilic atoms of the ligand and protein. Such functions are implicitly attempting to model the free energy gain upon displacement of waters by a given ligand atom, which is presumed to depend upon the hydrophobicity of the protein environment at the location of the ligand atom. Reasonable results can be obtained in a fraction of cases with such an approximation. However, as we have recently pointed out, the simple atom-atom pair term fails to take into account the specific positioning of the hydrophobic groups of the active site.1,2 In particular, regions that exhibit “hydrophobic enclosure”, i.e. are surrounded by hydrophobic protein atoms, provide a much less favorable environment for water molecules than is reflected in additive pair scoring. This argument applies not only to purely hydrophobic cavities, but also to regions in which the ligand must make a small number of hydrogen bonds but otherwise is hydrophobically enclosed by protein groups. A new empirical scoring function, implemented in the Glide docking program as Glide XP1, incorporates these geometrical factors and has been shown to substantially improve the ability of the scoring function to separate active and inactive compounds.
While the Glide XP model represented a significant improvement as compared to previous empirical approaches, it should be possible to achieve a higher level of detail, and numerical precision, by mapping out the thermodynamics of water molecules in the active site, using explicit solvent simulations and appropriate approximations for the thermodynamic functions. In ref. 2, we presented an initial effort in this direction, demonstrating that regions of the active site identified by Glide XP as hydrophobically enclosed dramatically affected the structure and thermodynamic properties of solvating water molecules. In one case, the active site of cyclooxygenase-2 (COX-2), the active site cavity dewetted; in a second, the active site of streptavidin, the solvating water molecules formed an ice-like five membered ring, incurring a large entropic penalty in order to avoid loss of hydrogen bonds. The thermodynamics of the solvating water in these cases was analyzed via inhomogeneous solvation theory, proposed originally by Lazaridis3, which provides an approximate description of the hydration thermodynamics using data from relatively short (~10ns) molecular dynamics simulations.
In the present paper, we continue the line of research described in ref. 2 by applying the inhomogenous solvation theory approach to study ligand binding in factor Xa (fXa), an important drug target in the thrombosis pathway, several inhibitors of which are currently in Phase III clinical trials4. We use a clustering technique to build a map of water occupancy in the fXa active site, and assign chemical potentials to the water sites using the inhomogenous solvation theory discussed above.2 We then construct a semiempirical extension of the model which enables computation of free energy differences (ΔΔG values) for selected pairs of fXa ligands, and compare the success of this approach with the more standard technique, MM-GBSA5,6. The free energy differences calculated from our semiempirical model are shown to correlate exceptionally well with experimental data (R2 = 0.81, reduced to 0.80 after leave-one-out validation) via the use of only three adjustable parameters, and substantially out perform the analogous MM-GBSA calculations (R2 = 0.29). We investigated 31 pairs of ligands using data from only a single 10 ns MD simulation, illustrating the high computational efficiency of our methodology. Furthermore, the solvent chemical potential map produced here appears to elucidate features of the known fXa structure activity relationship (SAR), and would very likely provide a useful starting point for efforts to design novel compounds. An effort to calculate absolute binding free energies for highly diverse ligands displays less accuracy and some overfitting (as would be expected, since the displacement of water molecules is not the only factor determining binding affinity), but still shows a significant correlation with experimental data for this challenging data set.
Results and Discussion
1. Mapping of the thermodynamic properties of the active site solvent
When a ligand binds to a protein, the water solvating the active site is expelled into the bulk fluid. This expulsion of the active site solvent makes enthalpic and entropic contributions to the binding free energy of the complex. The less energetically or entropically favorable the expelled water, the more favorable its contributions to the binding free energy. The active sites of proteins provide very diverse environments for solvating water. Water solvating narrow hydrophobic enclosures such as the COX-2 binding cavity is energetically unfavorable because it cannot form a full complement of hydrogen bonds.2 Similarly, water molecules solvating enclosed protein hydrogen bonding sites are entropically unfavorable since the number of configurations they can adapt while simultaneously forming hydrogen bonds with the protein and their water neighbors is severely reduced.2 The expulsion of water from such enclosed regions has been shown to lead to enhancements in protein binding affinity.1 From these observations we wanted to determine if a computationally derived map of the thermodynamic properties of the active site solvent could be used to rank the binding affinities of congeneric compounds. We hypothesized that the contributions to the binding free energy of adding a complementary chemical group — i.e., chemical groups that make hydrogen bonds where appropriate and hydrophobic contacts otherwise — to a given ligand scaffold could largely be understood by an analysis of the solvent alone.
Testing this hypothesis requires a method to compute the local thermodynamic properties of the fXa active site solvent. We utilized data from 10 ns of explicitly solvated molecular dynamics simulations of fXa to sample the active site solvent distribution for this receptor. We then clustered the active site solvent distribution into high occupancy 1 Å spheres, which we denoted as the “hydration sites” of the active site cavity. Using inhomogeneous solvation theory, we then computed the average system interaction energy and excess entropy terms for the water in each hydration site. Comparing the system interaction energy of the hydration sites with the bulk reference value allowed us to estimate the enthalpic cost of transferring the water in the hydration site from the active site to the bulk fluid. The excess entropy calculated here can be used similarly. We also computed several other descriptors of the hydration site’s local environment. More details of these procedures and measurements are given in the Methods section. The data for each hydration site is presented in Table 1. Fig. 1 shows the calculated energies and excess entropies for each of the hydration sites in the fXa binding cavity. Relative to other hydration sites, the hydration sites circled in gray had poor system interaction energies, the hydration sites circled in green had unfavorable excess entropies, and the hydration sites circled in purple had both relatively poor system interaction energies and entropies. We show the resulting 3-dimensional active site hydration map with this same color coding in Fig. 2.
Table 1.
Calculated thermodynamic and local water structure data for each of the 43 hydration sites we identified by clustering the factor Xa active site solvent density distribution. The occupancy was the number of water oxygen atoms found occupying a given hydration site during the 10ns of molecular dynamics simulation, −TSe is the excess entropic contribution to the free energy calculated from a truncated expansion of the excess entropy in terms of correlations in the single particle translational and rotational density, E is average energy of interaction of the water molecules in a given hydration site with the rest of the system, the #nbrs value is the average number of neighboring waters found within a 3.5Å oxygen atom to oxygen atom distance from a water occupying the specified hydrations site, the #HBnbrs value is the is the average number of neighboring water oxygens found within a 3.5Å distance from the water oxygen occupying the specified hydrations site that make a less than 30° oxygen-oxygen-hydrogen hydrogen bonding angle with this water, the %HB value is the #HBnbrs/#nbrs fraction, and the exposure value is the #nbrs value divided by the bulk #nbrs value found in the bulk fluid.
| Hyd. Site | Occupancy | −TSe (kcal/mol) | E (kcal/mol) | #nbrs | #HBnbrs | % HB | Exposure |
|---|---|---|---|---|---|---|---|
| Neat | 1385.00 | N/A* | −19.67 | 5.09 | 3.53 | 0.69 | 1.00 |
| 1.00 | 9347.00 | 4.00 | −20.34 | 1.54 | 1.30 | 0.84 | 0.30 |
| 2.00 | 9062.00 | 3.91 | −22.59 | 3.13 | 1.99 | 0.64 | 0.61 |
| 3.00 | 8425.00 | 2.61 | −20.85 | 3.45 | 2.27 | 0.66 | 0.68 |
| 4.00 | 8383.00 | 2.93 | −19.55 | 3.12 | 2.79 | 0.89 | 0.61 |
| 5.00 | 8157.00 | 3.24 | −23.18 | 2.52 | 1.88 | 0.75 | 0.50 |
| 6.00 | 8123.00 | 3.20 | −21.86 | 3.62 | 2.24 | 0.62 | 0.71 |
| 7.00 | 8116.00 | 3.37 | −21.82 | 3.22 | 2.12 | 0.66 | 0.63 |
| 8.00 | 8081.00 | 2.74 | −22.73 | 3.05 | 2.39 | 0.78 | 0.60 |
| 9.00 | 7257.00 | 2.13 | −19.38 | 4.30 | 2.76 | 0.64 | 0.84 |
| 10.00 | 7172.00 | 2.52 | −21.04 | 3.75 | 2.85 | 0.76 | 0.74 |
| 11.00 | 6886.00 | 2.05 | −20.71 | 3.41 | 2.24 | 0.66 | 0.67 |
| 12.00 | 6815.00 | 2.28 | −16.93 | 1.62 | 1.49 | 0.92 | 0.32 |
| 13.00 | 6238.00 | 1.72 | −17.88 | 2.72 | 2.05 | 0.75 | 0.53 |
| 14.00 | 6081.00 | 1.95 | −19.89 | 2.58 | 2.11 | 0.82 | 0.51 |
| 15.00 | 5441.00 | 1.83 | −22.62 | 4.66 | 3.63 | 0.78 | 0.92 |
| 16.00 | 5078.00 | 1.51 | −20.01 | 3.30 | 2.56 | 0.78 | 0.65 |
| 17.00 | 4919.00 | 1.33 | −17.04 | 2.45 | 1.78 | 0.73 | 0.48 |
| 18.00 | 4887.00 | 1.35 | −17.74 | 3.38 | 2.46 | 0.73 | 0.66 |
| 19.00 | 4466.00 | 1.20 | −19.48 | 4.11 | 2.77 | 0.67 | 0.81 |
| 20.00 | 4386 .00 | 1.37 | −22.14 | 3.69 | 2.79 | 0.76 | 0.72 |
| 21.00 | 4356.00 | 1.23 | −18.50 | 3.75 | 2.67 | 0.71 | 0.74 |
| 22.00 | 4241.00 | 1.22 | −20.27 | 3.72 | 2.63 | 0.71 | 0.73 |
| 23.00 | 4189.00 | 1.13 | −19.58 | 3.87 | 2.84 | 0.73 | 0.76 |
| 24.00 | 4170.00 | 1.17 | −19.64 | 3.69 | 2.51 | 0.68 | 0.72 |
| 25.00 | 4137.00 | 1.12 | −20.85 | 4.61 | 2.59 | 0.56 | 0.91 |
| 26.00 | 4067.00 | 1.07 | −20.19 | 4.23 | 3.09 | 0.73 | 0.83 |
| 27.00 | 4046.00 | 1.03 | −20.72 | 4.37 | 3.48 | 0.80 | 0.86 |
| 28.00 | 3921.00 | 1.10 | −16.74 | 2.66 | 2.00 | 0.75 | 0.52 |
| 29.00 | 3833.00 | 1.03 | −21.44 | 4.27 | 2.57 | 0.60 | 0.84 |
| 30.00 | 3793.00 | 1.04 | −21.97 | 4.05 | 2.68 | 0.66 | 0.80 |
| 31.00 | 3786.00 | 0.99 | −20.00 | 4.70 | 3.39 | 0.72 | 0.92 |
| 32.00 | 3686.00 | 0.99 | −22.61 | 4.48 | 2.69 | 0.60 | 0.88 |
| 33.00 | 3618.00 | 1.00 | −20.46 | 4.34 | 2.56 | 0.59 | 0.85 |
| 34.00 | 3570.00 | 0.95 | −19.75 | 4.36 | 2.92 | 0.67 | 0.86 |
| 35.00 | 3312.00 | 0.90 | −24.24 | 4.41 | 2.74 | 0.62 | 0.87 |
| 36.00 | 3296.00 | 0.84 | −19.66 | 4.06 | 2.66 | 0.66 | 0.80 |
| 37.00 | 3152.00 | 0.79 | −18.87 | 4.57 | 3.15 | 0.69 | 0.90 |
| 38.00 | 3094.00 | 0.73 | −19.09 | 4.70 | 3.25 | 0.69 | 0.92 |
| 39.00 | 3089.00 | 0.92 | −21.61 | 3.55 | 2.55 | 0.72 | 0.70 |
| 40.00 | 3007.00 | 0.79 | −19.96 | 4.20 | 2.79 | 0.67 | 0.82 |
| 41.00 | 3003.00 | 0.78 | −20.41 | 3.71 | 2.70 | 0.73 | 0.73 |
| 42.00 | 2862.00 | 0.73 | −19.26 | 4.72 | 3.28 | 0.69 | 0.93 |
| 43.00 | 2791.00 | 0.75 | −20.93 | 3.98 | 2.84 | 0.71 | 0.78 |
The truncated expansion of the excess entropy used included only the first order terms. The first order excess entropic term for all neat fluids is strictly zero, however the second order and larger terms will be quite large.
Figure 1.
System interaction energies (E) and the excess entropic contribution to the free energy (−TSe) of water molecules in the principal hydration sites of the factor Xa active site. The system interaction energy is the average energy of interaction of the water molecules in a given hydration site with the rest of the system and the excess entropic contribution to the free energy is calculated from a truncated expansion of the excess entropy in terms of correlation functions. Those hydration sites that were expected to make large energetic contributions when evacuated by the ligand are circled in gray, those expected to make large entropic contributions are circled in green, and those expected to make both entropic and enthalpic contributions are circled in purple.
Figure 2.
Those hydration sites expected to contribute favorably to binding when evacuated by the ligand are here shown within the factor Xa active site in wire frame. Those expected to contribute energetically are shown in gray, those expected to contribute entropically are shown in green, and those expected to contribute energetically and entropically are shown in purple. The S1 and S4 pockets are labeled in yellow, as are several hydration sites discussed in the text.
The hydration site map depicted in Fig. 2 elucidated several features of the experimentally known SAR of the fXa ligands. Factor Xa inhibitors generally bind in an L shaped conformation, where one group of the ligand occupies the anionic S1 pocket lined by residues Asp189, Ser195, and Tyr228 and another group of the ligand occupies the aromatic S4 pocket lined by residues Tyr99, Phe174, and Trp215. Typically a fairly rigid linker group will bridge these two interaction sites. The solvent analysis identified three enthalpically unfavorable hydration sites, sites 13, 18, and 21, solvating the fXa S4 pocket. This finding agreed with the experimental result that the S4 pocket has an exceptionally high affinity for hydrophobic groups.7,8 We also identified a single very high excess chemical potential hydration site, site 12, solvating Tyr228 in the S1 pocket. Several studies have found that introducing a ligand chlorine atom at this location, and hence displacing the water from this site, makes a large favorable contribution to the binding affinity.9-12 Additionally, we identified an energetically depleted hydration site, site 17, solvating the disulfide bridge between Cys191 and Cys220. We expect displacement of water from this site would make favorable contributions to the binding free energy. This agrees with several reported chemical series targeting this site.13-15
We compared this hydration map with the locations of active site crystallographic waters from the fXa apo-structure, crystal structure 1HCG.16 Of the 11 crystallographic waters that resolve within the fXa active site, 9 of these crystallographic waters are within 1.5 Å of a hydration site, and all of the crystallographic waters are within 2.5 Å of a hydration site. One difficulty in the comparison is that we identified in the active site many more hydration sites than crystallographically resolved waters. However, this discrepancy is expected since the 1HCG crystal structure was only solved to a resolution of 2.2 Å, and it has been noted that the number of crystallographic water molecules identified in X-ray crystallography of proteins is quite sensitive to resolution (an average of 1.0 crystal waters per protein residue is expected at a resolution of 2 Å, but an average of 1.6-1.7 crystal waters per residue is expected at a resolution of 1 Å).17 The number and location of crystallographic waters identified in X-ray crystallography of proteins has also been found to be sensitive to temperature, pH, solvent conditions, and the crystal packing configuration.18,19 Given these sources of noise, we found our agreement was satisfactory and in line with other similar comparisons of the solvent distributions obtained from molecular dynamics simulations with those obtained from X-ray crystallography.20
To better quantify the visual correlation of the known SAR of fXa binding compounds and thermodynamic properties of the hydration sites, we constructed a simple 5-parameter scoring function based upon the hydration map of the fXa active site that attempts to rank the relative binding affinities of congeneric fXa ligands. This scoring function was based on the following physical principles: (1) if a heavy atom of a ligand overlapped with a hydration site, it displaced the water from that site; and (2) the less energetically or entropically favorable the expelled water, the more favorable its contributions to the binding free energy. A hydration site would contribute to the binding free energy if its excess entropy or system interaction energy were beyond the fitted entropy and energy cutoff parameters Sco and Eco , respectively. A flat reward was given for any hydration site that had excess entropies or system interaction energies that were beyond these values. The amplitude of the reward values, Srwd and Erwd, were fit accordingly. A fit cutoff distance (Rco) was used to determine whether a heavy atom of the ligand displaced water from a hydration site. If the ligand heavy atom had the same position as the hydration site, the full value of Srwd and Erwd would be awarded. The reward was then linearly reduced to zero over the distance Rco. This scoring function was implemented as
| (1) |
where ΔGbind was the predicted binding free energy of the ligand, Ehs is the system interaction energy of a hydration site, Sehs is the excess entropy of a hydration site, and Θ is the Heaviside step function. We will refer to this implementation as the “displaced-solvent functional”. Implementing this displaced-solvent functional was particularly simple since it is merely a sum over the ligand heavy atoms and a restricted sum over the entropically structured and energetically depleted hydration sites, with a linear function of the hydration-site-ligand-atom approach distance as its argument. Note that some hydration sites contributed in both the entropic and energetic sums. We also constructed a 3-parameter scoring function based on the same principles as the 5-parameter scoring function, where the value of Rco was set to 2.8 Å and the values of Srwd and Erwd were forced to be equal; and an “ab initio” parameter free form of the scoring function where contributions from all of the hydration sites were included, the Srwd and Erwd values were taken to be Srwd=Sehs and Erwd= Ebulk-Ehs, and an approximate value Rco=2.24 Å was deduced from physical arguments (see Methods). One minor technical point was that in the ab intio form of the scoring function, the maximum contribution from any given hydration was capped to never exceed ΔGhs=(Ebulk−Ehs)−TSe, ie the total computed transfer free energy of a hydration site into the bulk fluid. For the 3-parameter and 5-parameter functionals, we determined the optimal values of parameters Rco, Eco, Erwd, Sco, and Srwd by fitting to the binding thermodynamics of a set of 31 congeneric ligand pairs and a set of 28 ligands found in crystal structures (see Methods).
It is important to note that this functional was not intended to compute the absolute binding affinity of a given ligand and receptor. Computing absolute binding affinities would require terms that describe the loss of entropy of binding the ligand, the strength of the interaction energy between the ligand and the protein, and the reorganization free energy of the protein in addition to the contributions of solvent expulsion described here. However, for congeneric ligands that differ by only small chemical modifications, these additional contributions are likely quite small (given that those modifications are complementary to the protein surface). The ability of the proposed form of the scoring function to describe free energy differences between such congeneric ligand pairs tests if the thermodynamic consequences to the binding free energy of these small modifications can be largely understood from only the properties of the excluded solvent.
2. Development and testing of the displaced-solvent functional on the set of the congeneric inhibitor pairs
We prepared a dataset of 31 congeneric inhibitor pairs of fXa (see Methods) (Table 2). These 31 congeneric inhibitor pairs were pairs of fXa ligands that differed by at most three chemical groups. We expected that excluded solvent density effects would dominate this dataset since the other terms — the protein reorganization free energy, ligand conformational entropy, etc. — would be largely a consequence of the ligand scaffold shared by both members of the pair. We optimized the parameters of the displaced-solvent functionals to reproduce the experimentally measured differences in binding affinity between each of these congeneric ligand pairs. We also estimated the error of the resulting functionals with leave-one-out cross validation. The resulting values of the parameters can be found in the supplementary table 1 and plots of the predicted differences in binding free energy versus the experimental values are shown in Figs. 3 , 4, and Sup. Fig. 1. The agreement of the predictions of the functionals with the experimental data was quite striking: the Pearson correlation coefficient (R2) was 0.81 for both the 5-parameter and 3-parameter functionals, and 0.63 for the ab initio functional. Under leave-one-out cross-validation, the R2 values of the 5-parameter and 3-parameter functionals only degraded to 0.75 and 0.80, respectively. From the good numerical agreement observed over the 6 kcal/mol free energy range of modifications plotted in Figs. 3, 4, and Sup. Fig 1 we found this technique well differentiated modifications that make large contributions to the binding affinity from modifications that make only small modifications to the binding affinity for this fXa test system. The excellent predictive ability of the displaced solvent functional on this series confirms that the effect on the binding free energy of small complementary chemical modifications to existing leads can largely be understood by an analysis of the molecular properties of the solvent alone.
Table 2.
Inhibition data for the congeneric ligand pairs binding to factor Xa. Our predicted activity differences from the trained 3-parameter and 5-parameter displaced-solvent functionals and MM-GBSA method. When a ligand was taken from a solved crystal structure, the ligand was designated “(pdb id):(ligand residue name)”; and when the ligand was built from congeneric series data, the ligand was designated “(first author of the reporting publication):(molecule number in the reporting publication)”.
| Initial Ligand | Final Ligand | ΔΔGexp (kcal/mol) |
ΔΔG3p (kcal/mol) |
ΔΔG5p (kcal/mol) |
ΔΔGab initio (kcal/mol) |
ΔΔGMM-GBSA (kcal/mol) |
Ref. |
|---|---|---|---|---|---|---|---|
| 1MQ5:XLC | 1MQ6:XLD | −2.94 | −2.85 | −2.54 | −2.97 | −4.22 | Adler02 |
| 1NFU:RRP | 1NFY:RTR | −1.56 | −2.56 | −2.98 | −3.23 | −0.47 | Maignan03 |
| 1NFX:RDR | 1NFW:RRR | −0.59 | 1.35 | 0.94 | 0.21 | 2.01 | Maignan03 |
| Matter:25 | Matter:28 | −0.62 | −0.61 | −0.62 | −2.17 | −0.48 | Matter05 |
| Matter:25 | 2BMG:I1H | −1.05 | −1.31 | −1.31 | −3.52 | −3.8 | Matter05 |
| Matter:28 | 2BMG:I1H | −0.43 | −0.70 | −0.69 | −1.35 | −3.32 | Matter05 |
| Mueller:3 | Mueller:2 | −0.90 | −2.05 | −2.34 | −4.53 | −8.35 | Mueller02 |
| Haginoya:56 | Haginoya:57 | −0.59 | −1.15 | −1.12 | −0.23 | 1.41 | Haginoya04 |
| Haginoya:60 | Haginoya:56 | −0.19 | 0.00 | 0.00 | 0.08 | 0.81 | Haginoya04 |
| Haginoya:56 | 1V3X:D76 | −0.54 | −1.15 | −1.12 | −0.31 | −5.04 | Haginoya04 |
| Haginoya:60 | Haginoya:57 | −0.79 | −1.15 | −1.12 | −0.15 | 2.22 | Haginoya04 |
| 1V3X:D76 | Haginoya:57 | −0.05 | 0.00 | 0.00 | 0.08 | 6.45 | Haginoya04 |
| Haginoya:60 | 1V3X:D76 | −0.74 | −1.15 | −1.12 | −0.23 | −4.23 | Haginoya04 |
| 2BQ7:IID | 2BQW:IIE | −2.01 | −1.73 | −1.95 | −5.42 | −8.81 | Nazare05 |
| 2BQ7:IID | 2BOH:IIA | −2.01 | −1.80 | −2.09 | −1.98 | −6.7 | Nazare05 |
| 2BQW:IIE | 2BOH:IIA | 0.00 | −0.07 | −0.13 | 3.44 | 2.11 | Nazare05 |
| Quan:11a | Quan:43 | −0.09 | 0.04 | 0.39 | 0.27 | 0.3 | Quan05 |
| Quan:43 | 1Z6E:IK8 | −0.68 | −0.04 | −0.45 | −0.49 | −3.47 | Quan05 |
| Quan:11a | 1Z6E:IK8 | −0.77 | 0.00 | −0.06 | −0.22 | −3.17 | Quan05 |
| 1G2L:T87 | 1G2M:R11 | −0.21 | 0.79 | 0.20 | −0.32 | 10.1 | Nar01 |
| Guertin:5c | 1KSN:FXV | −0.13 | −0.87 | −0.78 | −0.68 | −4.42 | Guertin02 |
| 2FZZ:4QC | 2G00:5QC | −1.06 | −1.70 | −1.68 | 0.06 | 0.31 | Pinto06 |
| Matter:107 | 1LQD:CMI | −3.93 | −2.52 | −2.48 | −2.84 | −15.71 | Matter02 |
| Matter:108 | Matter:46 | −3.09 | −2.52 | −2.48 | −2.78 | −11.49 | Matter02 |
| 1F0R:815 | 1F0S:PR2 | −0.12 | 0.09 | 0.14 | −1.06 | −3.53 | Maignan00 |
| Young:33 | 2J4I:GSJ | −0.82 | 0.00 | 0.00 | 0.36 | −7.53 | Young06 |
| Young:32 | 2J4I:GSJ | −4.93 | −4.87 | −4.83 | −5.31 | −15.25 | Young06 |
| Young:38 | 2J4I:GSJ | −6.26 | −4.87 | −4.83 | −5.67 | −7.27 | Young06 |
| Young:32 | Young:33 | −4.11 | −4.87 | −4.83 | −5.67 | −7.72 | Young06 |
| Young:38 | Young:33 | −5.44 | −4.87 | −4.83 | −6.03 | 0.26 | Young06 |
| Young:38 | Young:32 | −1.33 | 0.00 | 0.00 | −0.36 | 7.98 | Young06 |
Figure 3.
Computed relative activities using the 5-parameter form of eq. 1 versus experimental relative activities of the 31 congeneric inhibitor pairs with factor Xa. Note the stability of this fit under leave-one-out cross validation.
Figure 4.
Computed relative activities using the 3-parameter form of eq. 1 versus experimental relative activities of the 31 congeneric inhibitor pairs with factor Xa. Note the stability of this fit under leave-one-out cross validation.
Despite the effectiveness of the displaced-solvent functional describing the binding thermodynamics of this set, it is difficult to judge the success of the method in describing novel solvation physics without direct comparison to the results of more commonly used continuum theories of solvation. Toward this end, we performed MM-GBSA calculations for each of the congeneric ligand pairs (see Methods). The agreement of the MM-GBSA calculations with the experimental data was fair (R2=0.29), but substantially worse than the results obtained by the displaced-solvent functional. The plot of the data in Fig. 5 shows that although the MM-GBSA results did correlate with the experimentally measured binding affinities, the binding thermodynamics of several of the congeneric pairs was poorly described by the MM-GBSA methodology. These results suggest that the set of congeneric pairs is a challenging test set for state-of-the-art continuum methodologies, and that the displaced-solvent functional captures molecular length scale solvation physics relevant to the binding thermodynamics of these compounds that may be missing from other continuum and electrostatic theories of solvation.
Figure 5.
Computed relative activities using the MM-GBSA methodology versus experimental relative activities of the 31 congeneric inhibitor pairs with factor Xa.
3. Characterization of the contributions of the evacuated hydration site as predicted by the functionals with direct comparison with experiment for selected congeneric pairs
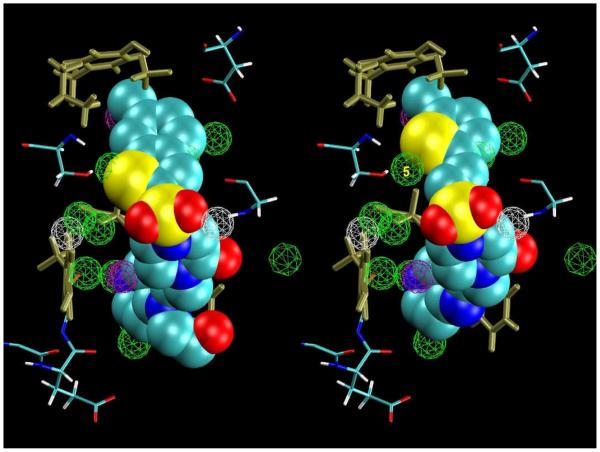
Young:38-2J4I:GSJ
Congeneric ligands Young:38 and 2J4I:GSJ, depicted in Fig. 6, were representative of the types of modifications we correctly predicted would contribute most strongly to the binding affinity. These ligands differ in that GSJ has an additional isopropyl group located in the S4 pocket. This isopropyl group fills a portion of the S4 pocket that is lined by the side chains of residues Tyr99, Phe174, and Trp215 and, in the absence of the ligand, is principally solvated by hydration sites 13, 18, and 21. Hydration site 13 is in close contact (<4.5 Å) with each of these three aromatic side chains and has a very low exposure parameter of 0.53. Water molecules in this hydration site cannot form hydrogen bonds with the hydrophobic protein and maintain only an average of 2.05 water-water hydrogen bonds, which leads to relatively unfavorable system interaction energies. The hydrogen bonds that it does form are mainly donated by hydration sites 18 and 21 and very rarely by hydration site 1. The orientational and translational restrictions necessary to maintain this hydrogen bonding profile result in relatively unfavorable excess entropies for water at this hydration site. The hydrophobic enclosure for hydration sites 18 and 21 is not as tight (exposure parameters of 0.66 and 0.74, respectively); however, the environment is otherwise qualitatively similar. Both these hydration sites have above average system interaction energies due to the hydrophobic bulk of the protein enclosing them, and hydration site 18 was also identified by our empirical criteria to be entropically unfavorable, although it was a borderline case. GSJ’s additional isopropyl group expels water from all three of the above described hydration sites: hydration sites 13 and 18 were predicted by the optimized displaced-solvent functionals to make both energetic and entropic contributions to binding, and hydration site 21 was predicted to make only energetic contributions.
Figure 6.
Ligand Young:38 (left) and ligand 2J4I:GSJ (right) in the factor Xa active site. The hydration sites that receive an energetic score in equation 1 are depicted in gray wireframe, the hydration sites that receive an entropic score are depicted in green wireframe, and the hydration sites that receive both energetic and entropic scores are depicted in purple wireframe. Several hydration sites discussed in the text are labeled in yellow. The experimentally measured affinity difference between these two compounds is ΔΔGexp=−6.26kcal/mol. The optimized 3- and 5-parameter functionals predicted ΔΔG3p=−4.87 and ΔΔG5p=−4.83 respectively. The isopropyl group of ligand 2J4I:GSJ displaces three energetically depleted hydration sites, two of which are predicted to also be entropically structured, which resulted in a large predicted contribution to the binding affinity of the complex.
The experimentally measured affinity difference between these two compounds is ΔΔGexp = −6.26 kcal/mol. The optimized 3-parameter, 5-parameter, and ab initio functionals predicted ΔΔG3p = −4.87, ΔΔG5p = −4.83, and ΔΔGab initio = −5.67 respectively. This agreed with the experimental finding that adding an isopropyl group to ligand Young:38 at this location makes a large and favorable contribution to the binding free energy. The MM-GBSA ΔΔG for this pair of ligands is predicted to be −7.27 kcal/mol, which also agrees well with ΔΔGexp. The congeneric ligands Young:32/Young:33 (ΔΔGexp = −4.11 kcal/mol) has precisely the same hydrogen/isopropyl substitution as the Young:38/2J4I:GSJ pair, and therefore the same values for ΔΔG3p and ΔΔG5p of −4.87 and −4.83 kcal/mol, respectively, which matches very well with ΔΔGexp. However, for this pair of ligands, the MM-GBSA predicted ΔΔG is −7.72 kcal/mol, which is more negative than the experimental value by 3.61 kcal/mol. Visual inspection of the MM-GBSA structure does not reveal the origin of this discrepancy.
1MQ5:XLC-1MQ6:XLD
The congeneric ligands 1MQ5:XLC and 1MQ6:XLD are depicted in Fig. 7. This pair has a more subtle modification of the group binding the S4 pocket than the Young:38-2J4I:GSJ congeneric pair described above. For this pair, the S4 binding group found in ligand 1MQ6:XLD overlapped with hydration sites 13 and 20 whereas the S4 binding group of ligand 1MQ5:XLC did not. As noted above, expulsion of water from hydration site 13 is expected to make both favorable energetic and entropic contributions to binding. Water in hydration site 20 has favorable energetic interactions due to several well formed hydrogen bonds: water molecules occupying this the site predominately donate a hydrogen bond to the backbone carbonyl group of Glu97, nearly always receive a hydrogen bond from hydration site 4, and have good hydrogen bonding interactions with hydration site 35. Hydration site 20 though also incurred unfavorable contributions to its excess entropy due to the structuring required to maintain these favorable interactions. When displaced by the S4 binding group of ligand 1MQ6:XLD, an electropositive carbon (the carbon is bound to an oxygen) comes into close contact with the backbone carbonyl group of Glu97. This electropositive carbon likely recaptures much of the interaction energy between the protein carbonyl group and the water in hydration site 20 without the associated entropic cost. From these water thermodynamics considerations, the optimized 3-parameter, 5-parameter, and ab initio displaced-solvent functionals predict affinity differences of ΔΔG3p = −2.85, ΔΔG5p = −2.54, and ΔΔGab initio = −2.97 kcal/mol respectively. The experimental difference binding affinity between the two ligands is ΔΔGexp = −2.94 kcal/mol. The MM-GBSA predicted ΔΔG for this pair of ligands is −4.22 kcal/mol.
Figure 7.
Ligand 1MQ5:XLC (left) and ligand 1MQ6:XLD (right) in the factor Xa active site. The hydration sites that receive an energetic score in equation 1 are depicted in gray wireframe, the hydration sites that receive an entropic score are depicted in green wireframe, and the hydration sites that receive both energetic and entropic scores are depicted in purple wireframe. Several hydration sites discussed in the text are labeled in yellow. The experimentally measured affinity difference between these two compounds is ΔΔGexp=−2.94kcal/mol. The optimized 3- and 5-parameter functionals predicted ΔΔG3p=−2.85 and ΔΔG5p=−2.54 respectively. Unlike the S4 group of ligand 1MQ5:XLC, the S4 pocket group of ligand 1MQ6:XLD displaced the energetically depleted and entropically structured hydration site 13 and partially displaced entropically structured hydration sites 20, which resulted in a large solvent related contribution to the binding affinity quantitatively predicted by our theory.
2BQ7:IID-2BQW:IIE
The congeneric ligands 2BQ7:IID and 2BQW:IIE are depicted in Fig. 8. This congeneric pair isolates the contribution of inserting a ligand chlorine atom into the region of the S1 pocket lined by the side chains of residues Ala190, Val213, and Tyr228. The chlorine atom on 2BQW:IIE displaces water from hydration site 12, which is tightly enclosed by the side chains of residues Ala190, Val213, and Tyr228. The exposure parameter of this hydration site is only 0.32. This extremely tight enclosure by hydrophobic groups caused the system interaction energy of water in this hydration site to be several kcal/mol less favorable than in the neat fluid. Water molecules in this site maintained hydrogen bonds with its few water neighbors 92% of the simulation time, which made unfavorable contributions to its excess entropy. The location of this hydration site coincided with the location of a structurally conserved water molecule that several studies have shown is favorable to displace.10,11 Several studies have suggested the free energy contribution of expelling this structurally conserved water should be close to the theoretical maximum of 2.0 kcal/mol derived by Dunitz from the thermodynamics of inorganic hydrates.10,12,21 The Dunitz upper bound, however, is inappropriate here since it only includes entropic contributions. Since water in this region suffers from both poor energetic interactions as well as entropic penalties due to structuring, the contribution to the binding free energy from displacing this water molecule may be much greater. The experimentally measured affinity difference between these two compounds is ΔΔGexp = −2.01 kcal/mol, whereas the optimized 3-parameter, 5-parameter, and ab initio functionals predicted ΔΔG3p = −1.73, ΔΔG5p = −1.95, and ΔΔGab initio = −5.42 respectively. In contrast, the MM-GBSA predicted ΔΔG is −8.81 kcal/mol.
Figure 8.
Ligand 2BQ7:IID (left) and ligand 2BQW:IIE (right) in the factor Xa active site. The hydration sites that receive an energetic score in equation 1 are depicted in gray wireframe, the hydration sites that receive an entropic score are depicted in green wireframe, and the hydration sites that receive both energetic and entropic scores are depicted in purple wireframe. Several hydration sites discussed in the text are labeled in yellow. The experimentally measured affinity difference between these two compounds is ΔΔGexp=−2.01 kcal/mol. The optimized 3- and 5-parameter functionals predicted ΔΔG3p = −1.73 and ΔΔG5p = −1.95 respectively. Unlike the S1 group of ligand 2BQ7:IID, the S1 pocket group of ligand 2BQW:IIE displaces the energetically depleted and entropically structured hydration site 12 found within the S1 subgroove. The contribution to the binding affinity predicted by the 3-parameter and 5-parameter displaced-solvent functionals agreed with experiment.
1V3X:D76-Haginoya:57
The congeneric ligands 1V3X:D76 and Haginoya:57 are depicted in Fig. 9. Ligand Haginoya:57 has an additional amide group which is oriented away from the protein in the linker region of the complex. The displaced-solvent functionals correctly predicted that the addition of this group has a marginal contribution to the binding affinity. This is because the amide group does not displace water from any contributing hydration site. It is interesting to note that the size of this added group is approximately equal to that of the isopropyl group added in pair Young:38-2J4I:GSJ. This underscored that the displaced-solvent functional evaluated a weighted shape complementarity — i.e., it rewarded the introduction of complementary groups where predicted to make large contributions from the solvent properties and does not reward shape complementarity away from these regions. The experimentally measured affinity difference between these two compounds is ΔΔGexp = −0.05 kcal/mol. The optimized 3-parameter, 5-parameter, and ab initio functionals all predict no significant affinity difference between the two compounds, consistent with the experimental ΔΔG. In contrast, MM-GBSA predicts a ΔΔG of +6.45 kcal/mol, and therefore appears to be over-predicting the contribution of the amide group to the binding of ligand 1V3X:D76.
Figure 9.
Ligand 1V3X:D76 (left) and ligand Haginoya:57 (right) in the factor Xa active site. The hydration sites that receive an energetic score in equation 1 are depicted in gray wireframe, the hydration sites that receive an entropic score are depicted in green wireframe, and the hydration sites that receive both energetic and entropic scores are depicted in purple wireframe. Several hydration sites discussed in the text are labeled in yellow. The experimentally measured affinity difference between these two compounds is ΔΔGexp=−0.05kcal/mol. The optimized 3- and 5-parameter functionals predicted ΔΔG3p=0.0 and ΔΔG5p=0.0 respectively. The addition of the amide group to ligand D76 contribute negligibly to the binding affinity of the complex, which the method predicted from the the location of the amide group away from any structured or energetically depleted hydration sites.
1NFX:RDR-1NFW:RRR
Congeneric ligands 1NFX:RDR and 1NFW:RRR are depicted in Fig. 10. These ligands differ by a substantial modification to the ring that binds the S1 pocket. They also differ by the removal of an ethanol group that is distant from any contributing hydration sites. The S1 binding group of ligand 1NFX:RDR has a sulfur atom in close contact with Ser195. This sulfur atom displaces water from hydration site 5 whereas ligand 1NFW:RRR does not displace water from this site. Water molecules in this hydration site have favorable interactions with the protein and the surrounding waters but are entropically structured. The structuring and corresponding entropic penalties come from the large degree of enclosure (exposure parameter of 0.5) in combination with the energetic demands of maintaining favorable hydrogen bonding interactions with the protein and surrounding water; most notably a persistent hydrogen bond is donated from Ser195 to the water molecules in this site. The displacement of water leads the optimized 3-parameter, 5-parameter, and ab initio functionals to predict ΔΔG3p = +1.94, ΔΔG5p = +1.53, and ΔΔGab initio = +0.21 respectively. However, the experimentally measured difference in binding affinities is ΔGexp = −0.59 kcal/mol. We believe the scoring function preformed poorly for this inhibitor pair because the sulfur atom in the benzothiophene group of ligand 1NFX:RDR and Ser195 breaks our underlying assumption that the added chemical groups must be complementary to the protein surface. Thus, though the displacement of water from hydration site 5 should contribute favorably to the binding free energy, it is more than offset by the loss of hydrogen bonding energy between the water and Ser195. This resulted in the displaced-solvent functional predicting 1NFX:RDR would be the tighter binding ligand, in disagreement with the experimental data. Interestingly, MM-GBSA also over-predicts the stability of ligand 1NFX:RDR relative to ligand 1NFW:RRR; the MM-GBSA ΔΔG is +2.01 kcal/mol. The minimized MM-GBSA complex associated with ligand 1NFX:RDR incorrectly produces a strong hydrogen bond between the Ser195 side chain and the sulfur atom in the benzothiophene group of the ligand, which is the result of erroneous conformational change in the side chain of Ser195.
Figure 10.
Ligand 1NFX:RDR (left) and ligand 1NFU:RRR (right) in the factor Xa active site. The hydration sites that receive an energetic score in equation 1 are depicted in gray wireframe, the hydration sites that receive an entropic score are depicted in green wireframe, and the hydration sites that receive both energetic and entropic scores are depicted in purple wireframe. Several hydration sites discussed in the text are labeled in yellow. The experimentally measured affinity difference between these two compounds is ΔΔGexp=−0.59kcal/mol. The optimized 3- and 5-parameter functionals predicted ΔΔG3p=+1.94 and ΔΔG5p=+1.53 respectively. The poor agreement of the theory with experiment here is due to the poor interaction energy of the S1 pocket sulfur atom of 1NFX:RDR with Ser195 compared with hydration 5, which is not displaced when ligand 1NFU:RRR docks with the receptor.
4. Development and testing of the displaced-solvent functional on the set of 28 factor Xa crystal structure ligands
In addition to the set of 31 congeneric pairs, we prepared a dataset of 28 inhibitors taken from solved fXa crystal structures (see Methods) (Table 3). These fXa ligands belonged to many different congeneric series, and typically did not share a common chemical scaffold with each other. In the previous section we hypothesized that the contributions to the free energy of binding from changes in conformational entropy, protein-ligand interaction energy, and protein reorganization free energy would be similar for ligand pairs that shared a common chemical scaffold. If this was the case, we posited that the differences in the binding free energies of congeneric pairs could be understood mainly by an analysis of the displaced solvent alone. The success of the displaced-solvent functionals outlined in the previous section supports the validity of this hypothesis. However, for ligand pairs that do not share a common scaffold, we would expect that differences in these contributions would not be small and that predictions based solely on an analysis of the solvent would be less successful. Despite this concern, since the functional performed well over the set of congeneric pair, we were interested in determining how much of the binding affinities of these ligands could be understood from only the contributions described by the displaced-solvent functional, as measured by the RMSD, absolute average error, and R2 values. To study this question, we optimized the 3- and 5-parameter displaced-solvent functionals to reproduce the experimentally measured differences in binding affinities between 378 unique ligand pairs (all combinations) of this 28 ligand set, and we performed leave-one-out (LOO) cross validation to better estimate the error of the functionals. The optimal values of the parameters can be found in supplementary table 2 and the agreement of the fit functionals and the ab initio functional with the experimental data can be found in Figs. 11, 12 and Sup. Fig. 2. Although the 3- and 5-parameter functionals could be tuned to correlate reasonably well with the experimental data (R2 of 0.50 and 0.48, respectively), the performance under leave-one-out cross validation suggested substantial over-fitting of the 5-parameter functional (LOO R2 = 0.11). Notably though, the cross validated R2 of 0.30 (p-value of 0.24% as determined by a Monte Carlo permutation test) for the 3-parameter fit indicated terms of the type described by the displaced-solvent functional are likely important to understanding the absolute binding thermodynamics of fXa ligand, but also clearly indicated that more traditional terms will also be needed to quantitatively predict absolute binding free energies with desired accuracies.
Table 3.
Inhibition data for the 28 ligands extracted from solved crystal structures binding to factor Xa and our predicted activity differences from the trained 3-parameter and 5-parameter displaced-solvent functionals. Each ligand was designated “(pdb id):(ligand residue name)”.
| Ligand | ΔGexp (kcal/mol) | ΔG3p (kcal/mol) | ΔG5p (kcal/mol) | ΔGab initio (kcal/mol) |
|---|---|---|---|---|
| 2BOK:784 | −9.39 | −6.12 | −7.24 | 0.00 |
| 2J2U:GSQ | −9.61 | −7.26 | −8.60 | 3.34 |
| 2BQ7:IID | −9.62 | −7.78 | −8.77 | 3.00 |
| 1G2L:T87 | −9.88 | −6.86 | −8.93 | −0.03 |
| 2J34:GS5 | −10.00 | −6.73 | −7.80 | 1.57 |
| 1G2M:R11 | −10.09 | −6.54 | −8.42 | 0.29 |
| 1KYE:RUP | −10.37 | −7.47 | −8.88 | −0.03 |
| 1F0R:815 | −10.45 | −6.26 | −7.53 | −6.91 |
| 1F0S:PR2 | −10.57 | −6.39 | −7.77 | −5.85 |
| 2BMG:I1H | −10.57 | −8.49 | −9.34 | 5.49 |
| 1NFU:RRP | −10.57 | −6.98 | −8.50 | −2.21 |
| 2J38:GS6 | −10.67 | −6.94 | −8.06 | 2.15 |
| 1LQD:CMI | −10.98 | −8.22 | −9.30 | 4.31 |
| 2CJI:GSK | −11.22 | −7.48 | −8.52 | 1.76 |
| 2BQW:IIE | −11.63 | −8.07 | −9.16 | 8.42 |
| 1NFX:RDR | −11.63 | −7.58 | −8.97 | 0.69 |
| 2BOH:IIA | −11.63 | −8.61 | −9.72 | 4.98 |
| 1NFY:RTR | −12.12 | −7.47 | −8.89 | 1.01 |
| 1NFW:RRR | −12.22 | −7.21 | −8.46 | 0.48 |
| 1MQ5:XLC | −12.28 | −8.53 | −9.58 | 3.77 |
| 2J4I:GSJ | −12.28 | −7.98 | −9.33 | 2.01 |
| 1EZQ:RPR | −12.34 | −8.41 | −9.91 | −1.99 |
| 1KSN:FXV | −12.82 | −8.10 | −9.39 | −2.59 |
| 1Z6E:IK8 | −13.26 | −9.90 | −11.55 | 5.22 |
| 2FZZ:4QC | −13.29 | −9.93 | −11.33 | 4.94 |
| 1FJS:Z34 | −13.59 | −7.04 | −8.75 | −0.05 |
| 2G00:5QC | −14.36 | −9.98 | −11.44 | 4.88 |
| 1MQ6:XLD | −15.22 | −8.66 | −9.75 | 6.74 |
Figure 11.
Computed activities using the 5-parameter form of eq. 1 versus experimental activities for the for the set of 28 inhibitors with factor Xa. The poor stability of the fit under cross validation suggested substantial over fitting.
Figure 12.
Computed activities using the 5-parameter form of eq 1 versus experimental activities for the set of 28 inhibitors with factor Xa. The moderate stability of the fit under cross validation suggested the problems associated with over fitting were reduced when the three parameter form of eq. 1 was used.
In both the 3- and 5-parameter fits of the displaced-solvent functional to the set of 28 crystal structure ligands, two particular ligands, 1MQ6:XLD and 1FJS:Z34, were consistently the worst outliers in the set. Both of these ligands have excellent overlap with contributing hydration sites, but were out scored by ligands that placed larger aromatic groups at similar positions in the binding pocket, such as ligands 1Z6E:IK8, 2FZZ:4QC, and 2G00:5QC. This error was expected because our pairwise reward of atoms in close contact with the hydration sites approximated to what degree the contributing hydration sites were displaced by the surface of the ligand. Thus, when an aromatic group displaced a hydration site, a disproportionately large number of ligand atoms contributed in the displaced-solvent functional since the tighter covalent bonding in these groups placed many ligand atoms closer in space to the hydration site than could be seen otherwise. We should note that this systematic error was likely much less problematic in the set of congeneric pairs because the pathological bulky aromatic groups typically appeared in both congeners, leading to an exact cancellation of this error. When ligands 1MQ6:XLD and 1FJS:Z34 were excluded from the fit, the leave-one-out cross validation of the 3- and 5-parameter functionals yield R2 values of 0.40 and 0.55, respectively. This dramatic improvement of the stability and quality of the fit underscores how poor the linear pairwise approximation of the excluded volume of the ligand was for inhibitors 1MQ6:XLD and 1FJS:Z34. It is also possible that the known favorable electrostatic interaction between 1FJS:Z34 and the fXa S4 pocket, which was not described by the displaced-solvent functional, contributed to 1FJS:Z34 being an outlier in this data set.22
5. Cross testing of the trained displaced solvent density functionals
It was interesting to check the transferability of the parameters trained on the set of 31 congeneric inhibitor pairs to the set of 28 crystal structure ligands (supplementary table 3). The optimized 3- and 5-parameter functionals trained on the set of 31 congeneric inhibitor pairs each had R2 values of 0.17 when predicting the relative binding affinities of the 28 crystal structure ligands to fXa. The functionals performed poorly because the values of the parameters we obtained from training to the set of congeneric pairs typically predicted the difference in binding affinity between crystal structure pairs to be much too large (often greater than 10 kcal/mol). The reason for this may be subtle: typically only the tightest binding compound of a series will be crystallized, and even then it is typically only crystallized if it binds with a submicromolar affinity. Thus, if a ligand displaces a sub optimal portion of the active site solvent density, then it, by construction, only becomes a crystallized ligand if it is possible to tune the other contributions to the free energy (ligand entropy, ligand desolvation free energy, protein ligand interaction energy, etc) to offset this suboptimal active-site-solvent evacuation, resulting in the needed submicromolar affinity. So the magnitude of the contributions predicted by the displaced-solvent functionals may be qualitatively correct, but the other terms not described by the functional systematically offset them.
We found an interesting contrast to this result when we used the 3- and 5-parameter functionals trained on the set of 28 crystal structure ligands to predict the binding affinity differences of the set of 31 congeneric inhibitor pairs. We found the 3- and 5-parameter functionals trained on set of crystal structure ligands predicted the binding affinity differences of the set of 31 congeneric inhibitor pairs with R2 values of 0.53 and 0.59, respectively. This result suggested the functional form of the displaced-solvent functional may have fundamental features that lend themselves to ranking the binding affinities of compounds that differ by deletions of atoms — i.e., as long as the chosen parameters are physically reasonable, the performance of the functional over congeneric sets of this kind may be quite good.
Conclusions
Our results suggest that the expulsion of active site water strongly impacts protein-ligand binding affinities in 2 ways: (1) hydrophobic ligand groups that displace water from energetically unfavorable (hydrophobically enclosed) environments contribute enthalpically since the water molecules will make more favorable interactions in the bulk fluid; and (2) ligand groups that displace entropically structured solvent contribute even when the solvent interacts favorably with the protein since well designed ligands will recapture the protein-water interaction energy. Congeneric inhibitor pair Young:38-2J4I:GSJ is particularly clear example where the expulsion of active site water that solvates an energetically unfavorable environment lead to large favorable contributions to the binding free energy. In contrast, congeneric pair 1MQ5:XLC-1MQ6:XLD offered an interesting example of the expulsion of water from a hydration site with a favorable interaction energy and unfavorable excess entropy. The expulsion of water from this hydration site was found to be favorable, by our empirical criteria, presumably because the ligand group that displaces this water does a reasonably good job recapturing the interaction energy of the solvent with the protein with less entropic cost. Congeneric inhibitor pair 2BQ7:IID-2BQW:IIE illustrated that these two solvent categories, energetically unfavorable and entropically unfavorable, are by no means mutually exclusive and that the evacuation of solvent from the protein active site will often make both entropic and enthalpic contributions to the binding free energy. Instrumental to our analysis is the assumption of complementarity — that is, that the difference between the water-protein energetic interactions and the ligand-protein interactions was expected to be small. This assumption is valid when the ligands form hydrogen bonds with the protein where appropriate and hydrophobic contacts otherwise; however, the congeneric ligand pair 1NFX:RDR/1NFW:RRR illustrated ligands that violate this hypothesis will often be mistreated by the method. This has relevance to modern drug design since it suggests that it is misleading to look at particular crystal waters as favorable or unfavorable to displace, as is often done in structure based drug design. Instead, it may be more productive to consider how thermodynamically favorable displacing a crystal water will be when it is displaced by a complementary chemical group of a ligand.
The empirical functionals we developed were quite successful at quantifying the contributions to the free energy of binding due to the ligand evacuating energetically unfavorable and entropically structured solvent for the set of congeneric pairs. They were able to differentiate those modifications to an existing ligand scaffold that made small contributions to the binding affinity of the complex from those modifications that made large contributions over a 6 kcal/mol range. In their present form, the 3- and 5-parameter functionals may be useful to lead optimization, since the functionals appeared to well describe the thermodynamics of adding small chemical groups to a given ligand scaffold that are complementary to the protein surface. The performance of the functionals on the set of 28 crystal structure ligands suggests that terms of this type may make large contributions to binding; however, these functionals should not be used as a stand alone tool for computational screening of chemically diverse compounds. The reason for this was clear: the displaced-solvent functionals presented here neglect several terms which will vary considerably over sets of chemically diverse ligands. These terms include the protein-ligand electrostatic and Van der Waals interaction energies, ligand solvation free energy, ligand configurational entropy, and the protein-reorganization free energy. Thus, a functional designed for computational screening would have to include additional terms describing these types of contributions to the free energy in addition to those contributions captured by the displaced-solvent functional.
Methods
1. Structure preparation and simulation
We chose to use PDB crystal structure 1FJS as our initial model of the fXa protein.22 This structure was imported into the Maestro23 program, all crystallographic water were deleted, and hydrogens were added to the structure assuming a pH 7 environment. Chain L of the crystal structure was also deleted, since it contained no atoms within 20 Å of the fXa active site. We then used the protein preparation utility found in Maestro to run a restrained minimization of the protein in the presence of the 1FJS crystal structure ligand.24 This removed bad steric contacts and improved the quality of the protein-protein and protein-ligand hydrogen bonding without large rearrangements of the protein heavy atoms. Using the OPLSAA-200125 potential, we imported this model of the protein into a modified version of GROMACS26,27 prepared by Shirts et al. We then solvated the system in a cubic TIP4P28 water box where each boundary was greater than 10 Å away from the protein and added one chlorine ion to neutralize the system.
We minimized the energy of the system to relieve bad steric contacts between the protein and the water and equilibrated the system for 100 ps with the velocity version of the Verlet integrator29 and Berendsen30 temperature and pressure controls at 298 K and 1 bar, where a frame of the system was saved every 1 ps. The Lennard-Jones interactions were truncated at 9 Å, the electrostatic interactions were described exactly for pairs within 10 Å and by Particle-Mesh-Ewald31,32 for pairs outside of this radius, and all protein heavy atoms were harmonically restrained with spring constants of 1000 kJ/mol/nm2. We used the final 10 ps of equilibration data to seed 10 different 1 ns molecular dynamics trajectories with the velocity version of the Verlet integrator29, Andersen33 temperature controls, and Parrinello-Rahman34,35 pressure controls at 298 K and 1 bar. For these simulations the Lennard-Jones, electrostatic forces, and harmonic restraints on the heavy atoms of the protein were the same as in the equilibration simulations. Frames of this simulation were saved every picosecond.
The MM-GBSA5,6 calculations of the protein-ligand binding affinities were carried out using Prime 1.6 and were setup using the graphical user interface available in Maestro 8.0. The free energy of binding was estimated using the following equation: ΔGbinding= Ecomplex(minimized) − Eligand(from minimized complex) − Ereceptor(from minimized complex). The OPLS-AA-2001 potential25 was used to model the protein and the ligand, and the Surface Generalized Born49 model was used to describe the polar and nonpolar contributions of the solvent. The protein and ligand coordinates used for these calculations were taken directly from the reported crystal structure for the particular ligand. The energy of each protein/ligand complex was determined by minimizing the ligand and residues within 8 Å of the ligand. The energy of the ligand and receptor are from the minimized complex, so the estimated binding energy is the protein/ligand interaction energy without accounting for ligand or receptor strain. For each ligand pair that involved one co-crystallized ligand and one modified ligand (constructed as described in Methods section 3), the receptor that was used for the MM-GBSA was the prepared (as described Methods section 3) PDB structure associated with the co-cocrystallized ligand. For ligand pairs in which both ligands are co-crystallized, two MM-GBSA runs were conducted using both protein structures associated with each of the ligands and the results were reported for the receptor structure that yielded the smallest change in the ligand conformation following the MM-GBSA calculation.
2. Active site hydration analysis
In order to analyze the thermodynamic and structural properties of the water molecules hydrating the fXa active site, we needed to develop some sensible definition for when a solvating water should be considered within the fXa active site and when it should not.2 We used a set of 35 fXa crystal structures with bound inhibitors to define the volume of the active site (PDB structures 1EZQ13, 1F0R13, 1F0S13, 1FAX36, 1FJS22, 1G2L37, 1G2M37, 1IOE38, 1IQE38, 1IQF38, 1IQG38, 1IQH38, 1IQI38, 1IQJ38, 1IQK38, 1IQL38, 1IQM38, 1IQN38, 1KSN39, 1KYE14, 1MQ59, 1MQ69, 1NFU12, 1NFW12, 1NFX12, 1NFY12, 1V3X41, 1XKA41, 1XKB41, 2BOK42, 2CJI43, 2J2U44, 2J3444, 2J3844, 2J4I7). We computed a multiple structure alignment between the 35 fXa crystal structures containing inhibitors and our prepared fXa model structure. This alignment rotated the crystal structures onto our prepared fXa structure. This procedure also rotated the inhibitors found in these crystal structures into the active site of the our prepared model fXa structure. The results of these alignments were hand inspected for severe steric clashes and none were found. Using this set of aligned structures, we defined the active site as the volume containing all points in space that are within 3 Å of any ligand heavy atom. The position of the active site volume was constant throughout the simulation because the protein heavy atoms were harmonically restrained. The coordinates of all waters observed within this region of space during the 10 ns of simulation data were saved every picosecond. We considered this water distribution to be the equilibrium distribution of water within the fXa active site and we characterized its thermodynamic properties with inhomogeneous solvation theory along with several other measures of local water structure.
The application of inhomogeneous solvation theory to the heterogeneous surface of a protein active site where the solvating waters can exchange with the bulk fluid is highly nontrivial. Although the difficulty posed by waters exchanging with the bulk fluid is alleviated by our definition of the active site, the inhomogenous topography of the protein surface made the orientational distributions of the water molecules highly dependent on their position within the active site. Following procedures we previously developed2, we partitioned the active site volume into small subvolumes which we denote “hydration sites” and treated the angular distributions as independent of position in these subvolumes. We identified the subvolumes by applying a clustering algorithm to partition the solvent density distribution into a set of high water occupancy 1 Å radius spheres. This algorithm cycled through the positions of the oxygen atom of every water molecule found in the active site solvent density distribution and found the position that has the greatest number of water neighbors within a 1 Å radius. We denoted this position as a hydration site and removed it and all of the oxygen positions within 1 Å of it from the solvent density distribution. This process was then repeated, cycling through the remaining positions. This loop was terminated when the clustering algorithm identified a hydration site with a water-oxygen occupancy less than twice the expected value of a 1 Å radius sphere in the bulk fluid. These hydration sites are well defined subvolumes of the active site and have good convergence properties for the inhomogenous solvation theory machinery since they have sparse water density toward the edges of the clusters.
We performed an inhomogenous solvation theory analysis of the thermodynamic properties of each hydration site to elucidate how the properties of the solvating water may affect the thermodynamics of fXa inhibitor association. Consistent with our prior work, we defined the system interaction energy (Ehs) of each hydration site to be the average energy of interaction of the water molecules in a given hydration site with the rest of the system.2 We also computed the partial excess entropy (Se) of each hydration site by numerically integrating an expansion of the entropy in terms of orientational and spacial correlation functions.3,45,46 In this work we included only contributions from the first order term for each hydration site:
| (2) |
where r and ω are the Cartesian position and Euler angle orientation of a water molecule, gsw(r, ω) is the 1-body distribution of the water (w) at r and ω in the fixed reference frame of the solute protein (s), ρw is the density of the neat TIP4P system, kb is the Boltzmann constant, Ω is the total orientational space accessible to a water molecule, and NVW is total number of water oxygens found within a given hydration site of volume V. We numerically integrated the translational contribution to the excess entropy in spherical coordinates using a length of 0.03 Å along r, 15° along θ, and 30° along φ; and we numerically integrated the orientational contribution with 10° along each Euler angle.
We also calculated several measures of local water structure properties for the water molecules found within each hydration site. These were the average number of of water neighbors, the average number of hydrogen bonding water neighbors, the fraction of the water neighbors that were hydrogen bonding, and the water exposure of each hydration site. These averages are for all water molecules in each hydration site. The number of neighbors value is the average number of water molecules found within 3.5 Å, where the distance is measured water oxygen to water oxygen. We used a geometric definition of a hydrogen bond where two water molecules were deemed to be hydrogen bonded if their oxygens were within 3.5 Å of each other and at least one oxygen-oxygen-hydrogen angle was less than 30°. The exposure value quantifies to what degree a hydration site is surrounded by other water molecules: a value of unity suggests it is in a water environment similar to the bulk fluid, and a value of zero suggests the hydration site is occluded from any other solvent molecules. The exposure value is computed as the average number of neighbors that water molecules in a hydration site have divided by the average number of neighbors that a water molecule has in the bulk.
3. Construction of the factor Xa ligand binding affinity data sets
Within the PDB, we found 28 published crystal structures of fXa bound to various inhibitors with thermodynamic binding data reported in the associated publication (2BOK42, 2J2U44, 2BQ710, 1G2L37, 2J3844, 1G2M37, 1KYE14, 1F0R13, 1F0S13, 2BMG10, 1NFU12, 2J3444, 1LQD8, 2CJI43, 2BQW10, 1NFX12, 2BOH11, 1NFY12, 1NFW12, 1MQ59, 2J4I7, 1EZQ13, 1KSN39, 1Z6E15, 2G0047, 1FJS22, 2FZZ48, 1MQ69). We computed a multiple structure alignment between the 28 fXa crystal structures containing inhibitors and our prepared fXa model structure. This procedure rotated the 28 inhibitors found in these crystal structures into the active site of the our prepared model fXa structure. The results of these alignments were hand inspected for severe steric clashes and none were found. The orientations of each of these 28 inhibitors with respect to our prepared model fXa structure were saved and were referred to as the 28 crystal structure ligand set.
From this set of 28 crystal structure ligand set, we prepared a set of 31 congeneric inhibitor pairs. The goal of this set of inhibitor pairs was to isolate the effects of solvent displacement on the free energy of binding. Each congeneric pair was created by either noting that 2 of the crystal structure ligands reported in the prior set were congeneric or by building a congeneric pair from a single crystal structure ligand by deleting or swapping atoms of the crystal structure ligand. We devised several rules to construct this set. When any two members of the 28 crystal structure ligand set were reported in the same publication and differed by no more than 3 chemical groups, they were considered congeneric pairs. When the publication reporting the crystal structure ligand contained congeneric series data for structurally similar ligands, we followed three rules to build new congeneric pairs:
We would only delete atoms from a crystal structure ligand and not add them
We would not accept deletions of atoms that resulted in a group that could rotate around a single bond and donate hydrogen bonds
A congeneric pair that was built by changing the identity of a ligand atom (for instance changing a carbon atom to an oxygen atom) must have the change applied to both members of the pair.
These three rules were intended to minimize the error of assuming that the binding mode of the new inhibitor structures, which were built from deleting and swapping atoms of the crystallized inhibitors, would not change. These rules were also intended to minimize differences in contributions to binding affinity from non-solvent related terms for each inhibitor pair, such as the loss of entropy of docking the ligand, the strength of the interaction energy between the ligand and the protein, and the reorganization free energy of the protein. We expected that excluded solvent density effects would dominate this set since these other non-solvent related terms contributing to the free energy of binding would be relatively constant for each congeneric pair. We also chose only to compare binding affinities between pairs of ligands that were determined in the same publication, due to the variance in experimental methods commonly employed. We referred to the resulting set as the set of 31 congeneric inhibitor pairs (supplementary table 4).
4. Development and parameterization of the displaced-solvent functional
We devised a 5-parameter scoring function to determine if the relative binding affinities of the 28 crystal structure ligands and the binding affinity differences of the 31 congeneric inhibitor pairs correlated with the thermodynamic properties of the displaced active site solvent. Additional discussion of the physical motivation that lead us to this functional form can be found in the results section. The form of the functional was a sum over ligand heavy atoms and a sum over hydration sites. Each time a ligand heavy was found within some parameterized distance of a hydration site with an interaction energy or excess entropy predicted to be favorable to evacuate by some fit empirical criteria, an additive contribution was summed. The functional itself was
| (1) |
where ΔGbind was the predicted binding free energy of the ligand, Rco was the distance cutoff for a ligand atom beginning to displace a hydration site, Eco was the minimum Ehs of a hydration site that was considered energetically depleted, Erwd was the energetic contribution to ΔGbind for displacing an energetically depleted hydration site, Sco was the minimum Se term of a hydration site that was considered entropically structured, −TSrwd was the Entropic contribution to ΔGbind for displacing an entropically structured hydration site, and Θ was the Heaviside step function. We also considered a 3-parameter form of this equation where we fixed Rco = 2.8 Å and −TSrwd = Erwd.
The parameters were optimized by a Monte Carlo walk in parameter space. The error-function we used to train the parameters was the root-mean-square-deviation of the predicted relative binding free energies of the 28 crystal ligands and the root-mean-square-deviation of the differences in the binding free energies of the 31 congeneric pairs. For the training of the 3- and 5-parameter functionals on the 28 crystal structure ligand set, we chose an initial seed value of Rco = 2.8 Å, Erwd = −0.5 kcal/mol, −TSrwd = −0.5 kcal/mol, Eco = −18.5 kcal/mol, and TSco = 1.5 kcal/mol. Five separate 1000 step optimizations were run where the first move was always accepted and the lowest RMSD value encountered in these optimizations was taken to be the optimal parameter set. The initial seed values used to train the 3- and 5-parameter functionals on the set of 31 congeneric inhibitor pairs were Rco = 2.8 Å, Erwd = −1.0 kcal/mol, −TSrwd = −1.0 kcal/mol, Eco = −18.5 kcal/mol, and TSco = 1.5 kcal/mol. The parameters were then optimized in a procedure identical to that used for the 28 crystal structure ligands.
We also constructed an “ab initio” form of the displaced solvent functional Containing no fit parameters. The functional itself was
| (3) |
where ΔGbind was the predicted binding free energy of the ligand, Rco was the distance cutoff for a ligand atom beginning to displace a hydration site and ΔGhs was the computed free energy of transferring the solvent in a given hydration site from the active site to the bulk fluid. We also capped the contribution from each hydration site, such that it would never contribute more than ΔGhs to ΔGbind no matter how many ligand atoms were in close proximity to it. The value Rco might be considered a free parameter. However, an approximate value was adopted by noting that the radius of a carbon atom and a water oxygen atom are both approximately 1.4 Å, thus suggesting contact distances between a water oxygen atom and a ligand carbon atom less than 0.8*(1.4 Å+1.4 Å)=2.24 Å are statistically improbable due to the stiffness of the Van der Waals potential.50 Thus, we chose for the ab initio functional to specify Rco=2.24 Å. The sensitivity of the results reported here to this choice of the Rco parameter was quite low for adjustments of this value within a few tenths of an angstrom of the specified value.
We estimated the error of the resulting optimized functionals with leave-one-out cross validation. In this techniques a functional is trained to an N-1 point subset of data and then the value of point N is predicted with this functional. This is repeated N times, once for each data point, and the error of the functional is estimated by summing the error of the predictions for each of these points. The Pearson correlation coefficient (R2) computed in this procedure for the N data points is bounded between the R2 value found by training of the functional on all N data points and zero. A cross validation R2 value close to the R2 value found by training of the functional on all N data points suggests very little over-fitting has occurred when training the functional.
We performed a Monte Carlo permutation analysis to estimate the p-value of a given R2 value. The analysis proceeded by generating 5 million random permutations of the mapping between the predicted ΔG values and the experimentally measured ΔG values. The p-value of a given R2 value was taken to be the number of random permutations yielding and R2 value greater than or equal to the original R2 value divided by the total number of permutations generated. This allowed us to very accurately estimate the probability of attaining a given R2 value for our particular distribution without assuming any properties about the relationships between the predicted and experimentally measured distributions.
Supplementary Material
ACKNOWLEDGMENT
This work was supported in part by a grant from the NIH to RAF (GM-52018) and to BJB (GM-43340). This material is based upon work supported in part by a National Science Foundation Graduate Research Fellowship.
References
- 1.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49:6177–96. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 2.Young T, Abel R, Kim B, Berne BJ, Friesner RA. Motifs for molecular recognition exploiting hydrophobic enclosure in protein-ligand binding. Proc. Natl. Acad. Sci. USA. 2007;104:808–813. doi: 10.1073/pnas.0610202104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lazaridis T. Inhomogeneous Fluid Approach to Solvation Thermodynamics 1. Theory. J. Phys. Chem. B. 1998;102:3531–3541. [Google Scholar]
- 4.Turpie AG. Oral, direct factor Xa inhibitors in development for the prevention and treatment of thromboembolic diseases. Arterioscler Thromb. Vasc. Biol. 2007;27:1238–1247. doi: 10.1161/ATVBAHA.107.139402. [DOI] [PubMed] [Google Scholar]
- 5.Huang N, Kalyanaraman C, Bernacki K, Jacobson MP. Molecular mechanics methods for predicting protein-ligand binding. Phys. Chem. Chem. Phys. 2006;8:5166–5177. [Google Scholar]
- 6.Lyne PD, Lamb ML, Saeh JC. Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MM-GBSA scoring. J. Med. Chem. 2006;49:4805–4808. doi: 10.1021/jm060522a. [DOI] [PubMed] [Google Scholar]
- 7.Young RJ, Campbell M, Borthwick AD, Brown D, Burns-Kurtis CL, Chan C, Convery MA, Crowe MC, Dayal S, Diallo H, Kelly HA, King NP, Kleanthous S, Mason AM, Mordaunt JE, Patel C, Pateman AJ, Senger S, Shah GP, Smith PW, Watson NS, Weston HE, Zhou P. Structure- and property-based design of factor Xa inhibitors: pyrrolidin-2-ones with acyclic alanyl amides as P4 motifs. Bioorg. Med. Chem. Lett. 2006;16:5953–5957. doi: 10.1016/j.bmcl.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Matter H, Defossa E, Heinelt U, Blohm PM, Schneider D, Muller A, Herok S, Schreuder H, Liesum A, Brachvogel V, Lonze P, Walser A, Al-Obeidi F, Wildgoose P. Design and quantitative structure-activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa. J. Med. Chem. 2002;45:2749–2769. doi: 10.1021/jm0111346. [DOI] [PubMed] [Google Scholar]
- 9.Adler M, Kochanny MJ, Ye B, Rumennik G, Light DR, Biancalana S, Whitlow M. Crystal structures of two potent nonamidine inhibitors bound to factor Xa. Biochemistry. 2002;41:15514–15523. doi: 10.1021/bi0264061. [DOI] [PubMed] [Google Scholar]
- 10.Matter H, Will DW, Nazare M, Schreuder H, Laux V, Wehner V. Structural requirements for factor Xa inhibition by 3-oxybenzamides with neutral P1 substituents: combining X-ray crystallography, 3D-QSAR, and tailored scoring functions. J. Med. Chem. 2005;48:3290–3312. doi: 10.1021/jm049187l. [DOI] [PubMed] [Google Scholar]
- 11.Nazare M, Will DW, Matter H, Schreuder H, Ritter K, Urmann M, Essrich M, Bauer A, Wagner M, Czech J, Lorenz M, Laux V, Wehner V. Probing the subpockets of factor Xa reveals two binding modes for inhibitors based on a 2 carboxyindole scaffold: a study combining structure-activity relationship and X-ray crystallography. J. Med. Chem. 2005;48:4511–25. doi: 10.1021/jm0490540. [DOI] [PubMed] [Google Scholar]
- 12.Maignan S, Guilloteau JP, Choi-Sledeski YM, Becker MR, Ewing WR, Pauls HW, Spada AP, Mikol V. Molecular structures of human factor Xa complexed with ketopiperazine inhibitors: preference for a neutral group in the S1 pocket. J. Med. Chem. 2003;46:685–690. doi: 10.1021/jm0203837. [DOI] [PubMed] [Google Scholar]
- 13.Maignan S, Guilloteau JP, Pouzieux S, Choi-Sledeski YM, Becker MR, Klein SI, Ewing WR, Pauls HW, Spada AP, Mikol V. Crystal structures of human factor Xa complexed with potent inhibitors. J. Med. Chem. 2000;43:3226–32. doi: 10.1021/jm000940u. [DOI] [PubMed] [Google Scholar]
- 14.Mueller MM, Sperl S, Sturzebecher J, Bode W, Moroder L. (R)-3-Amidinophenylalanine-derived inhibitors of factor Xa with a novel active-site binding mode. Biol. Chem. 2002;383:1185–1191. doi: 10.1515/BC.2002.130. [DOI] [PubMed] [Google Scholar]
- 15.Quan ML, Lam PY, Han Q, Pinto DJ, He MY, Li R, Ellis CD, Clark CG, Teleha CA, Sun JH, Alexander RS, Bai S, Luettgen JM, Knabb RM, Wong PC, Wexler RR. Discovery of 1-(3′-aminobenzisoxazol-5′-yl)-3-trifluoromethyl-N-[2-fluoro-4- [(2′-dimethylaminomethyl)imidazol-1-yl]phenyl]-1H-pyrazole-5-carboxyamide hydrochloride (razaxaban), a highly potent, selective, and orally bioavailable factor Xa inhibitor. J. Med. Chem. 2005;48:1729–1744. doi: 10.1021/jm0497949. [DOI] [PubMed] [Google Scholar]
- 16.Padmanabhan K, Padmanabhan KP, Tulinsky A, Park CH, Bode W, Huber R, Blankenship DT, Cardin AD, Kisiel W. Structure of human des(1-45) factor Xa at 2.2 A resolution. J. Mol. Biol. 1993;232:947–966. doi: 10.1006/jmbi.1993.1441. [DOI] [PubMed] [Google Scholar]
- 17.Carugo O, Bordo D. How many water molecules can be detected by protein crystallography? Acta Crystallogr. D. Biol. Crystallogr. 1999;55:479–483. doi: 10.1107/s0907444998012086. [DOI] [PubMed] [Google Scholar]
- 18.Mattos C. Protein-water interactions in a dynamic world. Trends Biochem. Sci. 2002;27:203–208. doi: 10.1016/s0968-0004(02)02067-4. [DOI] [PubMed] [Google Scholar]
- 19.Nakasako M. Large-scale networks of hydration water molecules around bovine beta-trypsin revealed by cryogenic X-ray crystal structure analysis. J. Mol. Biol. 1999;289:547–64. doi: 10.1006/jmbi.1999.2795. [DOI] [PubMed] [Google Scholar]
- 20.Makarov VA, Andrews BK, Smith PE, Pettitt BM. Residence times of water molecules in the hydration sites of myoglobin. Biophys J. 2000;79:2966–74. doi: 10.1016/S0006-3495(00)76533-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunitz JD. The entropic cost of bound water in crystals and biomolecules. Science. 1994;264:670. doi: 10.1126/science.264.5159.670. [DOI] [PubMed] [Google Scholar]
- 22.Adler M, Davey DD, Phillips GB, Kim SH, Jancarik J, Rumennik G, Light DR, Whitlow M. Preparation, characterization, and the crystal structure of the inhibitor ZK-807834 (CI-1031) complexed with factor Xa. Biochemistry. 2000;39:12534–12542. doi: 10.1021/bi001477q. [DOI] [PubMed] [Google Scholar]
- 23.Banks JL, Beard HS, Cao YX, Cho AE, Damm W, Farid R, Felts AK, Halgren TA, Mainz DT, Maple JR, Murphy R, Philipp DM, Repasky MP, Zhang LY, Berne BJ, Friesner RA, Gallicchio E, Levy RM. Integrated modeling program, applied chemical theory (IMPACT) Journal of Computational Chemistry. 2005;26:1752–1780. doi: 10.1002/jcc.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 25.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B. 2001;105:6474–6487. [Google Scholar]
- 26.Lindahl E, Hess B, van der Spoel D. Gromacs 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Mod. 2001;7:306–317. [Google Scholar]
- 27.Shirts MR, Pande VS. Solvation free energies of amino acid side chain analogs for common molecular mechanics water models. J. Chem. Phys. 2005;122:134508–134508. doi: 10.1063/1.1877132. [DOI] [PubMed] [Google Scholar]
- 28.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein M. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 29.Swope WC, Anderson HC, Berens PH, Wilson KR. A computer simulation method for the calculation of equilibrium-constants for the formation of physical clusters of molecules- application to small water clusters. J. Chem. Phys. 1982;76:637–649. [Google Scholar]
- 30.Berendsen HJC, Postma JPM, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 31.Darden T, York D, Pedersen L. Particle mesh Ewald: An N-log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 32.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh ewald potential. J. Chem. Phys. 1995;103:8577–8592. [Google Scholar]
- 33.Andersen HC. Molecular-dynamics simulations at constant pressure and-or temperature. J. Chem. Phys. 1980;72:2384–2393. [Google Scholar]
- 34.Parrinello M, Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 35.Nose S, Klein ML. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983;50:1055–1076. [Google Scholar]
- 36.Brandstetter H, Kuhne A, Bode W, Huber R, von der Saal W, Wirthensohn K, Engh RA. X-ray structure of active site-inhibited clotting factor Xa. Implications for drug design and substrate recognition. J. Biol. Chem. 1996;271:29988–29992. doi: 10.1074/jbc.271.47.29988. [DOI] [PubMed] [Google Scholar]
- 37.Nar H, Bauer M, Schmid A, Stassen JM, Wienen W, Priepke HW, Kauffmann IK, Ries UJ, Hauel NH. Structural basis for inhibition promiscuity of dual specific thrombin and factor Xa blood coagulation inhibitors. Structure. 2001;9:29–38. doi: 10.1016/s0969-2126(00)00551-7. [DOI] [PubMed] [Google Scholar]
- 38.Matsusue T, Shiromizu I, Okamoto A, Nakayama K, Nishida H, Mukaihira T, Miyazaki Y, Saitou F, Morishita H, Ohnishi S, Mochizuki H. Factor Xa Specific Inhibitor That Induces the Novel Binding Model In Complex With Human FXa. To be Published.
- 39.Guertin KR, Gardner CJ, Klein SI, Zulli AL, Czekaj M, Gong Y, Spada AP, Cheney DL, Maignan S, Guilloteau JP, Brown KD, Colussi DJ, Chu V, Heran CL, Morgan SR, Bentley RG, Dunwiddie CT, Leadley RJ, Pauls HW. Optimization of the beta-aminoester class of factor Xa inhibitors. Part 2: Identification of FXV673 as a potent and selective inhibitor with excellent In vivo anticoagulant activity. Bioorg. Med. Chem. Lett. 2002;12:1671–1674. doi: 10.1016/s0960-894x(02)00213-5. [DOI] [PubMed] [Google Scholar]
- 40.Haginoya N, Kobayashi S, Komoriya S, Yoshino T, Suzuki M, Shimada T, Watanabe K, Hirokawa Y, Furugori T, Nagahara T. Synthesis and conformational analysis of a non-amidine factor Xa inhibitor that incorporates 5-methyl-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridine as S4 binding element. J. Med. Chem. 2004;47:5167–5182. doi: 10.1021/jm049884d. [DOI] [PubMed] [Google Scholar]
- 41.Kamata K, Kawamoto H, Honma T, Iwama T, Kim SH. Structural basis for chemical inhibition of human blood coagulation factor Xa. Proc. Natl. Acad. Sci. USA. 1998;95:6630–6635. doi: 10.1073/pnas.95.12.6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scharer K, Morgenthaler M, Paulini R, Obst-Sander U, Banner DW, Schlatter D, Benz J, Stihle M, Diederich F. Quantification of Cation-Pi Interactions in Protein-Ligand Complexes: Crystal-Structure Analysis of Factor Xa Bound to a Quaternary Ammonium Ion Ligand. Angew. Chem. Int. Ed. Engl. 2005;44:4400–4404. doi: 10.1002/anie.200500883. [DOI] [PubMed] [Google Scholar]
- 43.Watson NS, Brown D, Campbell M, Chan C, Chaudry L, Convery MA, Fenwick R, Hamblin JN, Haslam C, Kelly HA, King NP, Kurtis CL, Leach AR, Manchee GR, Mason AM, Mitchell C, Patel C, Patel VK, Senger S, Shah GP, Weston HE, Whitworth C, Young RJ. Design and Synthesis of Orally Active Pyrrolidin-2-One-Based Factor Xa Inhibitors. Bioorg. Med. Chem. Lett. 2006;16:3784–3788. doi: 10.1016/j.bmcl.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 44.Senger S, Convery MA, Chan C, Watson NS. Arylsulfonamides: A Study of the Relationship between Activity and Conformational Preferences for a Series of Factor Xa Inhibitors. Bioorg. Med. Chem. Lett. 2006;16:5731–5735. doi: 10.1016/j.bmcl.2006.08.092. [DOI] [PubMed] [Google Scholar]
- 45.Baranyai A, Evans DJ. Direct entropy calculation from computer simulation of liquids. Phys. Rev. A. 1989;40:3817–3822. doi: 10.1103/physreva.40.3817. [DOI] [PubMed] [Google Scholar]
- 46.Morita T, Hiroike K. A New Approach to the Theory of Classical Fluids. III— General Treatment of Classical Systems. Prog. of Theor. Phys. 1961;25:537–578. [Google Scholar]
- 47.Pinto DJ, Galemmo RA, Quan ML, Orwat MJ, Clark C, Li R, Wells B, Woerner F, Alexander RS, Rossi KA, Smallwood A, Wong PC, Luettgen JM, Rendina AR, Knabb RM, He K, Wexler RR, Lam PY. Discovery of potent, efficacious, and orally bioavailable inhibitors of blood coagulation factor Xa with neutral P1 moieties. Bioorg. Med. Chem. Lett. 2006;16:5584–5589. doi: 10.1016/j.bmcl.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 48.Pinto DJ, Orwat MJ, Quan ML, Han Q, Galemmo RA, Amparo E, Wells B, Ellis C, He MY, Alexander RS, Rossi KA, Smallwood A, Wong PC, Luettgen JM, Rendina AR, Knabb RM, Mersinger L, Kettner C, Bai S, He K, Wexler RR, Lam PY. 1-[3-Aminobenzisoxazol-5′-yl]-3-trifluoromethyl-6-[2′-(3-(R)-hydroxy-N-pyrrolidinyl)methyl-[1,1′]-biphen-4-yl]-1,4,5,6-tetrahydropyrazolo-[3,4-c]-pyridin-7-one (BMS-740808) a highly potent, selective, efficacious, and orally bioavailable inhibitor of blood coagulation factor Xa. Bioorg. Med. Chem. Lett. 2006;16:4141–4147. doi: 10.1016/j.bmcl.2006.02.069. [DOI] [PubMed] [Google Scholar]
- 49.Ghosh A, Rapp CS, Friesner RA. Generalized born model based on a surface integral formulation. J. Phys. Chem. B. 1998;102:10983–10990. [Google Scholar]
- 50.Jacobson MP, Pincus DL, Rapp CS, Day TJ, Honig B, Shaw DE, Friesner RA. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.