Abstract
Dynamic actin remodelling processes at the leading edge of migrating tumour cells are concerted events controlled by a fine-tuned temporal and spatial interplay of kinases and phosphatases. Actin severing is regulated by ADF/Cofilin which regulates stimulus-induced lamellipodia protrusion and directed cell motility. Cofilin is activated by dephosphorylation via phosphatases of the slingshot (SSH) family. SSH activity is strongly increased by its binding to filamentous actin (F-actin), however, other upstream regulators remain unknown. We show that in response to RhoA activation, Protein Kinase D1 (PKD1) phosphorylates the SSH enzyme SSH1L at a serine residue located in its actin binding motif. This generates a 14-3-3 binding motif, blocks the localization of SSH1L to F-actin-rich structures in the lamellipodium by sequestering it in the cytoplasm. Consequently, expression of constitutively-active PKD1 in invasive tumour cells enhanced phosphorylation of cofilin and effectively blocked the formation of free actin filament barbed ends and directed cell migration.
Migratory competence of tumour cells requires the activation of the “motile cycle”, the first step of which is actin remodelling, which drives the formation of cell protrusions, defines the direction of migration and initiates the growth of the lamellipodium. Dynamic actin remodelling processes at the leading edge of migrating tumour cells are complex, with areas showing increased actin filament severing, capping and dendritic branching 1, 2. The concerted regulation of these events is mediated by a complex temporal and spatial interplay of RhoGTPases, kinases and phosphatases.
A key regulator of polarized cell motility is the F-actin depolymerization and severing factor ADF/Cofilin (cofilin) 3, 4. By severing actin filaments, cofilin increases free barbed ends, which are the preferred substrate for dendritic nucleation by the Arp2/3 complex 3. Cofilin is highly expressed in multiple cancers including pancreatic cancer and invasive breast cancer 5, 6. The net effect of signalling cascades regulating cofilin activity mediates if a tumour cell migrates, or not 6, 7. The activity of cofilin is mainly regulated by phosphorylation and de-phosphorylation events, which allow rapid regulation of the enzyme in different regions of the migrating tumour cell 7, 8. Phosphorylation of cofilin at serine residue 3 (Ser3) is mediated by the LIM kinases (LIMK; Lin-11/Isl-1/Mec-3 kinases) LIMK1 or LIMK2 1, 9 and by TESK (testicular protein kinases) 10, 11, leads to loss of actin binding and severing activities and subsequently results in decreased directed cell motility 12.
The rapid actin remodelling events at the progressing leading edge of migrating cells require the rapid modulation of cofilin activity 6. The de-phosphorylation of Ser3 by slingshot (SSH) phosphatases reactivates cofilin 13, 14. The regulation of slingshots is not well understood, although recently a potential regulation by the PI3K pathway was implicated 15. Further, the SSH member SSH1L is regulated by association with filamentous actin, which increases its phosphatase activity 13. The re-localization of SSH1 at the leading edge of cells is controlled by the phosphorylation-dependent recruitment of 14-3-3 proteins, but functional consequences are not well defined 13.
Protein Kinase D1 is a serine/threonine kinase that so far was not implicated in the regulation of cofilin activity and actin remodelling at the lamellipodium of migrating tumor cells. Dependent on its subcellular localization PKD1 regulates a variety of cellular functions (reviewed in 7), including membrane receptor signalling 16, transport processes at the golgi 17, 18, protection from oxidative stress at the mitochondria 19 and transcriptional regulation in the nucleus 20. Recent studies suggest an involvement of PKD1 in the regulation of cell shape, motility and adhesion 21–23. However, conclusive molecular mechanisms linking PKD1 to cytoskeletal reorganization and also to mechanisms affecting cell motility remained elusive.
Here we describe the localized phosphorylation of substrates, which regulate actin organization, as a potential mechanism by which PKD1 exerts its effects on cell motility. Specifically we identify the SSH1L as a substrate, whose phosphorylation by PKD1 mediates re-localisation and inhibition, translating to altered cofilin activity, actin re-organization and decreased directed cell motility.
RESULTS
PKD1 co-localizes with F-actin and SSH1L
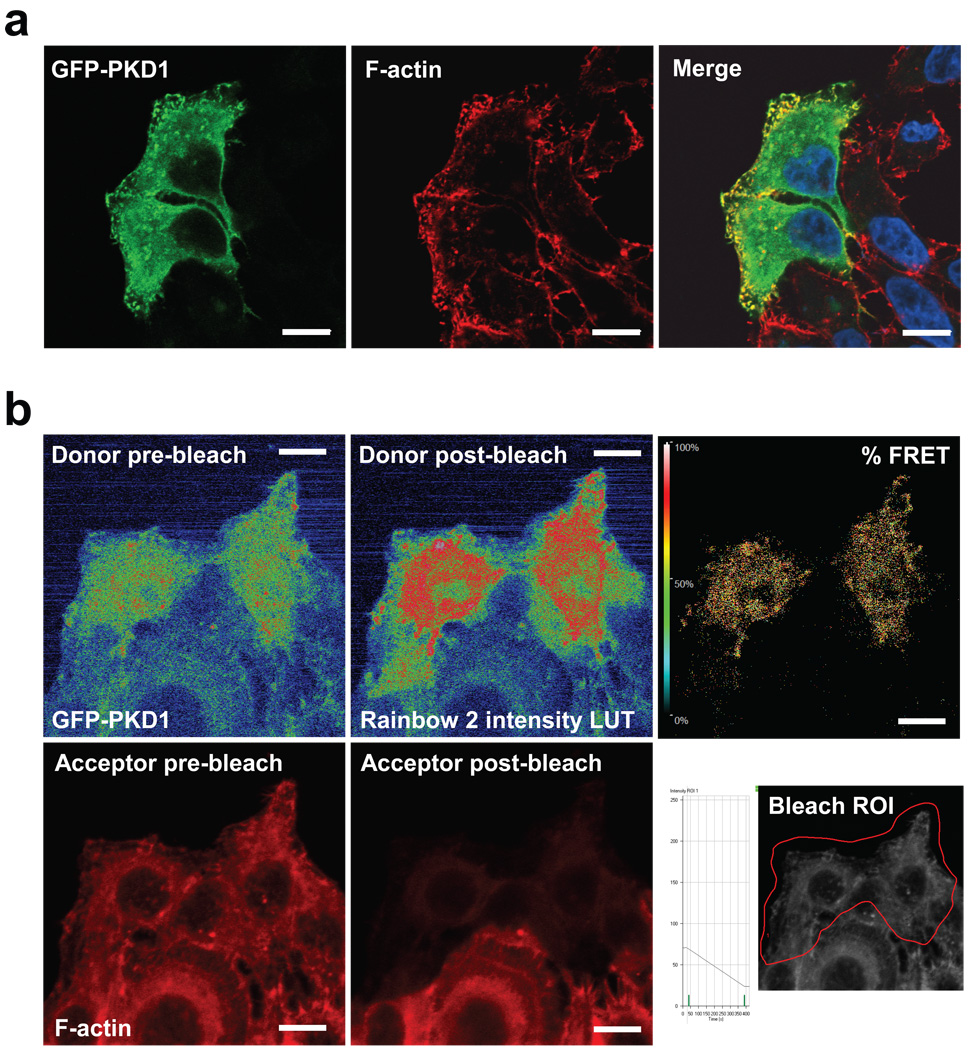
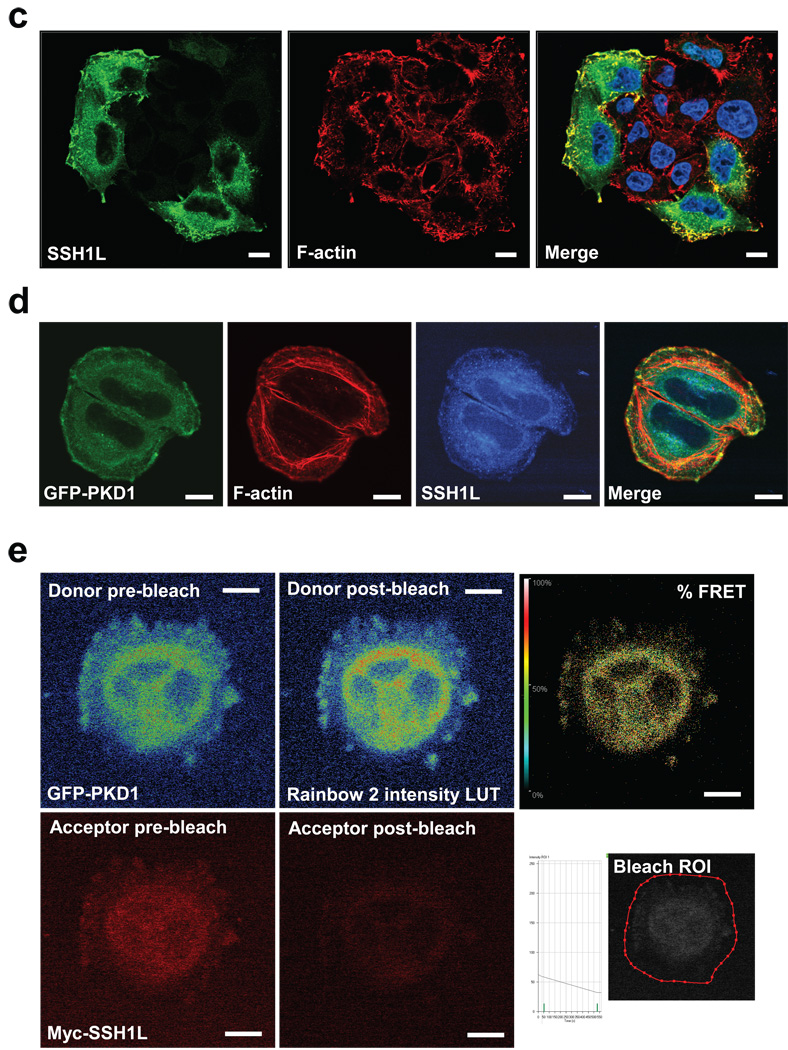
Immunohistochemistry of endogenous and overexpressed Protein Kinase D1 in cervical carcinoma cells revealed a co-localization with F-actin at peripheral F-actin-rich structures in membrane ruffles at the edge of lamellipodia (Fig. 1a, Supplementary information, Fig. S1A). This co-localization was independent of the PKD1 activity status (Supplementary information, Fig. S1B). To test whether PKD1 directly interacts with F-actin structures, we performed acceptor photobleach FRET studies in fixed samples of cells expressing GFP-tagged PKD1 (donor) and stained with Rhodamine-Phalloidin (F-actin: acceptor). FRET between the fluorophore-labelled proteins suggested binding or interaction of PKD1 and F-actin (Fig. 1b). These data are supported by recent in vitro studies showing that PKD1 binds to filamentous actin 21. Here we show in vivo evidence for such an interaction, further pointing to a novel regulatory function of PKD1 at the F-actin cytoskeletal compartment. Immunohistochemistry also revealed that SSH1L, a known F-actin binding protein 13, 24, similar to PKD1 localized to F-actin structures at the edge of lamellipodia and in membrane ruffles (Fig. 1c, Supplementary information, Fig. S1C). We therefore investigated the localization of PKD1 and SSH1L in relation to F-actin (Fig. 1d). Both proteins co-localize at the same filamentous actin structures at the periphery of membrane protrusions (Fig. 1d, merge). However, both also displayed considerable overlapping cytoplasmic localization, which is in line with single staining of both endogenous proteins (Supplementary information, Figs. S1A and S1C). Acceptor photobleach FRET analysis using GFP-tagged PKD1 as donor and Alexa Fluor 568-labeled myc-tagged SSH1L as acceptor showed direct interaction of both proteins in the cytoplasm as well as in membrane protrusions (Fig. 1e). In summary, this suggests a direct co-localization and interaction of PKD1 and SSH1L at F-actin-rich structures at the periphery of cells.
Figure 1. Co-localization of PKD1 with SSH1L at peripheral F-actin structures.
a, Immunofluorescence analysis of HeLa cells stained for PKD1 (GFP-PKD1) and F-actin (Rhodamine-Phalloidin) using confocal microscopy. Nuclei were stained with DAPI. b, PKD1 and F-actin are in proximity to facilitate Foerster energy transfer. HeLa cells were transfected with GFP-tagged PKD1 and stained for F-actin (Rhodamine-Phalloidin). A time bleach image series was acquired using a Zeiss LSM 510META confocal microscope with a Plan-Apochromat 63× /1.4 Dic oil immersion objective. Raw data was captured and analyzed using the LSM Software FRETplus Macro in Channel Mode. 4 pre- and post-bleach images of donor (GFP-PKD1) and acceptor (Rhodamine-Phalloidin) were recorded. The acceptor was photo-bleached using the 543 nm laser at 100 % transmission. The donor was exited at 488 nm using META-Detector Channels 490–543 for detection while the acceptor was exited at 543 nm using detection channels 560–635. The first pre- and post-bleach image of the series for donor and acceptor are shown. For better visualization of the donor fluorescence intensity a Rainbow2 LUT was applied to the images. The bleach ROI is shown below the % FRET image. c, Immunofluorescence analysis of HeLa cells stained for endogenous SSH1L and F-actin (Rhodamine-Phalloidin) using confocal microscopy. Nuclei were stained with DAPI. d, PKD1, SSH1L and F-actin co-localize at the edge of lamellipodia. HeLa cells were transfected with GFP-PKD1. Samples were stained with a α-SSH1L antibody directly labeled with Alexa Fluor 350 dye and with Rhodamine-Phalloidin. e, PKD1 and SSH1L-myc are in proximity to facilitate Foerster energy transfer. HeLa cells were transfected with GFP-PKD1 and Myc-SSH1L. SSH1L was indirectly labelled using α-myc and Alexa Fluor 568 antibodies. A time bleach image series was acquired. 3 pre- and 3 post-bleach images of donor (GFP-PKD1) and acceptor (Alexa Fluor 568) were recorded. The acceptor was photo-bleached using the 594 nm laser to reduce donor bleaching artefacts at 100 % transmission with a high number of iterations. FRET images were acquired and visualized as described above. In all experiments the scale bar represents 10 µm.
PKD1 regulates SSH1L by phosphorylation at S978
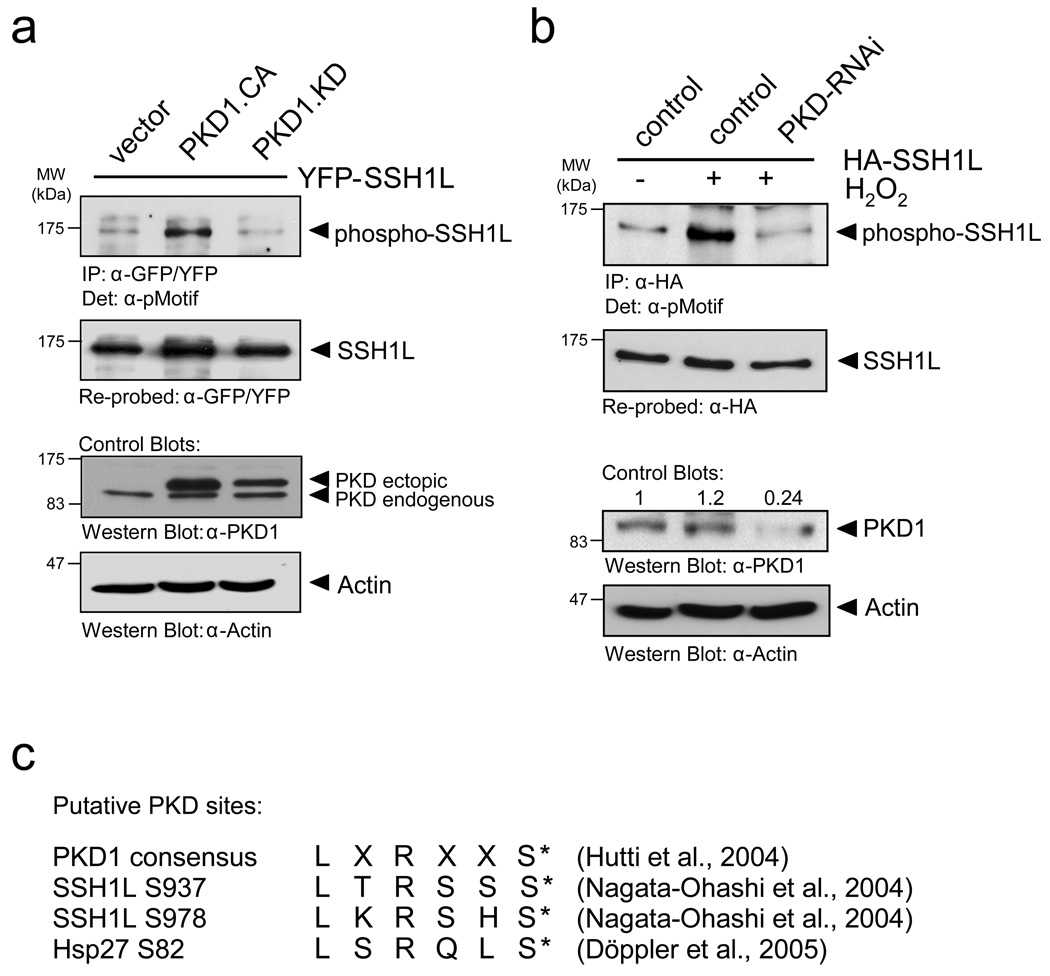
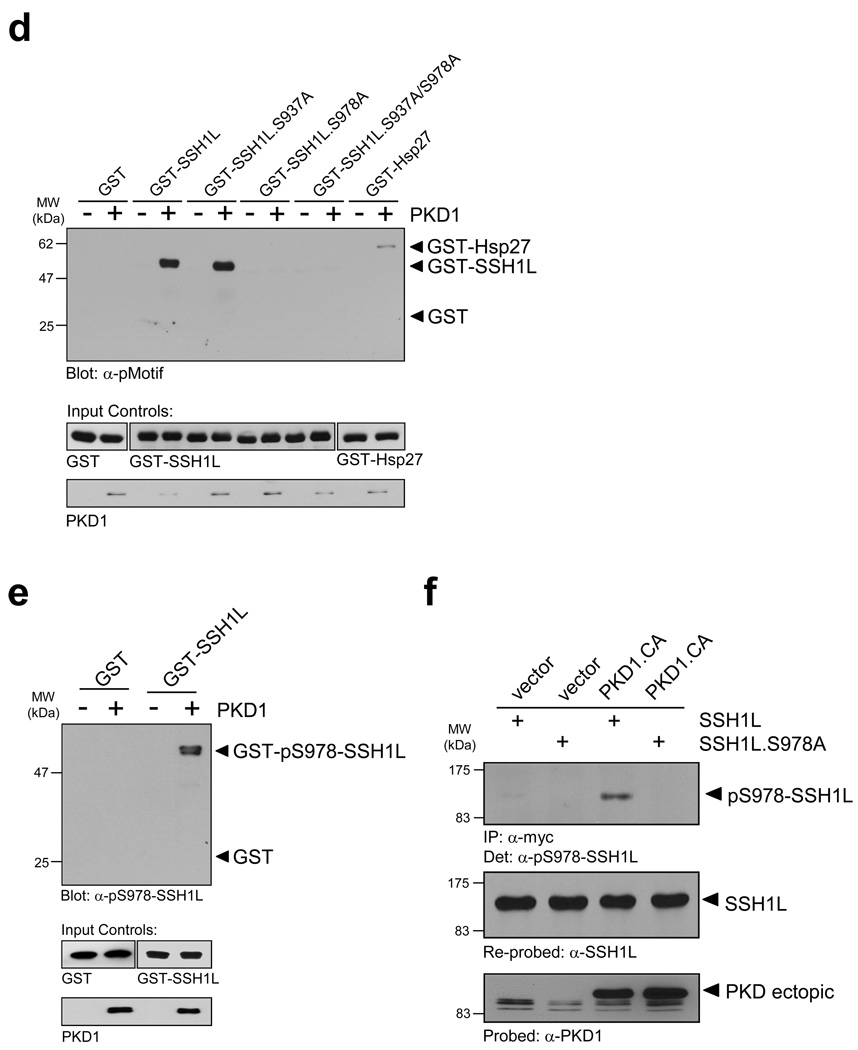
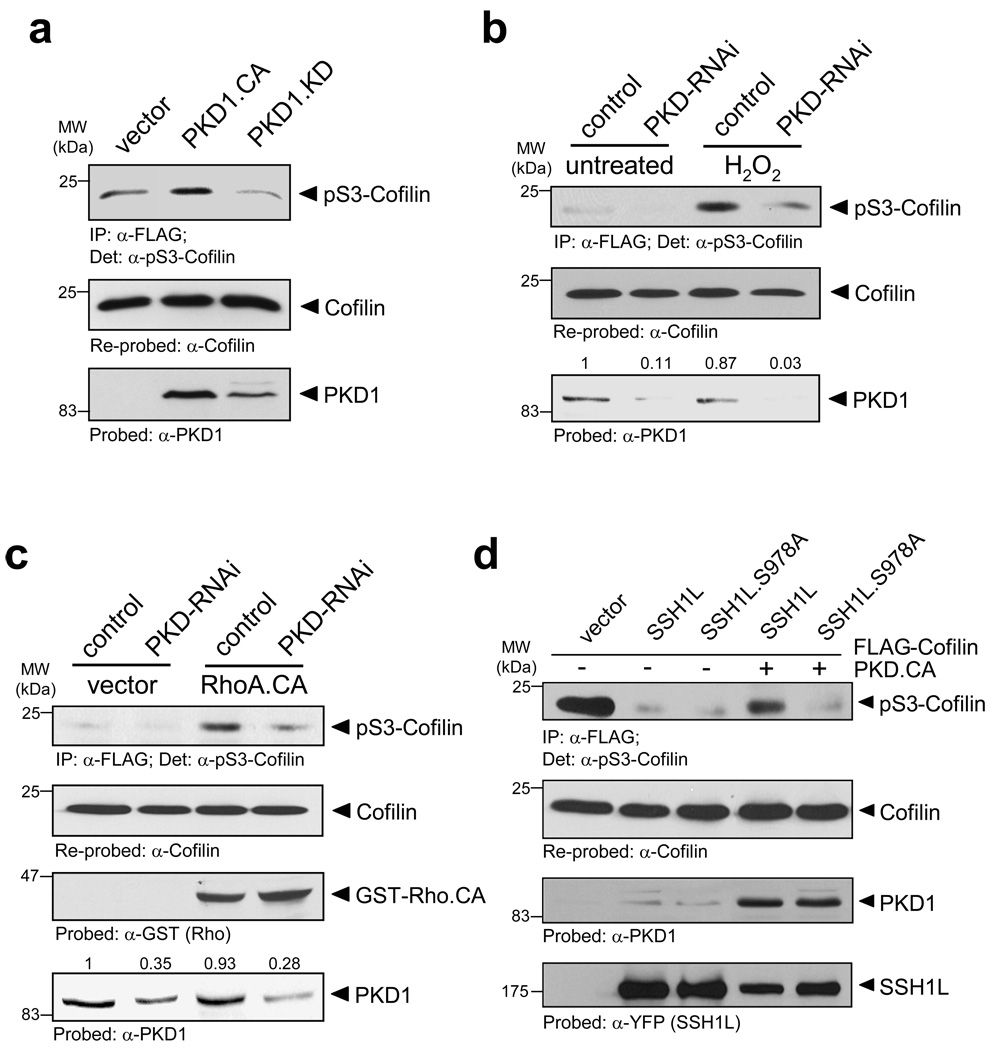
We next investigated if slingshot is a substrate for PKD1. To test this we utilized an antibody specifically designed to recognize PKD substrates (anti-pMotif), which has been characterized in detail before 25. We detected enhanced phosphorylation of SSH1L when co-expressed with active PKD1, and reduced phosphorylation when co-expressed with kinase-inactive PKD1, indicating that PKD1 mediates the phosphorylation of SSH1L in vivo (Fig. 2a). We also used reverse genetics where we knocked-down PKD with specific RNAi. To enhance PKD1 activity, cells were stimulated with hydrogen peroxide (H2O2), which is strong inducer of PKD1 activation 26–28. A knockdown of PKD1 reduced the H2O2-mediated phosphorylation of SSH1L as compared to the RNAi control, indicating that SSH1L indeed is a PKD1 substrate (Fig. 2b).
Figure 2. PKD1 phosphorylates SSH1L in vitro and in vivo.
a, HeLa cells were co-transfected with YFP-SSH1L and vector, constitutively-active PKD1 (PKD1.CA) or kinase-inactive PKD1 (PKD1.KD). SSH1L was immunoprecipitated with a α-GFP/YFP antibody and analyzed for phosphorylation by PKD1 using the α-pMotif PKD-substrate antibody. Blots were re-probed for SSH1L expression and control blots indicate PKD1 transgene expression (α-PKD1) or equal loading (α-actin). b, HeLa cells were transfected with RNAi control or PKD-RNAi. The next day cells were transfected with HA-SSH1L. After 48h, cells were stimulated with H2O2 (10 mM, 10 min) as indicated. SSH1L was immunoprecipitated (α-HA) and samples were analyzed using the α-pMotif PKD-substrate antibody. Blots were re-probed for SSH1L expression and control blots were probed with α-actin (loading control) or α-PKD1 (to show effective knockdown; numbers show knockdown of PKD1 relative to control). c, The PKD1 consensus motif indicates preference for serines with an arginine at -3 and leucine at -5 relative to the phosphorylated serine. Both serines, Ser937 and Ser978, in the serine-rich region of SSH1L fulfill the criteria of ideal phosphorylation consensus sequences. Also shown is the phosphorylation sequence of the known PKD1 substrate Hsp27. d, Purified GST-fusion proteins of SSH1L encompassing the putative PKD1 phosphorylation sites S937 and S978 as well as fusion proteins of SSH1L.S937A, SSH1L.S978A and SSH1L.S937A/S978A mutants were subjected a PKD1 kinase assay. Substrate phosphorylation was detected with the α-pMotif PKD-substrate antibody. Purified GST alone or GST-Hsp27 served as negative or positive controls. Control blots show substrate loading (α-GST) and PKD1 (α-PKD1). e, Purified GST (control) and GST-SSH1L were subjected to an in vitro kinase reaction with purified, active PKD1. Western blots of resolved proteins were probed with α-pS978-SSH1L. Control blots show substrate loading (α-GST) and PKD1 (α-PKD1). f, HeLa cells were co-transfected with Myc-tagged SSH1L or SSH1L.S978A and vector or constitutively-active PKD1 (PKD1.CA). SSH1L was immunoprecipitated (α-myc) and analyzed for phosphorylation at S978 (α-pS978-SSH1L). Blots were re-probed for SSH1L expression (α-SSH1L) and control blots were probed for PKD1 (α-PKD1). Uncropped images of Figs. 2a, 2d and 2f are shown in Supplementary information Fig. S6.
By analyzing the secondary structure of SSH1L for motifs that correspond to the optimal PKD1 consensus motif 29 we identified two putative phosphorylation sites (S937 and S978) (Fig. 2c). To analyze if PKD1 directly phosphorylates SSH1L at these sites, we performed in vitro kinase assays with purified, active PKD1 and bacterially-expressed, purified GST-tagged SSH1L encompassing the potential PKD1 phosphorylation sites as well as GST-SSH1L serine-to-alanine mutants of these residues (Fig. 2d). Our in vitro kinase assays implicated that PKD1 directly phosphorylates SSH1L at S978 but not S937. Following this initial mapping we confirmed the phosphorylation of S978 by PKD1 in vitro using a phosphosite-specific antibody directed against phospho-S978-SSH1L (Fig. 2e) 13. The phosphorylation of S978 in vivo was confirmed by co-expression of wildtype or SSH1L.S978A mutant SSH1L with active PKD1 (Fig. 2f). The S978A mutation of SSH1L abrogated the phosphorylation detected with the phospho-S978-SSH1L antibody, demonstrating both, the specificity of the used phospho-specific antibody and that S978 indeed is phosphorylated by PKD1 in vivo.
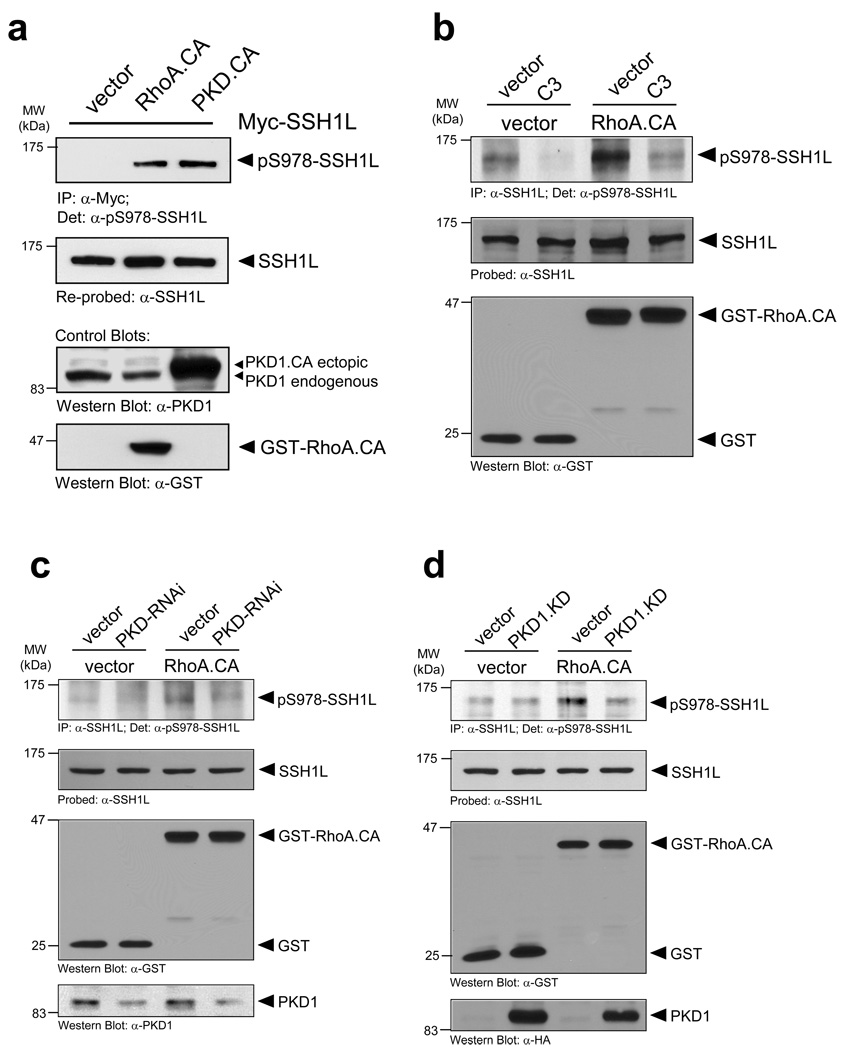
PKD1 regulates SSH1L downstream of RhoA
The Rho-GTPAse RhoA and slingshot both have been implicated in the regulation of directed cell migration 30. Further, PKD1 is activated downstream of RhoA 31, 32, but not RhoB or RhoC (Supplementary information, Fig. S2A). The expression of active RhoA and subsequent analysis of overexpressed SSH1L with the phospho-S978-SSH1L antibody revealed a RhoA-mediated regulation of the phosphorylation at this site (Fig. 3a). Next, we analyzed if RhoA-mediated activation of PKD1 regulates the phosphorylation of endogenous SSH1L at S978. Constitutively-active RhoA (RhoA.CA) clearly enhanced the phosphorylation of SSH1L at S978, which was reduced when cells were treated with the Rho-inhibitor C3 (Fig. 3b, Supplementary information, Fig. S2B). Similar results were obtained when SSH1L was overexpressed (Supplementary information, Fig. S2C). Interestingly treatment of cells with C3 also released PKD1 from the F-actin structures at the leading edge (Supplementary information, Fig. S2C). To directly link RhoA-mediated phosphorylation of SSH1L to PKD1, we expressed a kinase-dead PKD1 or knocked-down PKD in cells and tested their responsiveness to active RhoA using SSH1L phosphorylation as readout. We found that both, loss of PKD1 kinase activity or the knockdown of PKD blocked RhoA-mediated phosphorylation of SSH1L at S978 (Figs. 3c, 3d, Supplementary information, Figs. S2D, S2E). This implicates that SSH1L is a PKD1 substrate downstream of active RhoA.
Figure 3. PKD1 phosphorylates SSH1L downstream of RhoA.
a, Active RhoA and active PKD1 mediate SSH1L phosphorylation at S978. HeLa cells were co-transfected with myc-tagged SSH1L and vector, constitutively-active RhoA (RhoA.CA) or PKD1 (PKD1.CA). SSH1L was immunoprecipitated (α-myc) and samples were analyzed with α-pS978-SSH1L. Blots were re-probed for SSH1L (α-SSH1L). Control blots showing PKD1 and RhoA.CA transgene expression were performed by Western blotting using anti-PKD1 or anti-GST antibodies as indicated. b, Phosphorylation of endogenous SSH1L is dependent on RhoA. HeLa cells were transfected with vector control or active RhoA.CA and treated with C3 transferase as indicated. SSH1L was immunoprecipitated (α-SSH1L) and analyzed for phosphorylation at S978 (α-pS978). Blots were re-probed for SSH1L. Control blots showing RhoA.CA transgene expression were performed by Western blotting using α-GST antibodies. c, HeLa cells were transfected with RNAi control or PKD-RNAi. The next day cells were transfected with vector or constitutively-active RhoA (RhoA.CA). Endogenous SSH1L was immunoprecipitated (α-SSH1L) and analyzed for PKD1-mediated phosphorylation (α-pS978). Blots were re-probed for SSH1L (α-SSH1L). Control blots showing PKD1 (numbers show knockdown of PKD1 relative to control) and RhoA.CA transgene expression were performed by Western blotting using α-PKD1 or α-GST antibodies as indicated. A quantification of SSH1L phosphorylation at S978 from four different experiments is depicted in Fig. S6 (supplemental information). d, HeLa cells were co-transfected with vector control or PKD1.KD and constitutively-active RhoA (RhoA.CA). Endogenous SSH1L was immunoprecipitated (α-SSH1L) and analyzed for PKD1-mediated phosphorylation (α-pS978). Blots were re-probed for SSH1L (α-SSH1L). Control blots showing PKD1 and RhoA.CA transgene expression were performed by Western blotting using α-PKD1 or α-GST antibodies as indicated. Uncropped images of Fig. 3d are shown in Supplementary information Fig. S6.
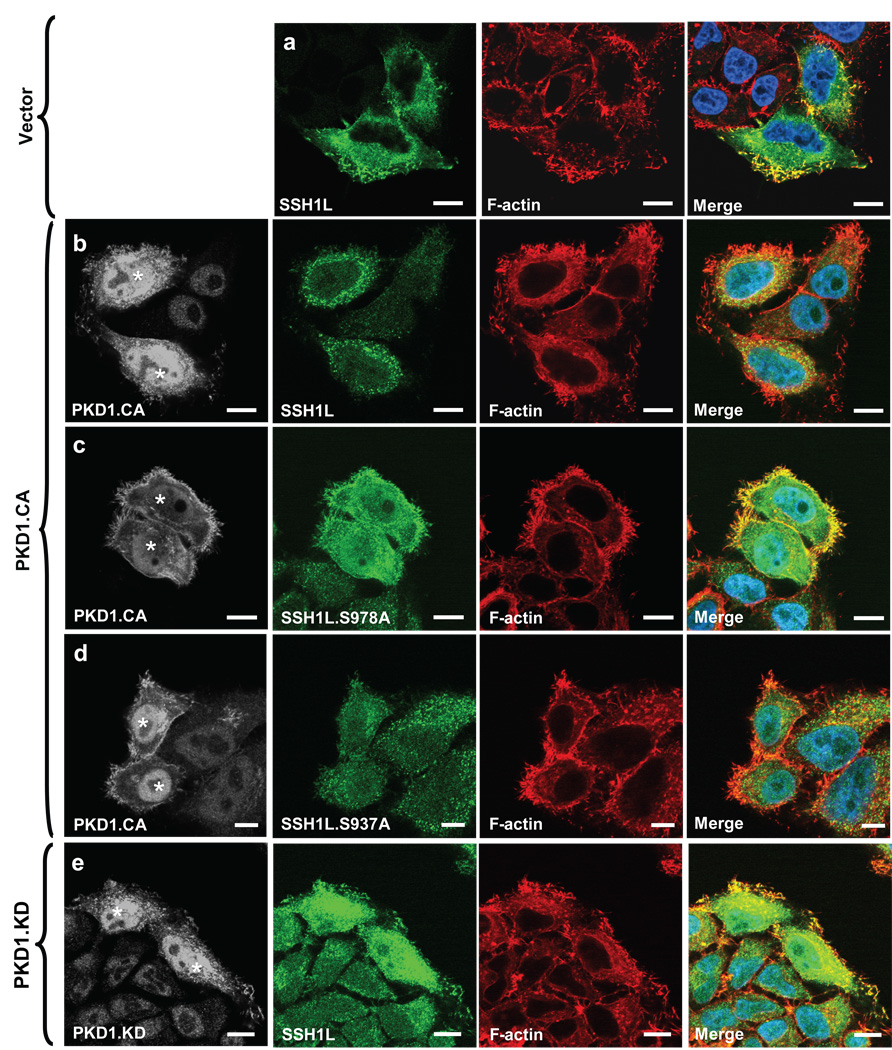
Active PKD1 regulates the dissociation of SSH1L from the actin cytoskeleton
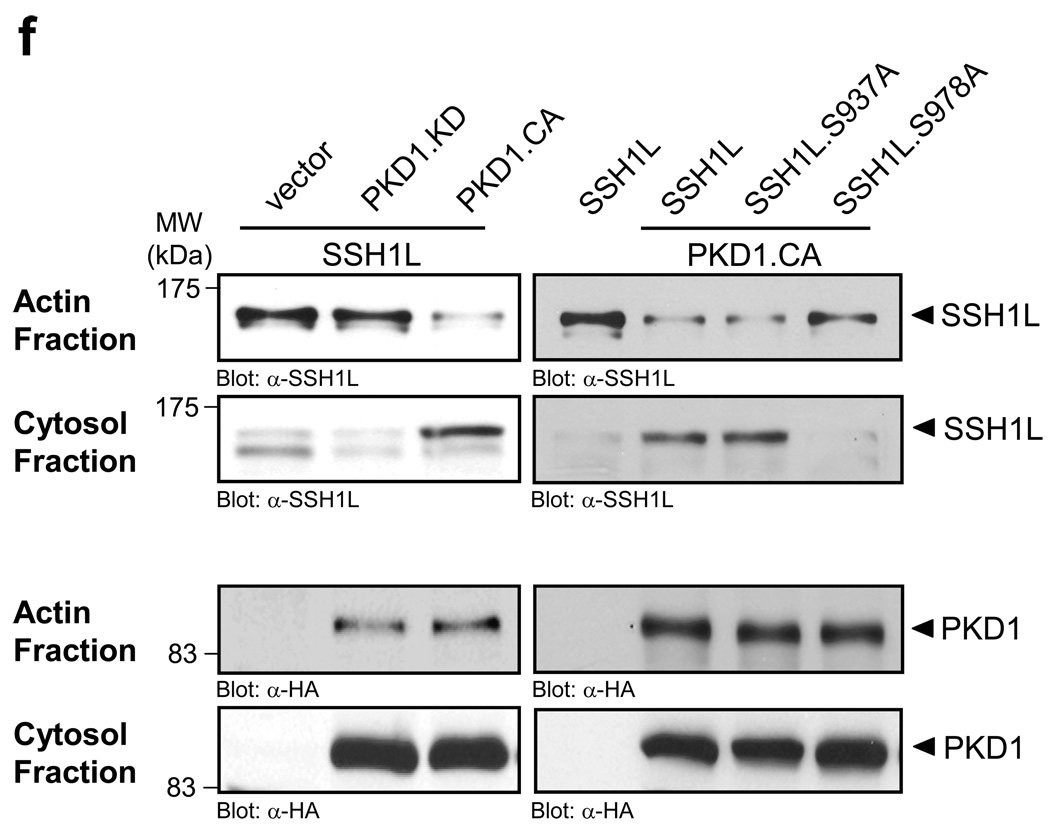
Next, we investigated the consequences of PKD1-mediated phosphorylation of SSH1L at S978. We co-expressed active PKD1 (PKD1.CA) with wildtype or S978A or S937A mutants of SSH1L to investigate their subcellular localization in relation to F-actin. In absence of active PKD1, wildtype SSH1L and the mutants strongly co-localized with F-actin-rich structures at the periphery of the cell (Supplementary information, Fig. S2A, Fig. 4a). However, following expression of active PKD1, SSH1L and the SSH1L.S738A mutant mostly displayed cytoplasmic staining (Figs. 4b, 4d). In contrast, the SSH1L.S978A mutant was still localized at F-actin structures at the cell periphery (Fig. 4c). The expression of kinase-inactive PKD1 (PKD1.KD) did not lead to a redistribution of wildtype SSH1L underlining the importance of PKD1 kinase activity for this regulation (Fig. 4e). We confirmed the obtained immunofluorescence data with a biochemical approach. Therefore, we analyzed the F-actin-containing and cytosolic fractions of cells. In control cells SSH1L was mainly localized in the F-actin fraction, whereas in presence of active PKD1, SSH1L was increasingly found in the cytosolic fraction (Fig. 4f). Further, active PKD1, also increased the abundance of an SSH1L.S937A mutant in the cytosolic fraction, whereas an SSH1L.S978A mutant, which is not targeted by PKD1, remained almost exclusively in the F-actin fraction (Fig. 4f). Taken together, PKD1, independent of its kinase activity, is localized at peripheral F-actin structures such as membrane ruffles or lamellipodia. The localization of SSH1L at these structures, however, is heavily dependent on its phosphorylation at serine residue S978, which is mediated by PKD1.
Figure 4. PKD1-mediated phosphorylation regulates SSH1L localization.
a–e, HeLa cells were co-transfected with vector control (a), constitutively-active PKD1 (PKD1.CA) (b–d) or kinase-dead PKD1 (PKD1.KD) (e) and Myc-tagged SSH1L, SSH1L.S937A orSSH1L.S978A (as indicated). Samples were subjected to indirect immunofluorescence analysis. SSH1L was detected using α-myc and Alexa Fluor 568 secondary antibodies. F-actin was stained with Alexa Fluor 633-Phalloidin and nuclei were stained with DAPI. PKD1 expression was detected using a HA-specific antibody and α-rabbit-Alexa Fluor 488 as a secondary antibody. The nuclear staining in the PKD1 samples is non-specific background. Samples were analyzed using a Zeiss LSM 510META confocal microscope with a Plan-Apochromat 63×/1.4 Dic oil immersion objective in Multi-Track-configuration. For visualization purposes, PKD1 images are presented in grayscale, SSH1L in green and Phalloidin in red. Double transfected cells (PKD1 and SSH1L) are marked with asterisks. Images shown depict single confocal sections. The scale bar represents 10 µm. f, HeLa cells were co-transfected with vector control, constitutively-active PKD1 (PKD1.CA) or kinase-dead PKD1 (PKD1.KD) and Myc-tagged SSH1L, SSH1L.S937A or SSH1L.S978A as indicated. Actin fraction or cytosolic fraction of cells were prepared and analyzed for SSH1L (α-SSH1L). Control blots for PKD1 expression were performed using α-HA antibodies. Uncropped images of Fig. 4f are shown in supplementary information Fig. S6. Control figures for 4b, 4c and 4d are shown in Supplementary information S3A.
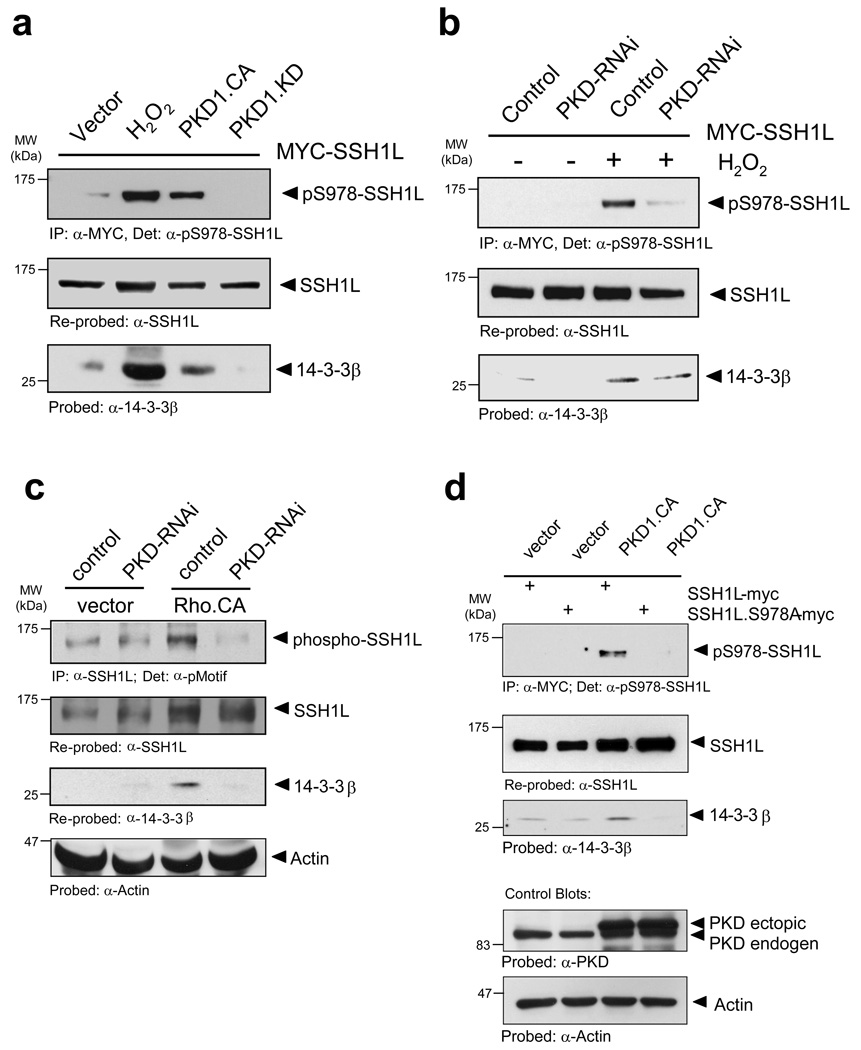
PKD1-mediated phosphorylation of SSH1L facilitates binding of 14-3-3
We next sought to identify the cellular mechanisms of how PKD1-mediated phosphorylation regulates SSH1L localization on a molecular level. Previously, Nagata-Ohasi et al. suggested that both S937 and S978 form the core motif of a binding site for 14-3-3β or 14-3-3ζ 13. Further, Yamamoto et al. described a F-actin binding motif (L-K-R) within the L-K-R-S-H-S978 phosphorylation motif surrounding S978 of SSH1L 24. We therefore hypothesized that PKD1-mediated phosphorylation of SSH1L at S978 leads to 14-3-3 binding and abrogates F-actin binding of this site, ultimately resulting in altered subcellular localization of SSH1L.
To test this we analyzed if the binding of SSH1L to 14-3-3 proteins is mediated by PKD1. Therefore, we expressed a constitutively-active PKD1, which enhanced S978 phosphorylation of SSH1L and also increased its binding to 14-3-3β (Fig. 5a). Oxidative stress, which potently activates PKD1, served as a positive control and also led to 14-3-3β binding to SSH1L. The expression of kinase-dead PKD1 acted in a dominant-negative manner and suppressed both S978 phosphorylation and binding of 14-3-3β proteins (Fig. 5a). Similar results were obtained with 14-3-3ζ, implicating that both 14-3-3 isoforms interact with SSH1L in a PKD1-mediated fashion (Supplementary information, Fig. S3B). Further, the depletion of PKD by RNA-interference reduced the phosphorylation of SSH1L at S978 as well as the binding of 14-3-3β in response to the potent PKD1 activator H2O2 or the expression of active RhoA (Figs. 5b, 5c). In order to demonstrate that 14-3-3 proteins indeed bind to SSH1L via the S978 residue, we compared PKD1-mediated binding of 14-3-3β proteins to wildtype SSH1L and SSH1L.S978A mutant. The S978A mutation abolished the co-precipitation of 14-3-3β indicating that 14-3-3β interaction with SSH1L is completely dependent on the 14-3-3 binding motif created at the PKD1 phosphorylation site (Fig. 5d).
Figure 5. Phosphorylation of SSH1L at S978 controls its association with 14-3-3 proteins.
a, HeLa cells were transfected with myc-SSH1L and vector, constitutively-active PKD1 (PKD1.CA) or kinase-dead PKD1 (PKD1.KD). Where indicated, cells were stimulated with H2O2 (10 mM, 10 min). SSH1L was immunoprecipitated (α-myc) and samples were stained for phosphorylation at S978 (α-pS978-SSH1L) and re-probed for total SSH1L (α-SSH1L). Co-precipitation of 14-3-3 was detected using a α-14-3-3β antibody. b, HeLa cells were transfected with control or PKD-RNAi. Next day, cells were transfected with Myc-SSH1L. After 48 h, cells were stimulated with H2O2 (10 mM, 10 min) as indicated. SSH1L was immunoprecipitated (α-myc) and samples were stained for phosphorylation at S978 (α-pS978-SSH1L) and re-probed for SSH1L (α-SSH1L). Co-precipitation of 14-3-3 was detected using a α-14-3-3βantibody. c, HeLa cells were transfected with RNAi control or PKD-RNAi. Next day, cells were transfected with vector or constitutively-active RhoA (RhoA.CA). Endogenous SSH1L was immunoprecipitated (α-SSH1L) and analyzed for co-immunopecipitation of 14-3-3β (α-14-3-3β). As control, samples were also stained for SSH1L phosphorylation at S978 (α-pMotif) and equal loading (α-actin). d, HeLa cells were co-transfected with Myc-tagged SSH1L or SSH1L.S978A and vector or constitutively-active PKD1 (PKD1.CA). SSH1L was immunoprecipitated (α-myc) and analyzed for co-immunopecipitation of 14-3-3 (α-14-3-3β). As control, samples were also stained for SSH1L phosphorylation at S978 (α-pS978-SSH1L) or equal loading (α-actin). e, HeLa cells were transfected with Myc-tagged SSH1L or SSH1L.S978A and HA-14-3-3ζ. 14-3-3ζ was stained using a α-HA (rat) antibody followed by detection with α-rat Alexa Fluor 488. SSH1L was detected using α-myc and Alexa Fluor 568 antibodies. F-actin was stained with Alexa Fluor 633-Phalloidin and nuclei with DAPI. Samples were analyzed using a Zeiss LSM 510META confocal microscope with a Plan-Apochromat 63×/1.4 Dic oil immersion objective in Multi-Track-configuration. E1–E4: Controls transfected with SSH1L and co-stained for F-actin. E5–E8: Cells transfected with SSH1L, 14-3-3ζ and co-stained for F-actin. E9–E12: Cells transfected with SSH1L.S978A and co-stained for F-actin. E13-E16: Cells transfected with SSH1L.S978A, 14-3-3ζ and co-stained for F-actin. Images show single confocal sections. The scale bar represents 10 µm. Uncropped images of Figs. 5c and 5d are shown in Supplementary information Fig. S6.
We then analyzed, if binding to 14-3-3ζ alters the cellular localization of SSH1L. In control-transfected cells, SSH1L or the SSH1L.S978A mutant displayed a broad co-localization with F-actin structures (Fig. 5e, E1–E4 and E9–E12). Cells expressing wildtype SSH1L and 14-3-3ζ, displayed a strong co-localization of both proteins and a sequestration of SSH1L in the cytoplasm away from F-actin structures (Fig. 5e E5–E8, middle row). In contrast the co-expression of SSH1L.S978A with 14-3-3ζ showed no such phenotype and SSH1L.S978A remained localized at F-actin-rich structures in the cell periphery, whereas 14-3-3ζ was mainly localized in the cytoplasm (Fig. 5e E13-E18, bottom row). These data suggest that binding of 14-3-3 proteins to the phosphorylated PKD1 consensus motif at S978 is the most likely mechanism, by which SSH1L subcellular localization is regulated via PKD1.
PKD1 regulates Cofilin phosphorylation
The localization of SSH1L to F-actin enhances SSH1L phosphatase activity towards cofilin 13. Therefore, we next determined if the regulation of SSH1L by PKD1 had an impact on its activity towards cofilin. Cofilin phosphorylation at serine residue 3 (S3) can be used as a direct measure for cofilin activity and decreased phosphorylation of this residue is due to increased activity of cofilin phosphatases such as SSH1L 13, 14.
Increased PKD activity as mimicked with a constitutively-active PKD1 mutant resulted in increased cofilin S3-phosphorylation, and the expression of a kinase-dead PKD1 allele decreased cofilin phosphorylation at this site (Fig. 6a). We also compared cells depleted from PKD (using specific RNAi) to cells treated with control RNAi and analyzed the cofilin phosphorylation status in response to PKD1 activation with oxidative stress or active RhoA (RhoA.CA). Both, oxidative stress as well as RhoA activation enhanced cofilin phosphorylation at S3, implicating an increased inhibition due to PKD1-mediated signalling events (Figs. 6b, 6c, Supplementary information, Fig. S4C). Our previous data show that both activators of PKD1, in a PKD1-dependent fashion, regulate SSH1L by phosphorylation. Subsequently, this data suggests that SSH1L once phosphorylated by PKD1 and sequestered by 14-3-3 proteins is inactive towards phospho-S3 cofilin. Finally, we tested if the effects on the cofilin S3 phosphorylation signal are indeed due to PKD1-mediated phosphorylation of SSH1L at S978. Therefore, we co-expressed active PKD1 with wildtype or S978A mutant SSH1L and analyzed cofilin phosphorylation at serine residue 3 as a measure of cofilin activity (Fig. 6d). Both, wildtype SSH1L and the SSH1L.S978A mutant decreased basal phosphorylation levels of cofilin. However, the expression of active PKD1 increased the phosphorylation of wildtype cofilin at S3, while there was no increase in cells expressing SSH1L.S978A. This indicates that the effects of PKD1 exerted on cofilin S3-phosphorylation are mediated via its phosphorylation of SSH1L at Ser978.
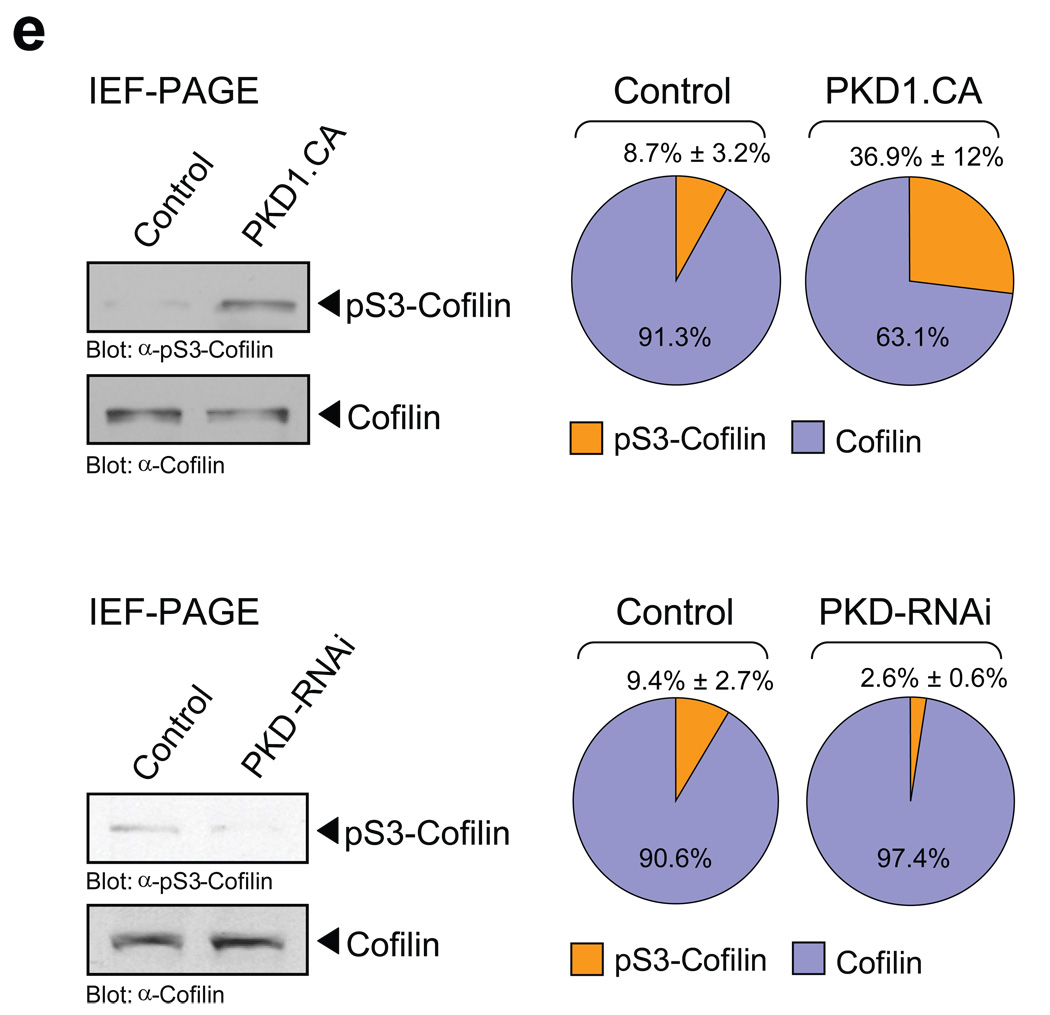
Figure 6. PKD1 regulates cofilin S3-phosphorylation.
a, HeLa cells were transfected with vector, constitutively-active PKD1 (PKD1.CA) or kinase-dead PKD1 (PKD1.KD) and FLAG-tagged cofilin. Cofilin was immunoprecipiated (α-FLAG), cofilin phosphorylation was detected with α-pS3-cofilin antibody and samples were re-probed for total cofilin (α-FLAG). The expression of PKD1 was verified by Western blotting (α-PKD1). b, HeLa cells were transfected with control RNAi or PKD-RNAi. The next day, cells were transfected with FLAG-tagged cofilin. Cofilin was immunoprecipiated (α-FLAG), cofilin phosphorylation was detected with α-pS3-cofilin antibody and samples were re-probed for total cofilin (α-FLAG). The knockdown of PKD1 was verified by Western blotting (α-PKD1) and numbers show knockdown of PKD1 relative to control. A quantification of cofilin phosphorylation at S3 from three different experiments is depicted in Fig. S9A (supplemental information). c, HeLa cells were transfected with control RNAi or PKD-RNAi. The next day, cells were transfected with FLAG-tagged cofilin and constitutively-active RhoA (RhoA.CA). Cofilin was immunoprecipiated (α-FLAG), cofilin phosphorylation was detected by probing with α-pS3-cofilin antibody and samples were re-probed for total cofilin (α-FLAG). The expression of RhoA and the knockdown of PKD1 were verified by Western blotting (α-GST and α-PKD1). Numbers show knockdown of PKD1 relative to control. A quantification of cofilin phosphorylation at S3 from three different experiments is depicted in Fig. S9B (supplemental information). d, HeLa cells were co-transfected with FLAG-tagged cofilin, vector control, SSH1L, SSH1L.S978A and constitutively-active PKD1 (PKD1.CA) as indicated. Cofilin was immunoprecipiated (α-FLAG), cofilin phosphorylation was detected by probing with α-pS3-cofilin antibody and samples were re-probed for total cofilin (α-FLAG). The expression of PKD1 and SSH1L was verified by Western blotting (α-PKD1 or α-YFP). e, HeLa cells were transfected with vector control, constitutively-active PKD1 (PKD1.CA), RNAi control or PKD1-RNAi as indicated. Cell lysates were subjected to IEF-PAGE to separate endogenous S3-phosphorylated and unphosphorylated cofilin. Samples were analyzed with α-S3-cofilin and α-cofilin antibodies. The Western blot show representative experiments. The ratio of phospho-cofilin/cofilin of three independent experiments is depicted in the right panel. Uncropped images of Fig. 6a are shown in Supplementary information Fig. S6.
Finally, we determined if PKD1 alters endogenous cofilin phosphorylation. To test this we subjected cell lysates of control cells or cells transfected with constitutively-active PKD1 to IEF-PAGE. Active PKD1 increased the levels of phospho-S3-cofilin (Fig. 6e). A quantitative analysis of the percent of phospho-cofilin determined from total cofilin suggests that active PKD1 increases endogenous phospho-cofilin levels in HeLa cells from 8.7% ± 3.2% of the total cofilin in untreated cells to 36.9% ± 12%. We also found that endogenous, basal phospho-S3-cofilin levels are decreased, when cells are depleted from PKD. The knockdown of PKD decreased phospho-S3-cofilin levels as compared to control cells from 9.4% ± 2.7% of the total cofilin in control cells to 2.6% ± 0.6% (Fig. 6e).
PKD1 inhibits barbed end formation and directed cell migration
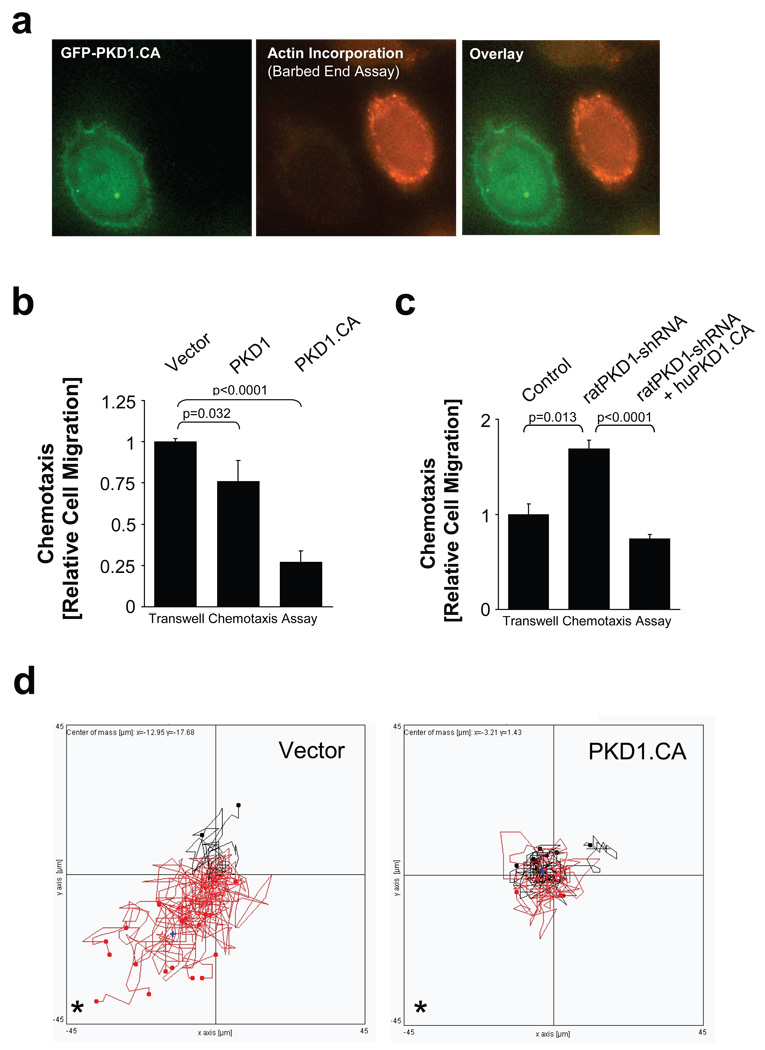
The activity status of the cofilin pathway and the subsequent generation of barbed ends correlate with the ability of cells to migrate towards a chemotactic stimulus 4, 33. Since PKD1 negatively regulates the SSH1L-mediated dephosphorylation and activation of cofilin, we hypothesized that this also has an impact on the cells’ ability to migrate. To test this we first determined the effect of active PKD1 on free actin barbed end filament formation. We found that active PKD1 completely blocked free actin barbed end formation, implicating an important role of PKD1 in blocking cofilin-mediated cell migration (Fig. 7a; Supplementary information, Figs. S5A, S5B).
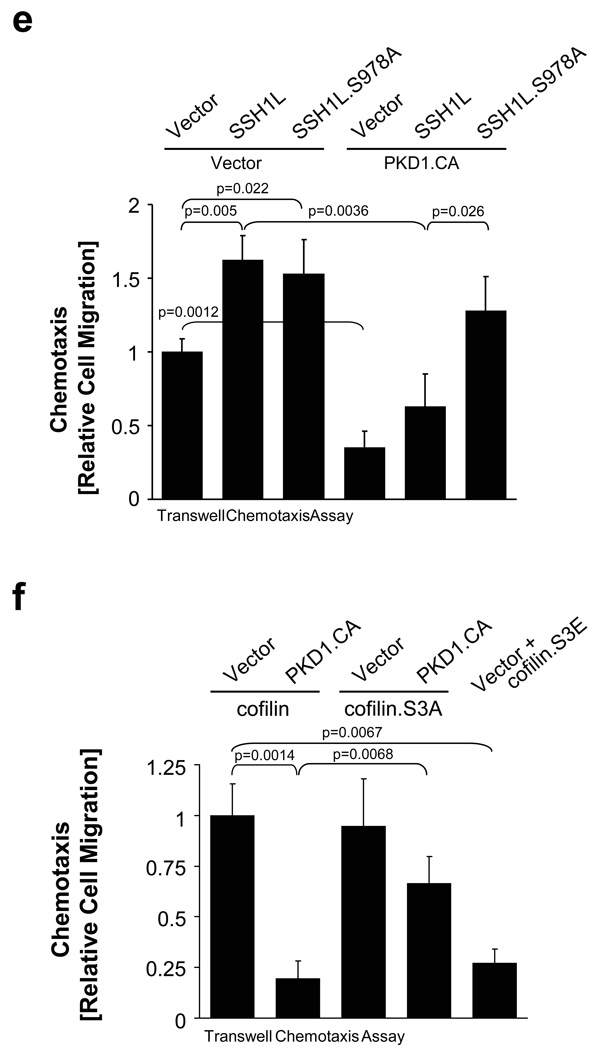
Figure 7. PKD1 regulates the formation of free actin filament barbed ends and cell migration.
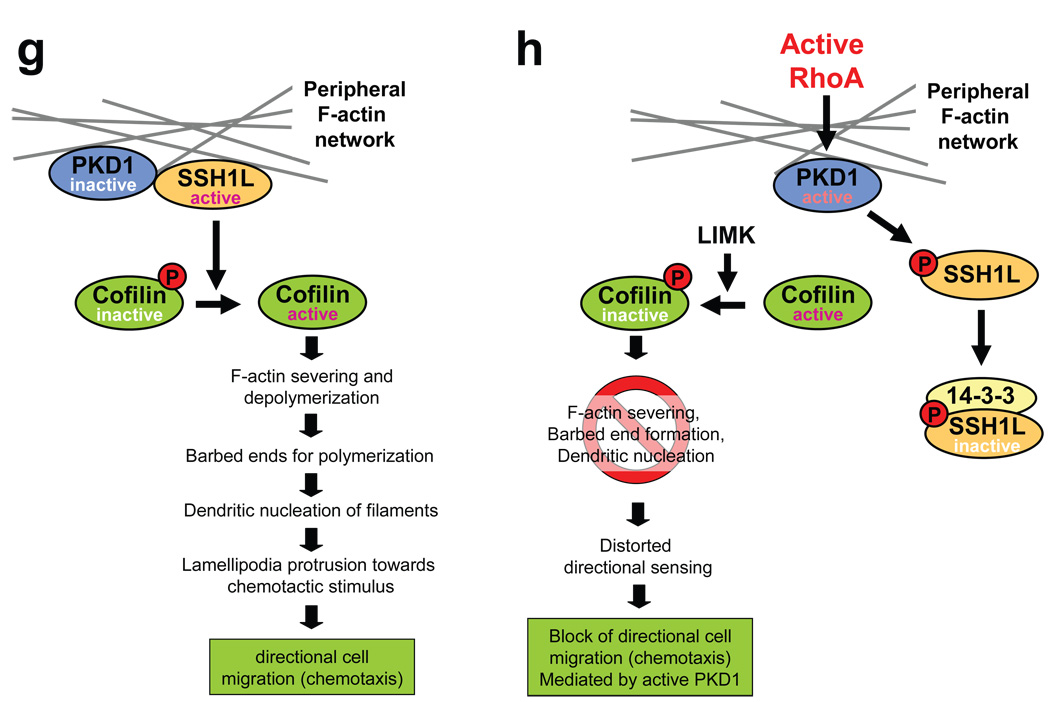
a, MTLn3 cells were transfected with constitutively-active GFP-PKD1 and a free actin filament barbed end assay performed. Depicted is a PKD1-transfected cell (green) next to a non-transfected cell (see also Fig.S10). Free barbed ends are shown in red. b, MTLn3 cells were transfected with vector, wildtype or PKD1.CA. Transwell chemotaxis assays were performed for 4 h. Statistically-significant changes (student T-Test) to the control are marked with asterisks. c, MTLn3 cells were transfected with control shRNA or shRNA specific for rat PKD1 as well as human PKD1.CA as indicated. Transwell chemotaxis assays were performed for 4 h. Statistically-significant changes (student T-Test) to control are marked with asterisks. d, MTLn3 cells were transfected with vector control or PKD1.CA. Cells were seeded on µ-Slides Chemotaxis chambers and induced to migrate for 6 h towards NIH-3T3-conditioned medium. Data obtained from several experiments were pooled. The asterisk marks the direction of the gradient. The blue cross marks the centre of mass (vector control:×= −12.95 µm, y = −17.68 µm; PKD1.CA (x = −3.21 µm, y = 1.43 µm). Tracks of cells overall migrating towards the gradient are shown in red, tracks of cells overall migrating in the other direction in black. e, MTLn3 cells were transfected with as indicated. Transwell chemotaxis assays were performed for 4 h. Statistically-significant changes (student T-Test) to the respective controls are marked with asterisks. f, MTLn3 cells were transfected as indicated. Transwell chemotaxis assays were performed for 4 h. Statistically-significant changes (student T-Test) to the respective controls are marked with asterisks. In 7b, 7c, 7e, 7f error bars represent standard deviation (n=3). g, h, In migrating cells active PKD1 co-localizes with active SSH1L at F-actin filaments. This allows SSH1L to dephosphorylate cofilin and keep it in an active state. Active cofilin induces actin severing and depolymerisation contributing to directional cell migration. After its activation PKD1 phosphorylates SSH1L at S978. This mediates 14-3-3 binding and inactivation of SSH1L. LIMK keeps cofilin in a phosphorylated, inactive state, blocking F-actin severing, barbed end formation and directional cell migration.
We next analyzed wildtype, constitutively-active PKD1 or inactive mutants of PKD1 for their effects on directional cell migration in Boyden chamber/Transwell chemotaxis assays. Constitutively-active PKD1 significantly decreased cell migration in both HeLa and MTLn3 cells (Fig. 7b; Supplementary information, Fig. S5C). Interestingly, shRNA directed against PKD1 or the expression of kinase-dead (PKD1.KD) or inactive PKD1 (PKD1.SSAA) mutants increased cell motility (Fig. 7c; Supplementary information, Fig. S5D). Additionally to the transwell chemotaxis assays, we performed cell tracking assays where we compared directed cell migration of MTLn3 cells expressing constitutively-active PKD1 or vector control towards NIH-3T3 conditioned medium using Chemotaxis μ-Slides (Fig. 7d). Control cells showed an average velocity of 24 µm/h and a directionality of 0.21, whereas cells transfected with constitutively-active PKD1 showed an average velocity of 18 µm/h and a directionality of 0.08. These effects on directed cell motility confirmed the results obtained with the transwell chemotaxis assays. Notably, PKD1 and mutants showed no significant effects on random cell migration (Supplementary information, Fig. S5E), suggesting a specific role of PKD1 in directed cell migration.
We next determined if PKD1 exerts its inhibitory function on directed cell migration through its activity on SSH1L. The co-expression of active PKD1 with SSH1L significantly decreased directed cell migration as compared to SSH1L alone. This is rescued by the SSH1L.S978A mutant that can not be phosphorylated by PKD1 (Fig. 7e). Since slingshots directly regulate phospho-cofilin levels, we then determined if constitutively-active PKD1 impacts cofilin-mediated cell migration (Fig. 7f). We found that cells expressing wildtype cofilin or a cofilin.S3A mutant, which can not be regulated by SSH1L and LIMK showed increased cell migration. The co-expression of active PKD1 inhibited the effects on cell migration mediated by the wildtype cofilin, but did not block effects mediated by the cofilin.S3A mutant (Fig. 7f). This is most likely due to PKD1-mediated regulation of SSH1L. As a control a cofilin.S3E mutant mimicking the phosphorylation of this site inhibited cell migration. Taken together, our data identify PKD1 as a key regulator of the SSH1L-cofilin signalling pathway and directed cell migration.
DISCUSSION
Rapid actin severing and polymerization at the leading edge of migrating or invading tumour cells require a cycling process of ADF/cofilin activation and inactivation 2, 6, 7, which is mediated by phosphorylation through LIMK and dephosphorylation by slingshot phosphatases 10, 11. Relatively little is known about the regulation of slingshot and by demonstrating its phosphorylation by PKD1 we here describe a new regulation mechanism for this phosphatase.
RhoGTPases are key-regulators of cell migration and control many processes of cell adhesion and actin dynamics. Cdc42 and Rac1 have been implicated in regulating the formation of filopodia and lamellipodia, whereas RhoA is important to inhibit actin severing at the leading edge 34. PKD1 is activated downstream of RhoA 32, however, the functions of this activation are not well defined. We found that RhoA-mediated activation of PKD1 leads to phosphorylation of SSH1L at S978 and subsequent inhibition of cofilin (Fig. 3, Fig 6). Interestingly, another PKD1 activation pathway, mediated by oxidative stress, also leads to similar SSH1L regulation (Fig. 3, Fig 6). Activation of PKD1 in response to both stimuli is mediated by novel PKC (nPKC) enzymes, which induce PKD1 activation loop phosphorylation 35. The activation of both nPKC and PKD1 is dependent on the lipid co-activator DAG, which targets these kinases to membrane structures within cells 36–38. Cleavage of PIP2 by PLC releases cofilin from its complex with PIP2 at the plasma membrane 39. This cleavage also generates DAG, which could recruit and activate PKD1 and its upstream kinases. Once activated, PKD1 then phosphorylates its substrate SSH1L, regulating its activity and subcellular localization.
Serine residue 978 is located within a potential actin binding motif in SSH1L 24. SSH1L activity is strongly increased after binding to F-actin resulting in the dephosphorylation and activation of cofilin 13. We therefore hypothesized that the phosphorylation of this residue by PKD1 regulates SSH1L-actin interactions and SSH1L activity towards cofilin. The PKD1-mediated phosphorylation of SSH1L at S978 generates a 14-3-3 binding motif (RSXpS), which is a common feature of PKD1 substrates 17. Our data indicate that 14-3-3 binding allows the sequestration of SSH1L away from filamentous actin (Fig. 5), resulting in decreased activity towards cofilin at the leading edge.
The activity status of cofilin directly relays to the motility of cancer cells 4, 6, 40. This prompted us to investigate whether PKD1 exerts effects on cofilin-mediated actin remodelling of the dendritic network. The expression of active PKD1 in MTLn3 cells, which are a bona fide model system for barbed end formation 6, demonstrated that PKD1 induced a global inhibition of actin incorporation (Fig. 7a, Supplementary information, Figs. S5A, S5B). As a mechanism our data suggest that PKD1-mediated SSH1L phosphorylation, effectively renders cofilin inactive, thereby failing to generate sufficient barbed ends for actin polymerization and Arp2/3-mediated branching 41. Similar dramatic effects on barbed end formation have been described following overexpression of LIMK 4, 7, 8, 42, 43. Given that LIMK is also activated downstream of a RhoA-ROCK signalling cascade 30, it is plausible that both enzymes LIMK and PKD1 synergize to facilitate the inactivation of cofilin. It is also possible that PKD1, additionally to regulating SSH1L activity, also activates LIMK. However, in HeLa cells LIMK showed basal activity and a knockdown of endogenous PKD1 had no significant effects on the activity of endogenous LIMK in control or RhoA-stimulated cells (data not shown).
The inhibition of barbed end formation by PKD1 directly translated to decreased directed cell migration of tumour cells (Figs. 7b–d and Supplementary information, Fig. S5C, S5D). This correlates with data obtained with pancreatic adenocarcinoma cell lines, where the stable overexpression of PKD1 also inhibited cell migration 21. Directed cell migration in response to a chemotactic stimulus requires cofilin activation at the leading edge. However, it also requires tight regulation or inactivation of cofilin in areas other than the lamellipodium, only allowing controlled and cyclic actin severing at the leading edge 6. Such global inactivation of cofilin is also mediated by RhoA and is required to prevent the uncontrolled formation of additional lamellipodia to ensure directed cell migration 8. Our data point to a role of PKD1-mediated regulation of SSH1L at the leading edge. We predict that PKD1, dependent on its intracellular localization, could have important in vivo functions during other stages of the cofilin cycling process. Our data also suggested clustering of subcellular pools of PKD1 and SSH1L at perinuclear structures (Fig. 1d) which are most likely parts of the TGN. A golgi-located pool of PKD1 has been shown to facilitate vesicular transport from the TGN to the plasma membrane 17. Our observation also points towards a potential function of PKD1/SSH1L signalling in actin remodelling events at the TGN complex involved in the scission of transport vesicles 18.
In summary, this study links PKD1 to cofilin-induced actin remodelling processes, which are essential for the initiation of polarized cell migration of cancer cells. We show that PKD1 regulates cofilin via phosphorylation of its activator SSH1L at S978, thereby creating a 14-3-3 binding motif. Binding of 14-3-3 to SSH1L results in the release of this phosphatase from F-actin and its sequestration in the cytoplasm. This translocation inhibits SSH1L activity towards cofilin and blocks barbed end formation and directed cell migration (Fig. 7 g, h).
METHODS
Cell Lines, Antibodies, and Reagents
HeLa cells were maintained in DMEM with 10% FBS, MTLn3 cells in MEM with 5 % FBS. Anti-GST, anti-PKD1 and anti-14-3-3β antibodies were from Santa Cruz (Santa Cruz, CA), anti-myc from Zymed (San Francisco, CA), anti-HA, anti-FLAG (M2), anti-actin from Sigma-Aldrich (St Louis, MO), anti-GFP/YFP (Ab290) from Abcam (Cambridge, MA), anti-SSH1L from ECM Biosciences (Versailles, KY), anti-pMotif (PKD substrate antibody) and anti-pS3-Cofilin from Cell Signaling Technology (Danvers, MA). The anti-pS978-SSH1L antibody was described previously 13. Secondary HRP-linked antibodies were from Roche (Indianapolis, IN). Secondary antibodies (donkey-anti-rat IgG Alexa Fluor 488; goat-anti-mouse Alexa Fluor 488 F(ab’)2; goat-anti-rabbit Alexa Fluor 488 F(ab’)2; goat-anti-mouse Alexa Fluor 568 F(ab’)2) were from Invitrogen (Carlsbad,CA). H2O2 was from Fisher Scientific. Mirus HeLa-Monster (Madison, WI) and Superfect (Qiagen, Valencia, CA) were used for transient transfection of HeLa and Lipofectamin 2000 (Invitrogen) for MTLn3 cells.
DNA Constructs
Expression plasmids including HA-tagged wildtype, PKD1.CA (constitutively-active) and PKD1.KD (kinase-dead, K612W mutation) or YFP-tagged wildtype, S3A or S3E cofilin were described before 19, 28, 35, 44. The use of PKD-RNAi was described elsewhere 17. The expression plasmids pUCD2-3xHA-SSH1L, pYFP-C1-SSH1L, pcDNA3.1-SSH1L-myc-His, pcDNA3.1-SSH1L-myc-His.S937A, pcDNA3.1-SSH1L-myc-His.S978A and pcDNA3.1-SSH1L-myc-His.S937A.S978A were described before 13. Mutagenesis to generate YFP-C1-SSH1L.S978A was carried out using the QuikChange kit (Stratagene, La Jolla, CA) and 5’-CTGAAGCGCTCACACGCTCTTGCCAAGCTGGGG-3’ and 5’-CCCCAGCTTGGCAAGAGCGTGTGAGCGCTTCAG3' as primers. GST-SSH1L fragments (S910-TGA) were generated by PCR using pcDNA3.1-SSH1L and mutants and 5’-CGCGGATCCAGTTTCTCAAAAGACTTT-3’ and 5’-CGCCTCGAGTCAGCTTTTGCTCATCCA-3’ as primers. Fragments were cloned into pGEX4-T1 via BamHI/XhoI. GST-Hsp27 was described previously 25. FLAG-tagged cofilin was generated by amplification of human cofilin from a MDA-MB-231 cDNA library using 5’-GCGGGATCCATGGCCTCCGGTGTGGCTGTCTCT-3’ and 5’-CGCCTCGAGTCATTTGTCATCATCGTCCTTATAGTCCAAAGGCTTGCCCTCCAGGG AGAT-3’ primers and cloning into pcDNA4/TO (Invitrogen) via BamHI/XhoI. The pCH110 β-galactosidase reporter construct was described before 28. The pcDNA3-14-3-3ζ-HA expression plasmid was from Dr. M. Yaffe (Massachusetts Institute of Technology, MA).
Immunoblotting, Immunoprecipitation and PAGE
Cells were washed twice with ice-cold PBS (140 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.2) and lysed with lysis buffer I (50 mM Tris-HCl pH7.4, 1% Triton X-100, 150 mM NaCl, 5 mM EDTA pH 7.4) plus Protease Inhibitor Cocktail (PIC, Sigma-Aldrich) to obtain the cytosolic fraction. Lysates were vortexed and incubated on ice for 30 min. Following centrifugation (13,000 rpm, 15 min, 4 °C) pellets were resolved in lysis buffer II and boiled to obtain the actin fraction. For other experiments, cells were lysed with lysis buffer II (50 mM Tris-HCl, pH7.6, 150 mM NaCl, 10 mM NaF, 1 mM Na3VO4, 1% Nonindet P-40, 10% glycerol) plus PIC. Proteins of interest were immunoprecipitated by 1 hr incubation with a specific antibody (2 µg) followed by 30 min incubation with protein G-Sepharose (Amersham Biosciences). Immune-complexes were washed 3 times with TBS (50 mM Tris-HCl pH 7.4, 150 mM NaCl) and resolved in 20 µl TBS and 2x Laemmli buffer. Samples were subjected to SDS-PAGE or IEF-PAGE (precast IEF pH 3–10 gels from Biorad). Following PAGE, proteins were transferred to nitrocellulose membranes and visualized by immunostaining.
In Vitro Kinase Assays
Kinase assays with GST fusion proteins were carried out by adding 250 ng of active, purified PKD1 (Upstate, Charlottesville, VA) to 2 µg of purified GST-fusion protein in a volume of 40 µl kinase buffer (50 mM Tris pH 7.4, 10 mM MgCl2 and 2 mM DTT) supplemented with 100 µM ATP. The kinase reaction (10 min, RT) was stopped by adding 2x Laemmli buffer.
Immunofluorescence Microscopy
Cells were plated on glass coverslips. Samples were washed twice with PBS, fixed with 3.5 % paraformaldehyde (15 min, 37 °C), washed thrice with PBS, permeabilized with 0.1% Triton X-100 in PBS (2 min, RT) and blocked with 3 % BSA and 0.05 % Tween 20 in PBS (30 min, RT). Coverslips were incubated with primary antibodies (1:2000 dilution, 2 h, RT), washed five times with PBS and incubated with secondary antibodies (1:500 dilution, 2 h, RT). F-actin was stained with Rhodamine-Phalloidin (Sigma) or Alexa Fluor 633-Phalloidin (Molecular Probes), nuclei with DAPI. Coverslips were mounted in Fluromount-G (Southern Biotech, Birmingham, AL). Samples were examined using a LSM 510META confocal laser scanning microscope (Zeiss, Jena, Germany) with a Plan-Apochromat 63x/1.4 Dic oil immersion objective. DAPI was exited at the 405 nm, Alexa Fluor 350 at the 405 nm laser, Alexa Fluor 488/GFP at the 488 nm, Rhodamine-Phalloidin/Alexa Fluor 568 at the 543 nm and Alexa Fluor 633-Phalloidin at the 633 nm laser line, respectively, in a Multi-Track configuration, switching Tracks after each line (DAPI: BP420-480, Alexa Fluor 488/GFP: BP505-530, Rhodamine/Alexa Fluor 568: META Chs 550–615, Alexa Fluor 633: META Chs 636–711)Images were processed using NIH ImageJ.
Acceptor Photobleach FRET
Transfection, plating, fixation, staining of cells and mounting of samples occurred as described above. Samples were examined using a LSM 510META confocal laser scanning microscope. For acceptor photobleach FRET imaging the Zeiss LSM Software FRETplus Macro in Channel Mode was used. Gain and Offset were adjusted to be within the lower linear range using intensity range settings. Lasers and detection channels were changed after every FRAME. All settings were adjusted using the FRETplus Macro. % FRET images were calculated using the FRETplus Macro acceptor photobleach analysis module. For presentation purposes donor and acceptor thresholds for calculation of % FRET images were adjusted to reduce background noise and enhance signal to noise ratio.
Analysis of free actin filament barbed ends
Cells were transfected and reseeded on a 8 well ibiTreat μ-Slide (Ibidi, Integrated BioDiagnostics, Martinsried, Germany). Cells were serum-starved (16 h). Further treatment occurred as described elsewhere 45. In short: The medium was removed, cells were washed with pre-warmed PBS, permeabilized and labelled with 0.4 µM Actin-Alexa Flour594 in permeabilization buffer (20 mM HEPES, 138 mM KCl, 4 mM MgCl2, 3 mM EGTA, 0.2 mg/ml saponin, 1 % BSA) plus 1 m M ATP for 30 second sat 37°C. The cells were fixed with 4 % paraformaldehyde in PBS (RT, 10 min).
Transwell Chemotaxis Assays
Assays were performed as previously described 46 using 24 well Transwell filters (Corning) with 8 µM pore diameter. In short: Cells were transfected with plasmids of interest and a β-galactosidase marker (pCH110) at a ratio of 3:1. After 24 h, cells were seeded on Transwell filters in DMEM + 0.1% BSA and induced to migrate towards NIH-3T3 conditioned medium. Cells were fixed with 3.7 % formaldehyde in PBS (30 min, RT). The inside of the Transwell insert was wiped and filters washed several times with PBS. Transfected cells on the bottom of the filter were stained for β-galactosidase expression and positive cells of triplicate filters per condition counted.
Cell Tracking/Time Laps studies
Cells were seeded in fibronectin-coated Chemotaxis μ-Slide (Ibidi) in the observation area in FBS-free medium and attached for 4 hours. Directed migration towards NIH-3T3 conditioned medium was analyzed. Migrating cells were tracked with a live-imaging inverted video microscope (DMIRE2, Leica) for 6 hours by recording phase contrast and fluorescence images every 10 minutes at multiple xy-positions. The xy-position of single cells in compiled xyt-image stacks were traced using NIH ImageJ and the Tracking plug-in provided by Fabrice Cordelieres. The results were further processed and statistically analyzed using the NIH ImageJ Chemotaxis tool (Ibidi).
ACKNOWLEDGEMENTS
We are grateful to L. Lewis-Tuffin for help with the video microscopy and R. Mooney (University of Rochester Medical Center, NY) for providing MTLn3 cells. This work was sponsored in part by funds from the Mayo Foundation and the Mayo Comprehensive Cancer Center, the Mayo Clinic Breast Cancer SPORE (CA116201-03DR4), a “Friends for an Earlier Breast Cancer Test” Grant - all to PS and by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to KM. The authors declare that they have no competing financial interests.
REFERENCES
- 1.Bamburg JR, McGough A, Ono S. Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol. 1999;9:364–370. doi: 10.1016/s0962-8924(99)01619-0. [DOI] [PubMed] [Google Scholar]
- 2.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 3.Yamaguchi H, Condeelis J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim Biophys Acta. 2007;1773:642–652. doi: 10.1016/j.bbamcr.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mouneimne G, et al. Spatial and temporal control of cofilin activity is required for directional sensing during chemotaxis. Curr Biol. 2006;16:2193–2205. doi: 10.1016/j.cub.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Sinha P, et al. Increased expression of epidermal fatty acid binding protein, cofilin, and 14-3-3-sigma (stratifin) detected by two-dimensional gel electrophoresis, mass spectrometry and microsequencing of drug-resistant human adenocarcinoma of the pancreas. Electrophoresis. 1999;20:2952–2960. doi: 10.1002/(SICI)1522-2683(19991001)20:14<2952::AID-ELPS2952>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 6.Wang W, Eddy R, Condeelis J. The cofilin pathway in breast cancer invasion and metastasis. Nat Rev Cancer. 2007;7:429–440. doi: 10.1038/nrc2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W, et al. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J Cell Biol. 2006;173:395–404. doi: 10.1083/jcb.200510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishita M, et al. Spatial and temporal regulation of cofilin activity by LIM kinase and Slingshot is critical for directional cell migration. J Cell Biol. 2005;171:349–359. doi: 10.1083/jcb.200504029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizuno K, et al. Identification of a human cDNA encoding a novel protein kinase with two repeats of the LIM/double zinc finger motif. Oncogene. 1994;9:1605–1612. [PubMed] [Google Scholar]
- 10.Huang TY, DerMardirossian C, Bokoch GM. Cofilin phosphatases and regulation of actin dynamics. Curr Opin Cell Biol. 2006;18:26–31. doi: 10.1016/j.ceb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Toshima J, et al. Cofilin phosphorylation by protein kinase testicular protein kinase 1 and its role in integrin-mediated actin reorganization and focal adhesion formation. Mol Biol Cell. 2001;12:1131–1145. doi: 10.1091/mbc.12.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agnew BJ, Minamide LS, Bamburg JR. Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J Biol Chem. 1995;270:17582–17587. doi: 10.1074/jbc.270.29.17582. [DOI] [PubMed] [Google Scholar]
- 13.Nagata-Ohashi K, et al. A pathway of neuregulin-induced activation of cofilin-phosphatase Slingshot and cofilin in lamellipodia. J Cell Biol. 2004;165:465–471. doi: 10.1083/jcb.200401136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilinc. Cell. 2002;108:233–246. doi: 10.1016/s0092-8674(01)00638-9. [DOI] [PubMed] [Google Scholar]
- 15.Nishita M, et al. Phosphoinositide 3-kinase-mediated activation of cofilin phosphatase Slingshot and its role for insulin-induced membrane protrusion. J Biol Chem. 2004;279:7193–7198. doi: 10.1074/jbc.M312591200. [DOI] [PubMed] [Google Scholar]
- 16.Zugaza JL, Waldron RT, Sinnett-Smith J, Rozengurt E. Bombesin, vasopressin, endothelin, bradykinin, and platelet-derived growth factor rapidly activate protein kinase D through a protein kinase C-dependent signal transduction pathway. J Biol Chem. 1997;272:23952–23960. doi: 10.1074/jbc.272.38.23952. [DOI] [PubMed] [Google Scholar]
- 17.Hausser A, et al. Protein kinase D regulates vesicular transport by phosphorylating and activating phosphatidylinositol-4 kinase IIIbeta at the Golgi complex. Nat Cell Biol. 2005;7:880–886. doi: 10.1038/ncb1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liljedahl M, et al. Protein kinase D regulates the fission of cell surface destined transport carriers from the trans-Golgi network. Cell. 2001;104:409–420. doi: 10.1016/s0092-8674(01)00228-8. [DOI] [PubMed] [Google Scholar]
- 19.Storz P, Doppler H, Toker A. Protein kinase D mediates mitochondrion-to-nucleus signaling and detoxification from mitochondrial reactive oxygen species. Mol Cell Biol. 2005;25:8520–8530. doi: 10.1128/MCB.25.19.8520-8530.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Blume J, et al. Phosphorylation at Ser244 by CK1 determines nuclear localization and substrate targeting of PKD2. Embo J. 2007;26:4619–4633. doi: 10.1038/sj.emboj.7601891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eiseler T, Schmid MA, Topbas F, Pfizenmaier K, Hausser A. PKD is recruited to sites of actin remodelling at the leading edge and negatively regulates cell migration. FEBS Lett. 2007;581:4279–4287. doi: 10.1016/j.febslet.2007.07.079. [DOI] [PubMed] [Google Scholar]
- 22.Jaggi M, et al. E-cadherin phosphorylation by protein kinase D1/protein kinase C{mu} is associated with altered cellular aggregation and motility in prostate cancer. Cancer Res. 2005;65:483–492. [PubMed] [Google Scholar]
- 23.Riol-Blanco L, et al. The neuronal protein Kidins220 localizes in a raft compartment at the leading edge of motile immature dendritic cells. Eur J Immunol. 2004;34:108–118. doi: 10.1002/eji.200324640. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto M, Nagata-Ohashi K, Ohta Y, Ohashi K, Mizuno K. Identification of multiple actin-binding sites in cofilin-phosphatase Slingshot-1L. FEBS Lett. 2006;580:1789–1794. doi: 10.1016/j.febslet.2006.02.034. [DOI] [PubMed] [Google Scholar]
- 25.Doppler H, Storz P, Li J, Comb MJ, Toker A. A phosphorylation state-specific antibody recognizes Hsp27, a novel substrate of protein kinase D. J Biol Chem. 2005;280:15013–15019. doi: 10.1074/jbc.C400575200. [DOI] [PubMed] [Google Scholar]
- 26.Doppler H, Storz P. A novel tyrosine phosphorylation site in protein kinase d contributes to oxidative stress-mediated activation. J Biol Chem. 2007 doi: 10.1074/jbc.M703584200. [DOI] [PubMed] [Google Scholar]
- 27.Song J, Li J, Lulla A, Evers BM, Chung DH. Protein kinase D protects against oxidative stress-induced intestinal epithelial cell injury via Rho/ROK/PKC-delta pathway activation. Am J Physiol Cell Physiol. 2006;290:C1469–C1476. doi: 10.1152/ajpcell.00486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Storz P, Toker A. Protein kinase D mediates a stress-induced NF-kappaB activation and survival pathway. Embo J. 2003;22:109–120. doi: 10.1093/emboj/cdg009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutti JE, et al. A rapid method for determining protein kinase phosphorylation specificity. Nat Methods. 2004;1:27–29. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- 30.Maekawa M, et al. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science. 1999;285:895–898. doi: 10.1126/science.285.5429.895. [DOI] [PubMed] [Google Scholar]
- 31.Mullin MJ, Lightfoot K, Marklund U, Cantrell DA. Differential requirement for RhoA GTPase depending on the cellular localization of protein kinase D. J Biol Chem. 2006;281:25089–25096. doi: 10.1074/jbc.M603591200. [DOI] [PubMed] [Google Scholar]
- 32.Yuan J, Slice LW, Rozengurt E. Activation of protein kinase D by signaling through Rho and the alpha subunit of the heterotrimeric G protein G13. J Biol Chem. 2001;276:38619–38627. doi: 10.1074/jbc.M105530200. [DOI] [PubMed] [Google Scholar]
- 33.Mouneimne G, et al. Phospholipase C and cofilin are required for carcinoma cell directionality in response to EGF stimulation. J Cell Biol. 2004;166:697–708. doi: 10.1083/jcb.200405156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Storz P, Doppler H, Toker A. Protein kinase Cdelta selectively regulates protein kinase D-dependent activation of NF-kappaB in oxidative stress signaling. Mol Cell Biol. 2004;24:2614–2626. doi: 10.1128/MCB.24.7.2614-2626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matthews SA, Rozengurt E, Cantrell D. Protein kinase D. A selective target for antigen receptors and a downstream target for protein kinase C in lymphocytes. J Exp Med. 2000;191:2075–2082. doi: 10.1084/jem.191.12.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rey O, Sinnett-Smith J, Zhukova E, Rozengurt E. Regulated nucleocytoplasmic transport of protein kinase D in response to G protein-coupled receptor activation. J Biol Chem. 2001;276:49228–49235. doi: 10.1074/jbc.M109395200. [DOI] [PubMed] [Google Scholar]
- 38.Baron CL, Malhotra V. Role of diacylglycerol in PKD recruitment to the TGN and protein transport to the plasma membrane. Science. 2002;295:325–328. doi: 10.1126/science.1066759. [DOI] [PubMed] [Google Scholar]
- 39.van Rheenen J, et al. EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J Cell Biol. 2007;179:1247–1259. doi: 10.1083/jcb.200706206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sidani M, et al. Cofilin determines the migration behavior and turning frequency of metastatic cancer cells. J Cell Biol. 2007;179:777–791. doi: 10.1083/jcb.200707009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ichetovkin I, Grant W, Condeelis J. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr Biol. 2002;12:79–84. doi: 10.1016/s0960-9822(01)00629-7. [DOI] [PubMed] [Google Scholar]
- 42.Dawe HR, Minamide LS, Bamburg JR, Cramer LP. ADF/cofilin controls cell polarity during fibroblast migration. Curr Biol. 2003;13:252–257. doi: 10.1016/s0960-9822(03)00040-x. [DOI] [PubMed] [Google Scholar]
- 43.Zebda N, et al. Phosphorylation of ADF/cofilin abolishes EGF-induced actin nucleation at the leading edge and subsequent lamellipod extension. J Cell Biol. 2000;151:1119–1128. doi: 10.1083/jcb.151.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Storz P, Doppler H, Johannes FJ, Toker A. Tyrosine phosphorylation of protein kinase D in the pleckstrin homology domain leads to activation. J Biol Chem. 2003;278:17969–17976. doi: 10.1074/jbc.M213224200. [DOI] [PubMed] [Google Scholar]
- 45.Cai L, Marshall TW, Uetrecht AC, Schafer DA, Bear JE. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell. 2007;128:915–929. doi: 10.1016/j.cell.2007.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jauliac S, et al. The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat Cell Biol. 2002;4:540–544. doi: 10.1038/ncb816. [DOI] [PubMed] [Google Scholar]