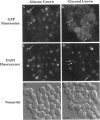
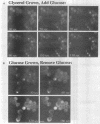
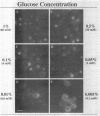
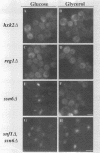
Abstract
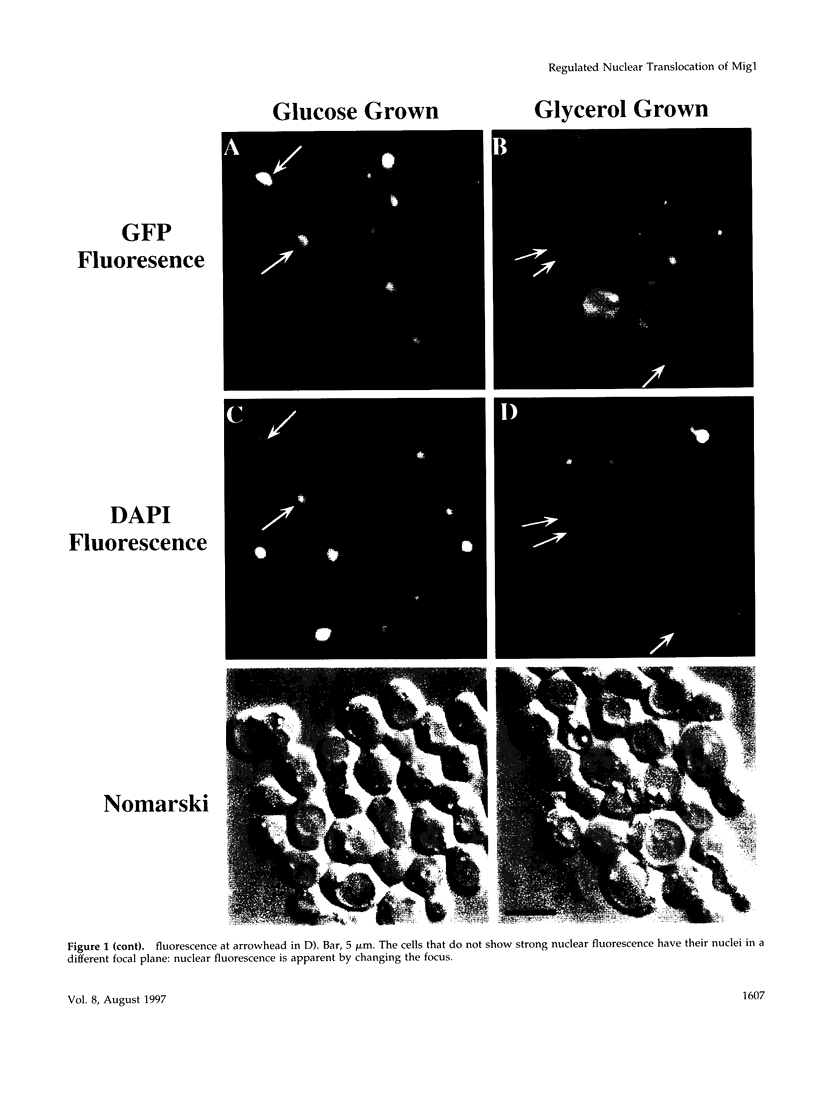
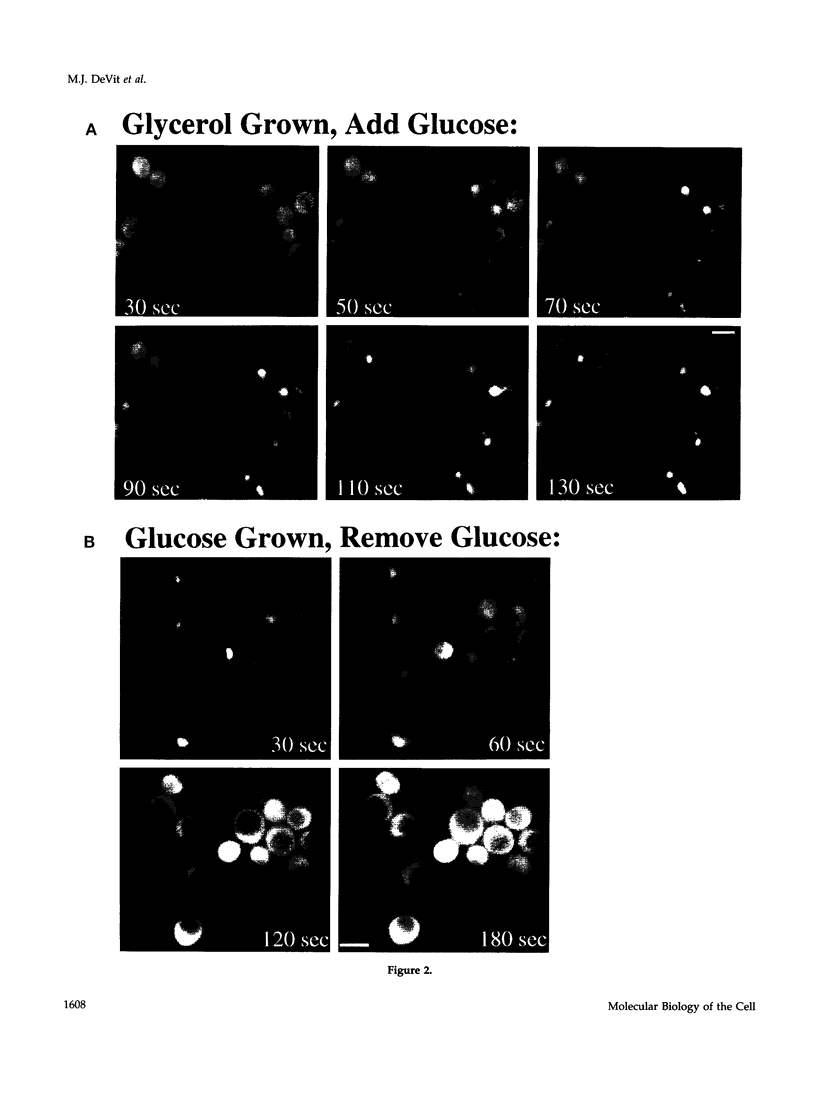
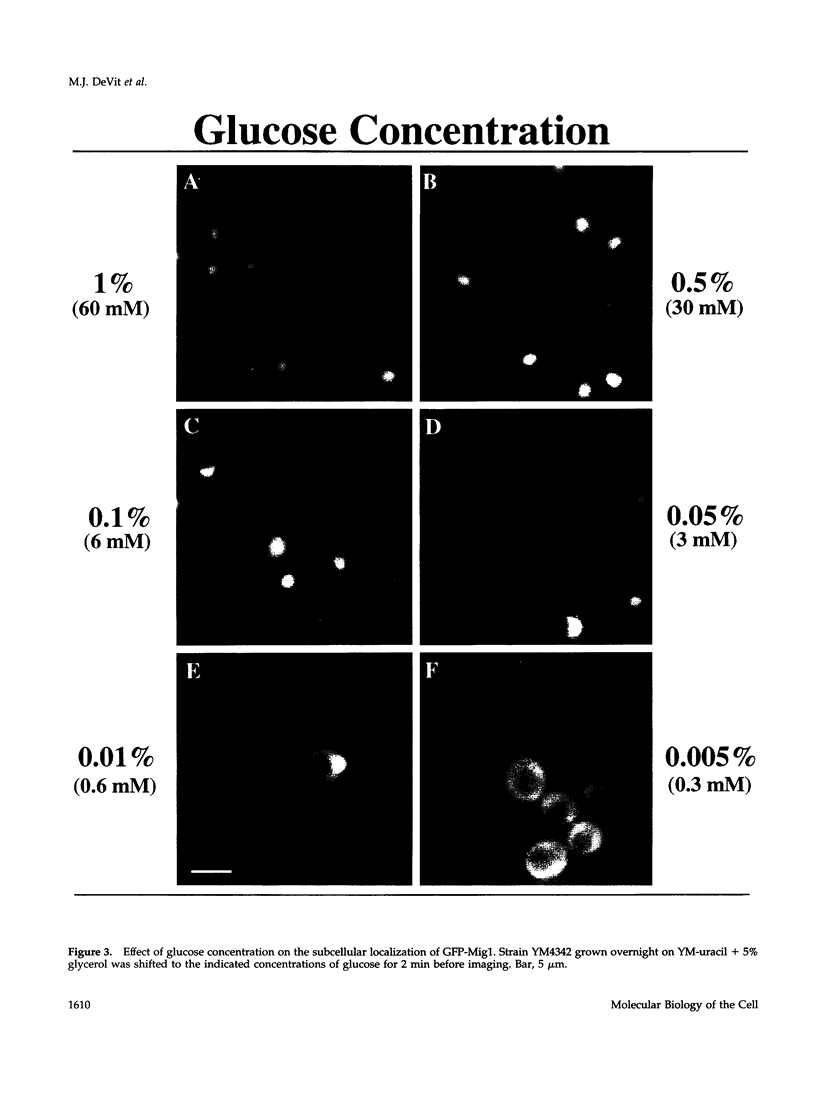
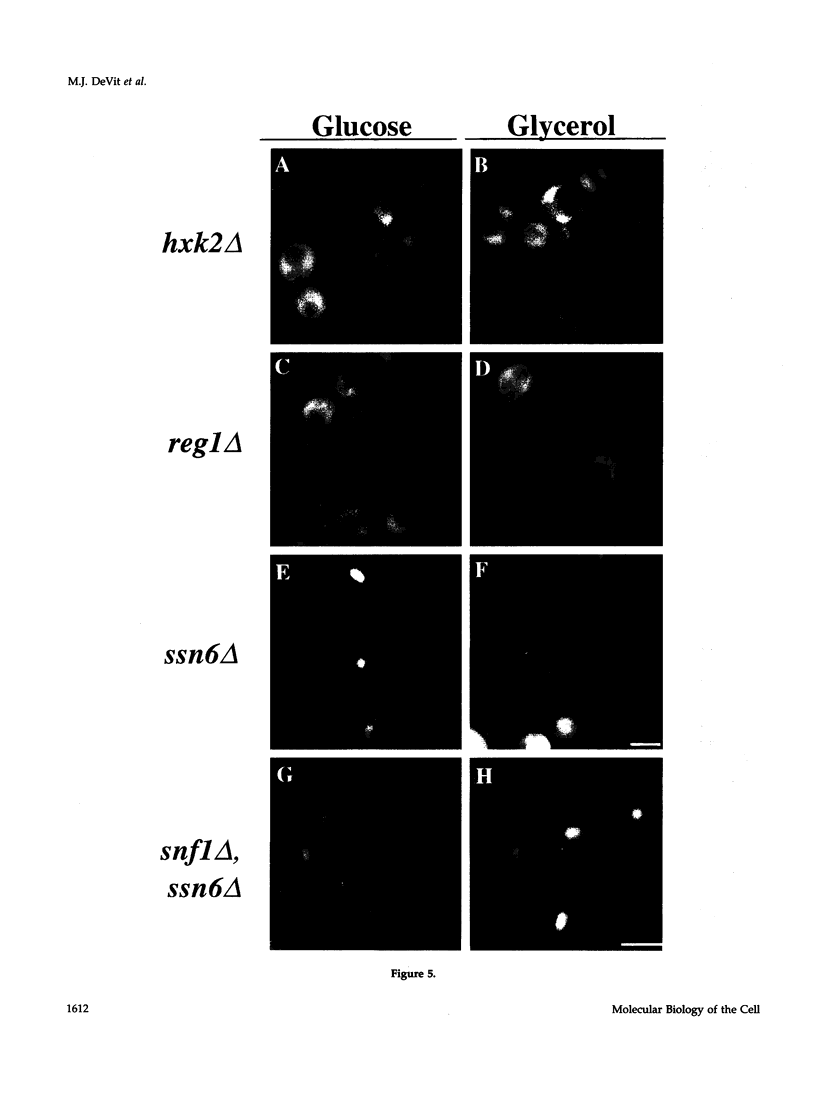
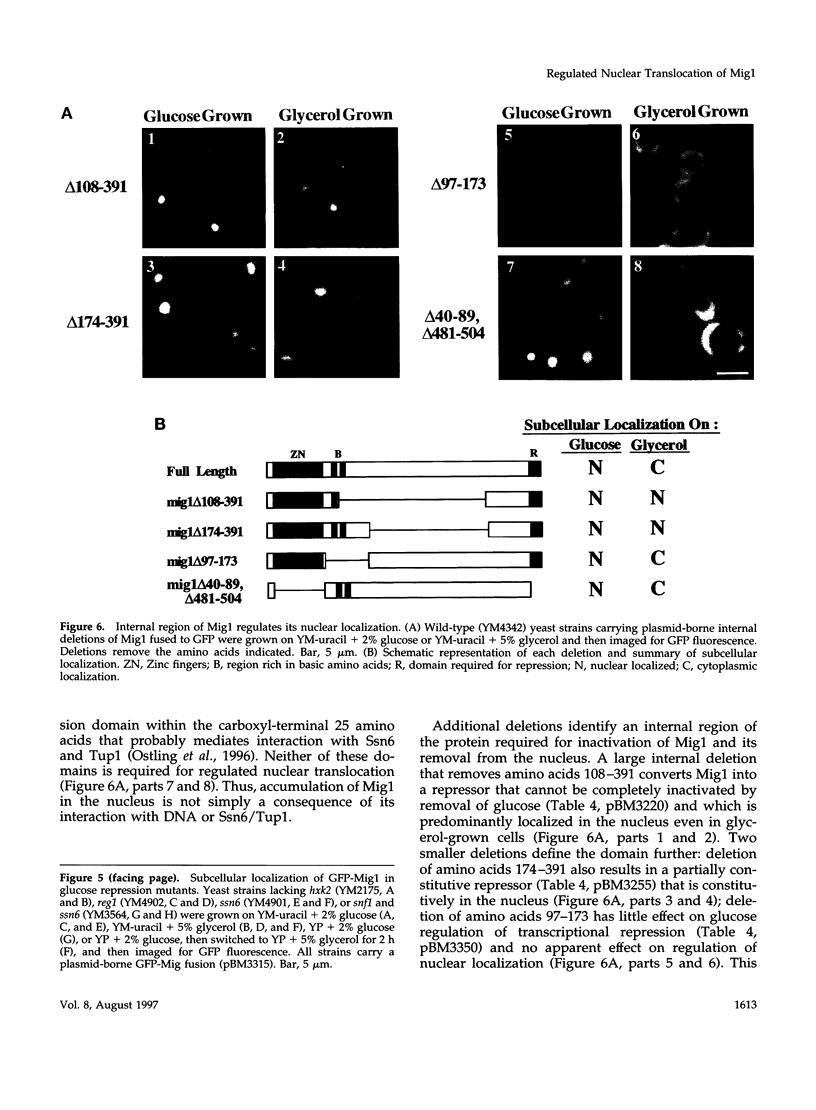
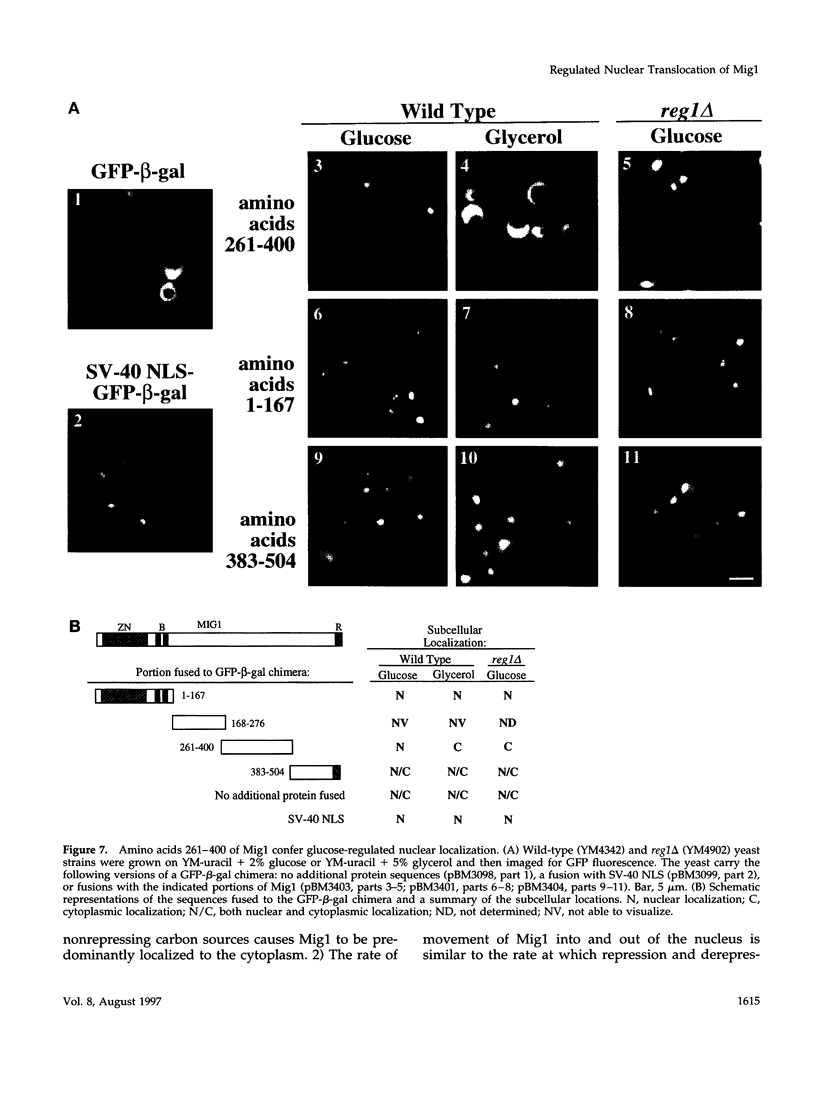
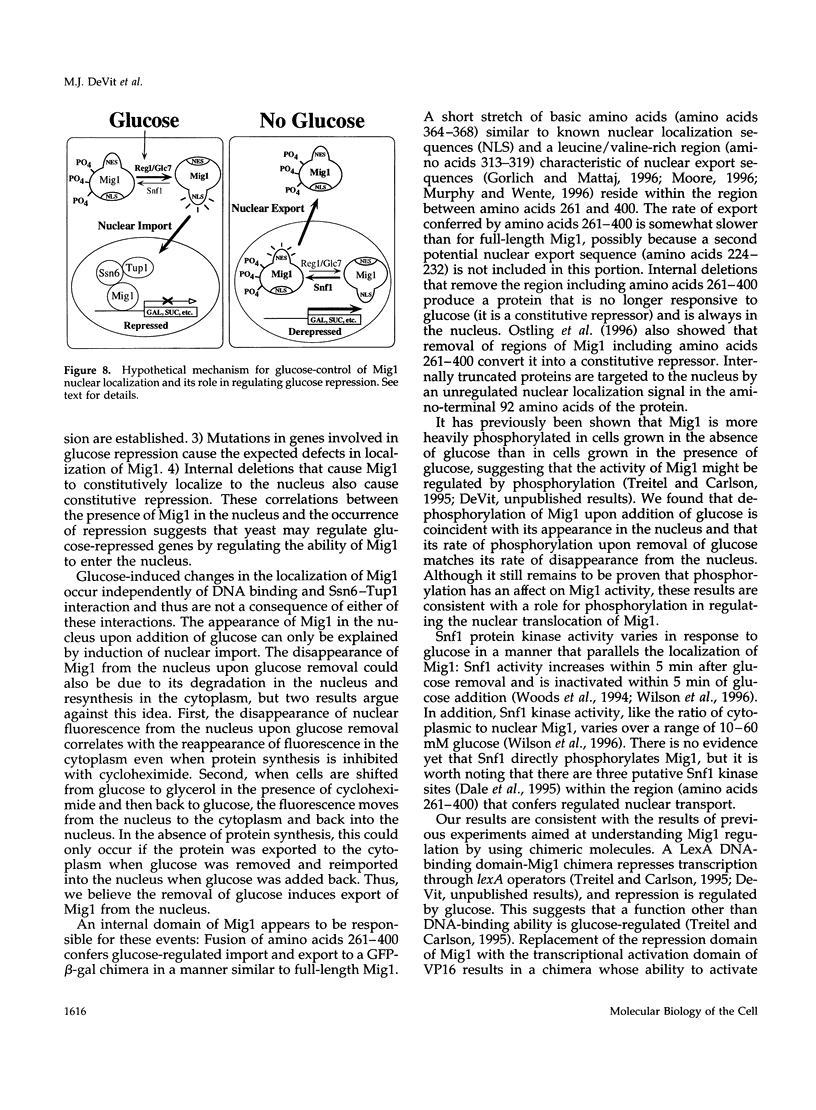
Glucose represses the transcription of many genes in bakers yeast (Saccharomyces cerevisiae). Mig1 is a Cys2-His2 zinc finger protein that mediates glucose repression of several genes by binding to their promoters and recruiting the general repression complex Ssn6-Tup1. We have found that the subcellular localization of Mig1 is regulated by glucose. Mig1 is imported into the nucleus within minutes after the addition of glucose and is just as rapidly transported back to the cytoplasm when glucose is removed. This regulated nuclear localization requires components of the glucose repression signal transduction pathway. An internal region of the protein separate from the DNA binding and repression domains is necessary and sufficient for glucose-regulated nuclear import and export. Changes in the phosphorylation status of Mig1 are coincident with the changes in its localization, suggesting a possible regulatory role for phosphorylation. Our results suggest that a glucose-regulated nuclear import and/or export mechanism controls the activity of Mig1.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cassart J. P., Georis I., Ostling J., Ronne H., Vandenhaute J. The MIG1 repressor from Kluyveromyces lactis: cloning, sequencing and functional analysis in Saccharomyces cerevisiae. FEBS Lett. 1995 Sep 4;371(2):191–194. doi: 10.1016/0014-5793(95)00909-s. [DOI] [PubMed] [Google Scholar]
- Celenza J. L., Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986 Sep 12;233(4769):1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- Chalfie M., Tu Y., Euskirchen G., Ward W. W., Prasher D. C. Green fluorescent protein as a marker for gene expression. Science. 1994 Feb 11;263(5148):802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Dale S., Wilson W. A., Edelman A. M., Hardie D. G. Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 1995 Mar 20;361(2-3):191–195. doi: 10.1016/0014-5793(95)00172-6. [DOI] [PubMed] [Google Scholar]
- Entian K. D., Zimmermann F. K. Glycolytic enzymes and intermediates in carbon catabolite repression mutants of Saccharomyces cerevisiae. Mol Gen Genet. 1980 Jan;177(2):345–350. doi: 10.1007/BF00267449. [DOI] [PubMed] [Google Scholar]
- Flick J. S., Johnston M. Analysis of URSG-mediated glucose repression of the GAL1 promoter of Saccharomyces cerevisiae. Genetics. 1992 Feb;130(2):295–304. doi: 10.1093/genetics/130.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick J. S., Johnston M. Two systems of glucose repression of the GAL1 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Sep;10(9):4757–4769. doi: 10.1128/mcb.10.9.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganchi P. A., Sun S. C., Greene W. C., Ballard D. W. I kappa B/MAD-3 masks the nuclear localization signal of NF-kappa B p65 and requires the transactivation domain to inhibit NF-kappa B p65 DNA binding. Mol Biol Cell. 1992 Dec;3(12):1339–1352. doi: 10.1091/mbc.3.12.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D., St Jean A., Woods R. A., Schiestl R. H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992 Mar 25;20(6):1425–1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs D. W., Johnston M. Regulated expression of the GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8597–8601. doi: 10.1073/pnas.88.19.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Mattaj I. W. Nucleocytoplasmic transport. Science. 1996 Mar 15;271(5255):1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Hu Z., Nehlin J. O., Ronne H., Michels C. A. MIG1-dependent and MIG1-independent glucose regulation of MAL gene expression in Saccharomyces cerevisiae. Curr Genet. 1995 Aug;28(3):258–266. doi: 10.1007/BF00309785. [DOI] [PubMed] [Google Scholar]
- Johnston M., Flick J. S., Pexton T. Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1994 Jun;14(6):3834–3841. doi: 10.1128/mcb.14.6.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher C. A., Redd M. J., Schultz J., Carlson M., Johnson A. D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992 Feb 21;68(4):709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- Lutfiyya L. L., Johnston M. Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol Cell Biol. 1996 Sep;16(9):4790–4797. doi: 10.1128/mcb.16.9.4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll T., Tebb G., Surana U., Robitsch H., Nasmyth K. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell. 1991 Aug 23;66(4):743–758. doi: 10.1016/0092-8674(91)90118-i. [DOI] [PubMed] [Google Scholar]
- Moore M. S. Protein translocation: nuclear export--out of the dark. Curr Biol. 1996 Feb 1;6(2):137–140. doi: 10.1016/s0960-9822(02)00444-x. [DOI] [PubMed] [Google Scholar]
- Murphy R., Wente S. R. An RNA-export mediator with an essential nuclear export signal. Nature. 1996 Sep 26;383(6598):357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- Nehlin J. O., Carlberg M., Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 1991 Nov;10(11):3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehlin J. O., Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 1990 Sep;9(9):2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostling J., Carlberg M., Ronne H. Functional domains in the Mig1 repressor. Mol Cell Biol. 1996 Mar;16(3):753–761. doi: 10.1128/mcb.16.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronne H. Glucose repression in fungi. Trends Genet. 1995 Jan;11(1):12–17. doi: 10.1016/s0168-9525(00)88980-5. [DOI] [PubMed] [Google Scholar]
- Schultz J., Carlson M. Molecular analysis of SSN6, a gene functionally related to the SNF1 protein kinase of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Oct;7(10):3637–3645. doi: 10.1128/mcb.7.10.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller H. J., Entian K. D. Extragenic suppressors of yeast glucose derepression mutants leading to constitutive synthesis of several glucose-repressible enzymes. J Bacteriol. 1991 Mar;173(6):2045–2052. doi: 10.1128/jb.173.6.2045-2052.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakawa F., Mizel S. B. In vitro activation and nuclear translocation of NF-kappa B catalyzed by cyclic AMP-dependent protein kinase and protein kinase C. Mol Cell Biol. 1989 Jun;9(6):2424–2430. doi: 10.1128/mcb.9.6.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierkstra L. N., Nouwen N. P., Verbakel J. M., Verrips C. T. Analysis of glucose repression in Saccharomyces cerevisiae by pulsing glucose to a galactose-limited continuous culture. Yeast. 1992 Dec;8(12):1077–1087. doi: 10.1002/yea.320081210. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treitel M. A., Carlson M. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci U S A. 1995 Apr 11;92(8):3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumbly R. J. Glucose repression in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1992 Jan;6(1):15–21. doi: 10.1111/j.1365-2958.1992.tb00832.x. [DOI] [PubMed] [Google Scholar]
- Tu J., Carlson M. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 1995 Dec 1;14(23):5939–5946. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzamarias D., Struhl K. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 corepressor complex to differentially regulated promoters. Genes Dev. 1995 Apr 1;9(7):821–831. doi: 10.1101/gad.9.7.821. [DOI] [PubMed] [Google Scholar]
- Tzamarias D., Struhl K. Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature. 1994 Jun 30;369(6483):758–761. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- Vallier L. G., Carlson M. Synergistic release from glucose repression by mig1 and ssn mutations in Saccharomyces cerevisiae. Genetics. 1994 May;137(1):49–54. doi: 10.1093/genetics/137.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet T., Dignard D., Thomas D. Y. A family of yeast expression vectors containing the phage f1 intergenic region. Gene. 1987;52(2-3):225–233. doi: 10.1016/0378-1119(87)90049-7. [DOI] [PubMed] [Google Scholar]
- Waddle J. A., Karpova T. S., Waterston R. H., Cooper J. A. Movement of cortical actin patches in yeast. J Cell Biol. 1996 Mar;132(5):861–870. doi: 10.1083/jcb.132.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Needleman R. Removal of Mig1p binding site converts a MAL63 constitutive mutant derived by interchromosomal gene conversion to glucose insensitivity. Genetics. 1996 Jan;142(1):51–63. doi: 10.1093/genetics/142.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W. A., Hawley S. A., Hardie D. G. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr Biol. 1996 Nov 1;6(11):1426–1434. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- Woods A., Munday M. R., Scott J., Yang X., Carlson M., Carling D. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J Biol Chem. 1994 Jul 29;269(30):19509–19515. [PubMed] [Google Scholar]
- Yaffe M. P., Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yocum R. R., Hanley S., West R., Jr, Ptashne M. Use of lacZ fusions to delimit regulatory elements of the inducible divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Oct;4(10):1985–1998. doi: 10.1128/mcb.4.10.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann F. K., Scheel I. Mutants of Saccharomyces cerevisiae resistant to carbon catabolite repression. Mol Gen Genet. 1977 Jul 7;154(1):75–82. doi: 10.1007/BF00265579. [DOI] [PubMed] [Google Scholar]