Abstract
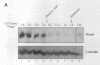
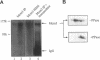
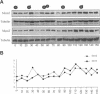
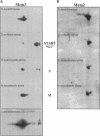
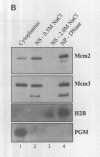
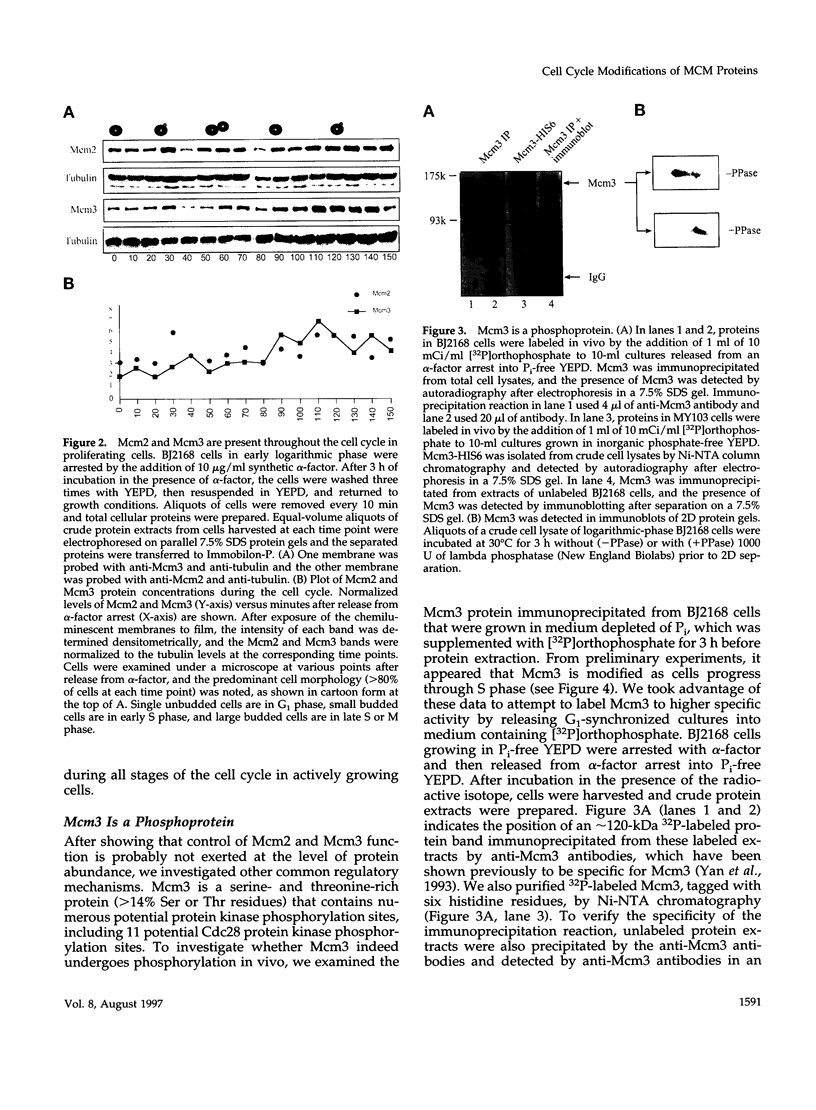
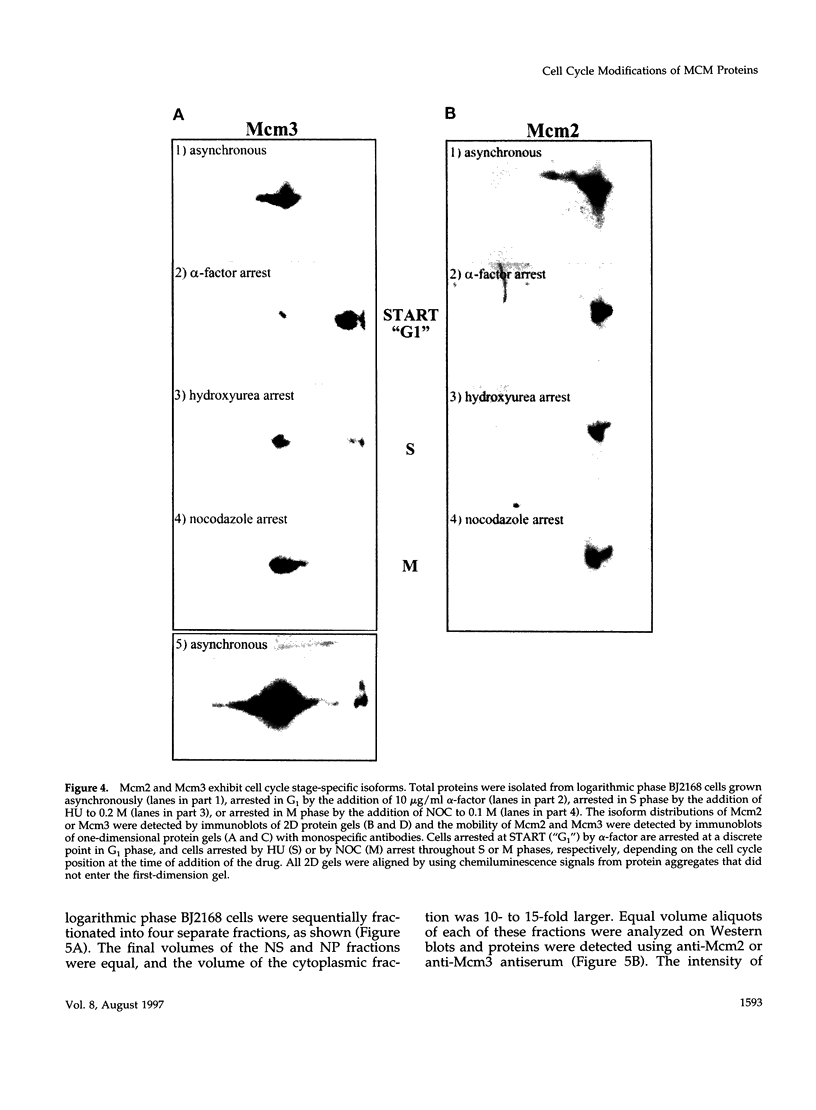
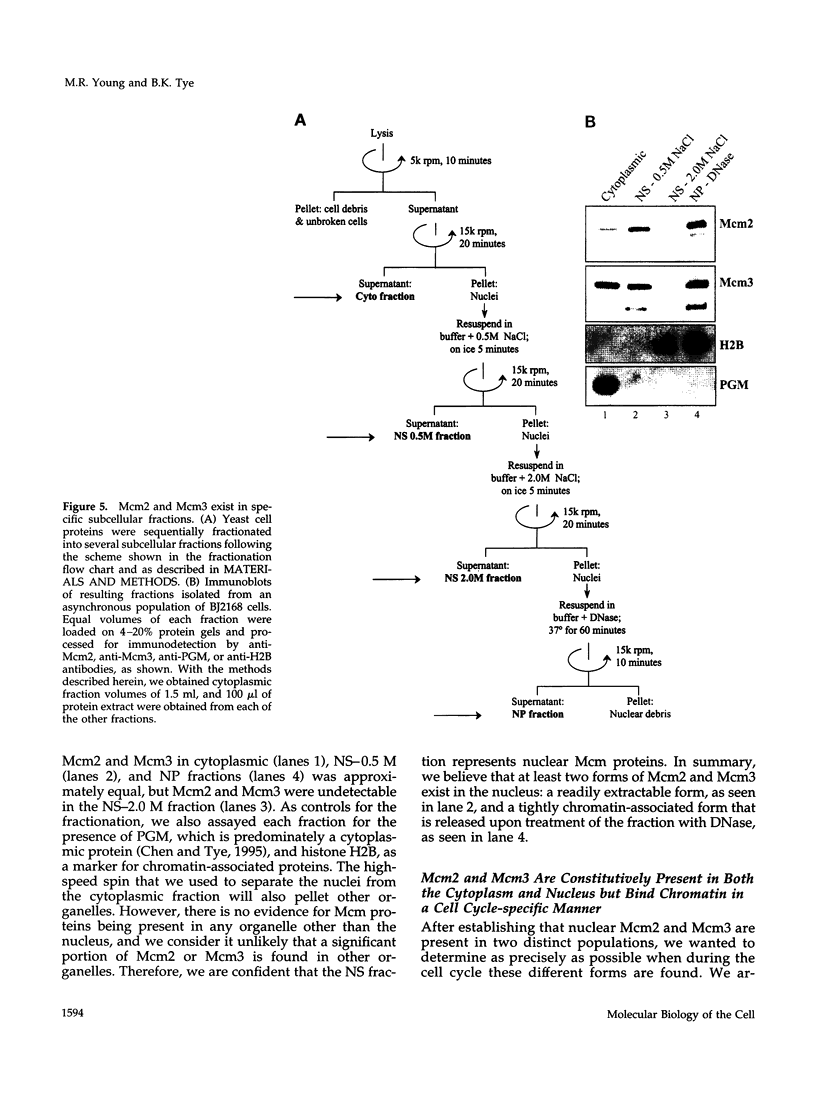
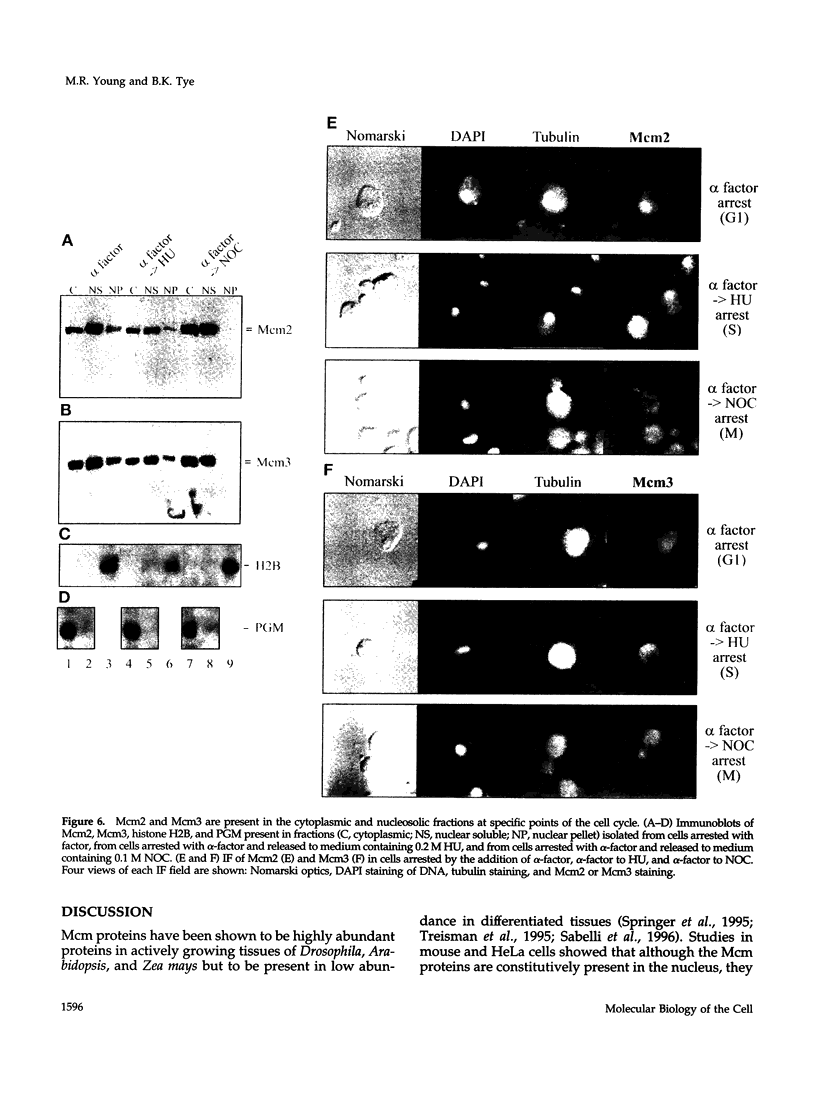
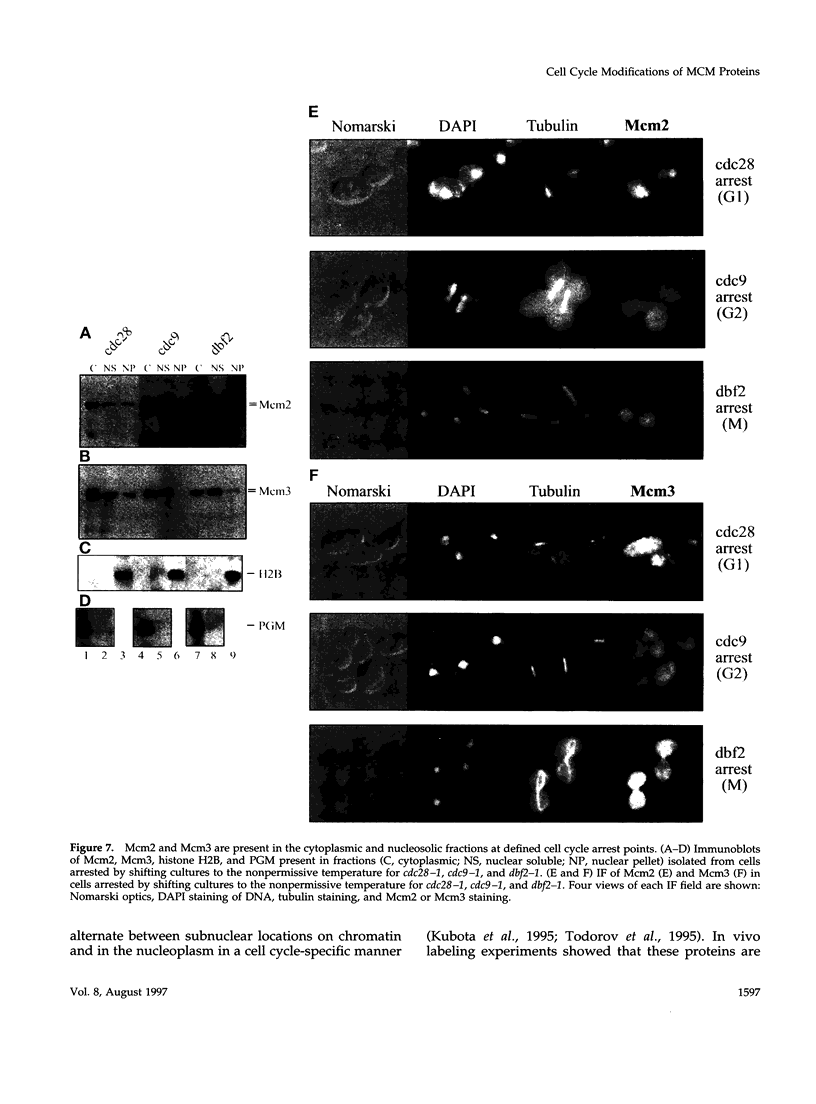
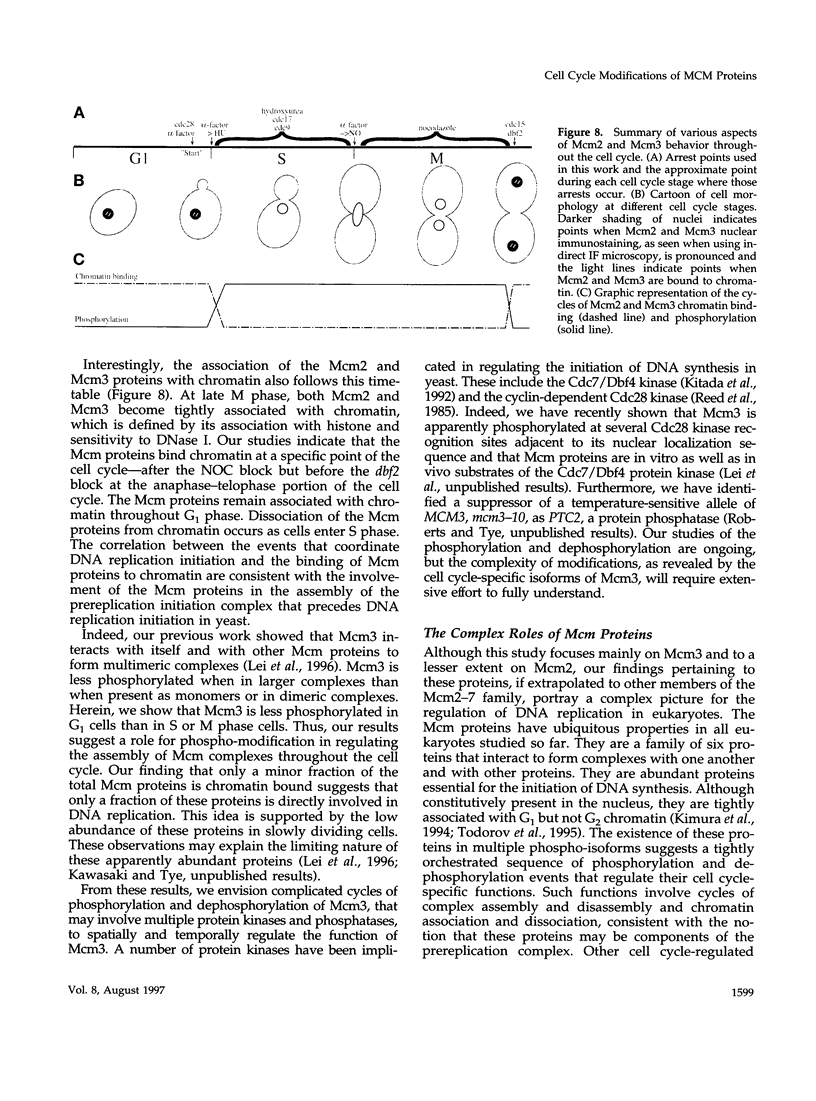
The Mcm2-7 proteins are a family of conserved proteins whose functions are essential for the initiation of DNA synthesis in all eukaryotes. These patients are constitutively present in high abundance in actively proliferating cells. In Saccharomyces cerevisiae, the intracellular concentrations of Mcms are between 100 and 500 times the number of replication origins. However, these proteins are limiting for the initiation of DNA synthesis at replication origins. Our studies indicate that only a small fraction of Mcm2 and Mcm3 tightly associates with chromatin, from late M phase to the beginning of the S phase. The rest of the Mcm2 and Mcm3 proteins are disturbed to both the cytoplasm and nucleoplasm in relatively constant levels throughout the cell cycle. We also show that S. cerevisiae Mcm3 is a phosphoprotein that exists in multiple isoforms and that distinct isoforms of Mcm2 and Mcm3 can be detected at specific stages of the cell cycle. These results suggest that the localization and function of the Mcm proteins are regulated by posttranslational phosphorylation in a manner that is consistent with a role for the Mcm proteins in restricting DNA replication to once per cell cycle.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell S. P., Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992 May 14;357(6374):128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Blow J. J., Laskey R. A. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988 Apr 7;332(6164):546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- Blow J. J. Preventing re-replication of DNA in a single cell cycle: evidence for a replication licensing factor. J Cell Biol. 1993 Sep;122(5):993–1002. doi: 10.1083/jcb.122.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Tye B. K. The yeast Mcm1 protein is regulated posttranscriptionally by the flux of glycolysis. Mol Cell Biol. 1995 Aug;15(8):4631–4639. doi: 10.1128/mcb.15.8.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J. P., Mahbubani H. M., Khoo C. Y., Blow J. J. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature. 1995 Jun 1;375(6530):418–421. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- Coué M., Kearsey S. E., Méchali M. Chromotin binding, nuclear localization and phosphorylation of Xenopus cdc21 are cell-cycle dependent and associated with the control of initiation of DNA replication. EMBO J. 1996 Mar 1;15(5):1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Coverley D., Downes C. S., Romanowski P., Laskey R. A. Reversible effects of nuclear membrane permeabilization on DNA replication: evidence for a positive licensing factor. J Cell Biol. 1993 Sep;122(5):985–992. doi: 10.1083/jcb.122.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton S., Whitbread L. Cell cycle-regulated nuclear import and export of Cdc47, a protein essential for initiation of DNA replication in budding yeast. Proc Natl Acad Sci U S A. 1995 Mar 28;92(7):2514–2518. doi: 10.1073/pnas.92.7.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Cocker J. H., Dowell S. J., Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994 Jul 29;78(2):303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Gibson S. I., Surosky R. T., Tye B. K. The phenotype of the minichromosome maintenance mutant mcm3 is characteristic of mutants defective in DNA replication. Mol Cell Biol. 1990 Nov;10(11):5707–5720. doi: 10.1128/mcb.10.11.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel T., Zabel U., van Zee K., Müller J. M., Fanning E., Baeuerle P. A. Intramolecular masking of the nuclear location signal and dimerization domain in the precursor for the p50 NF-kappa B subunit. Cell. 1992 Mar 20;68(6):1121–1133. doi: 10.1016/0092-8674(92)90083-o. [DOI] [PubMed] [Google Scholar]
- Hennessy K. M., Clark C. D., Botstein D. Subcellular localization of yeast CDC46 varies with the cell cycle. Genes Dev. 1990 Dec;4(12B):2252–2263. doi: 10.1101/gad.4.12b.2252. [DOI] [PubMed] [Google Scholar]
- Hennessy K. M., Lee A., Chen E., Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991 Jun;5(6):958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- Kimura H., Nozaki N., Sugimoto K. DNA polymerase alpha associated protein P1, a murine homolog of yeast MCM3, changes its intranuclear distribution during the DNA synthetic period. EMBO J. 1994 Sep 15;13(18):4311–4320. doi: 10.1002/j.1460-2075.1994.tb06751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Sakamoto H., Appella E., Siraganian R. P. Conformational changes induced in the protein tyrosine kinase p72syk by tyrosine phosphorylation or by binding of phosphorylated immunoreceptor tyrosine-based activation motif peptides. Mol Cell Biol. 1996 Apr;16(4):1471–1478. doi: 10.1128/mcb.16.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada K., Johnston L. H., Sugino T., Sugino A. Temperature-sensitive cdc7 mutations of Saccharomyces cerevisiae are suppressed by the DBF4 gene, which is required for the G1/S cell cycle transition. Genetics. 1992 May;131(1):21–29. doi: 10.1093/genetics/131.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y., Mimura S., Nishimoto S., Takisawa H., Nojima H. Identification of the yeast MCM3-related protein as a component of Xenopus DNA replication licensing factor. Cell. 1995 May 19;81(4):601–609. doi: 10.1016/0092-8674(95)90081-0. [DOI] [PubMed] [Google Scholar]
- Lei M., Kawasaki Y., Tye B. K. Physical interactions among Mcm proteins and effects of Mcm dosage on DNA replication in Saccharomyces cerevisiae. Mol Cell Biol. 1996 Sep;16(9):5081–5090. doi: 10.1128/mcb.16.9.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. J., Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993 Dec 17;262(5141):1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- Madine M. A., Khoo C. Y., Mills A. D., Musahl C., Laskey R. A. The nuclear envelope prevents reinitiation of replication by regulating the binding of MCM3 to chromatin in Xenopus egg extracts. Curr Biol. 1995 Nov 1;5(11):1270–1279. doi: 10.1016/s0960-9822(95)00253-3. [DOI] [PubMed] [Google Scholar]
- Maine G. T., Sinha P., Tye B. K. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics. 1984 Mar;106(3):365–385. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao P. N., Johnson R. T. Mammalian cell fusion: studies on the regulation of DNA synthesis and mitosis. Nature. 1970 Jan 10;225(5228):159–164. doi: 10.1038/225159a0. [DOI] [PubMed] [Google Scholar]
- Reed S. I., Hadwiger J. A., Lörincz A. T. Protein kinase activity associated with the product of the yeast cell division cycle gene CDC28. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4055–4059. doi: 10.1073/pnas.82.12.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S. I. The role of p34 kinases in the G1 to S-phase transition. Annu Rev Cell Biol. 1992;8:529–561. doi: 10.1146/annurev.cb.08.110192.002525. [DOI] [PubMed] [Google Scholar]
- Rubin G. M. The nucleotide sequence of Saccharomyces cerevisiae 5.8 S ribosomal ribonucleic acid. J Biol Chem. 1973 Jun 10;248(11):3860–3875. [PubMed] [Google Scholar]
- Sabelli P. A., Burgess S. R., Kush A. K., Young M. R., Shewry P. R. cDNA cloning and characterisation of a maize homologue of the MCM proteins required for the initiation of DNA replication. Mol Gen Genet. 1996 Aug 27;252(1-2):125–136. doi: 10.1007/BF02173212. [DOI] [PubMed] [Google Scholar]
- Schmidt-Zachmann M. S., Dargemont C., Kühn L. C., Nigg E. A. Nuclear export of proteins: the role of nuclear retention. Cell. 1993 Aug 13;74(3):493–504. doi: 10.1016/0092-8674(93)80051-f. [DOI] [PubMed] [Google Scholar]
- Springer P. S., McCombie W. R., Sundaresan V., Martienssen R. A. Gene trap tagging of PROLIFERA, an essential MCM2-3-5-like gene in Arabidopsis. Science. 1995 May 12;268(5212):877–880. doi: 10.1126/science.7754372. [DOI] [PubMed] [Google Scholar]
- Tan S., Richmond T. J. DNA binding-induced conformational change of the yeast transcriptional activator PRTF. Cell. 1990 Jul 27;62(2):367–377. doi: 10.1016/0092-8674(90)90373-m. [DOI] [PubMed] [Google Scholar]
- Todorov I. T., Attaran A., Kearsey S. E. BM28, a human member of the MCM2-3-5 family, is displaced from chromatin during DNA replication. J Cell Biol. 1995 Jun;129(6):1433–1445. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyn J. H., Johnston L. H. The Dbf2 and Dbf20 protein kinases of budding yeast are activated after the metaphase to anaphase cell cycle transition. EMBO J. 1994 Mar 1;13(5):1103–1113. doi: 10.1002/j.1460-2075.1994.tb06359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman J. E., Follette P. J., O'Farrell P. H., Rubin G. M. Cell proliferation and DNA replication defects in a Drosophila MCM2 mutant. Genes Dev. 1995 Jul 15;9(14):1709–1715. doi: 10.1101/gad.9.14.1709. [DOI] [PubMed] [Google Scholar]
- Whitebread L. A., Dalton S. Cdc54 belongs to the Cdc46/Mcm3 family of proteins which are essential for initiation of eukaryotic DNA replication. Gene. 1995 Mar 21;155(1):113–117. doi: 10.1016/0378-1119(94)00925-i. [DOI] [PubMed] [Google Scholar]
- Yan H., Gibson S., Tye B. K. Mcm2 and Mcm3, two proteins important for ARS activity, are related in structure and function. Genes Dev. 1991 Jun;5(6):944–957. doi: 10.1101/gad.5.6.944. [DOI] [PubMed] [Google Scholar]
- Yan H., Merchant A. M., Tye B. K. Cell cycle-regulated nuclear localization of MCM2 and MCM3, which are required for the initiation of DNA synthesis at chromosomal replication origins in yeast. Genes Dev. 1993 Nov;7(11):2149–2160. doi: 10.1101/gad.7.11.2149. [DOI] [PubMed] [Google Scholar]