Abstract
Many receptors that are employed for the engulfment of apoptotic cells are also used for the recognition and phagocytosis of bacteria. Tyro3, Axl, and Mertk (TAM) are important in the phagocytosis of apoptotic cells by macrophages. Animals lacking these receptors are hypersensitive to bacterial products. In this report, we examine whether the TAM receptors are involved in the phagocytosis of bacteria. We found that macrophages lacking Mertk, Axl, Tyro3 or all three receptors were equally efficient in the phagocytosis of Gram-negative E. coli. Similarly, the phagocytosis of E. coli and Gram-positive S. aureus bioparticles by macrophages lacking TAM receptors was equal to wild-type. In addition, we found that Mertk did not play a role in killing of extracellular E. coli or the replication status of intracellular F. tularensis. Thus, while TAM receptors may regulate signal transduction to bacterial components, they are not essential for the phagocytosis and killing of bacteria.
Keywords: Macrophage, E. coli, Mertk, Axl, Tyro3, phagocytosis
Introduction
During the course of a bacterial infection, innate immune cells such as neutrophils and macrophages sense bacteria and bacterial products via pathogen-associated molecular patterns (PAMPS) such as lipopolysaccharide (LPS) or peptidoglycan. PAMPs are recognized by several pattern recognition receptors on innate immune cells, including the Toll-like receptors (TLR)[1]. Following recognition, the resident cells release pro-inflammatory cytokines and chemokines to recruit neutrophils and additional macrophages to the lesion. These responding neutrophils and monocytes arrive equipped as professional phagocytes to quickly ingest and kill the invading bacteria. This rapid immune response and phagocytosis of bacteria is critical to the resolution of a bacterial infection.
Many other receptors are implicated in the recognition and phagocytosis of bacteria including: Scavenger receptor A, Mannose receptor, TLRs, integrins, as well as Fc receptors and complement receptors[2, 3]. Upon binding of bacteria, these receptors signal a rearrangement of the actin cytoskeleton that is mediated by the Rho family of GTPases, Rac, Rho, and Cdc42, ultimately leading to engulfment of the bacteria[4]. One indication of the importance of these molecules is evident in that some bacteria attempt to evade phagocytosis by producing molecules that modulate the signaling to and from the Rho GTPases [5, 6].
Phagocytosis is not only important for controlling infection, it is also key in down-regulating inflammation and limiting tissue damage by returning the infected area to proper homeostasis. This is partly accomplished by the phagocytosis of apoptotic cells. As bacteria grow and/or infect cells, these cells often succumb to the infection and die, either indirectly by toxic bacterial byproducts or directly by intracellular pathogens. For example, neutrophils have been shown to become apoptotic after treatment with E. coli[7]. Apoptosis is a form of cell death that has been well characterized and includes exposure of phosphatidylserine from the inner membrane to the outer surface of the dying cell. Subsequently, these apoptotic cells are cleared from the infected area typically by macrophages. Interestingly, many of the same receptors that facilitate phagocytosis of bacteria are also important for the phagocytosis of apoptotic cells including CD14, Scavenger receptor A, Fc Receptor, complement, and integrins[8, 9].
The Tyro3/Axl/Mertk (TAM) receptors have been shown to be required for efficient phagocytosis of apoptotic cells [10, 11]. All three receptors are expressed by macrophage and dendritic cells(DC), however, Mertk is also expressed by NK cells, but is not expressed on neutrophils or non-malignant lymphocytes[11–13]. These proteins are receptor tyrosine kinases that are similar in structure and function. Each family member has two Ig-like and two fibronectin III-like extracellular domains, as well as a characteristic KWAIAES sequence in the intracellular domain[12, 14]. In macrophages, Mertk is essential for rapid ingestion of apoptotic cells[10]; though Axl and Tyro3 are partly involved[11]. In contrast, Axl and Tyro3 appear to function in DC and Mertk has no role in phagocytosis although the process is much slower[11, 15].Knock-out mice have been generated for all three family members[16, 17]. The inability to properly clear apoptotic cells in these knock-out mice results in the development of a lupus-like autoimmunity characterized by autoantibody production as well as splenomegaly[10, 15, 18]. Gas6, a ligand for the TAM family members, binds these receptors with a relative affinity of Axl>Tyro3> >Mertk[19]. Gas6 is produced by several cell types including macrophages, DC and apoptotic thymocytes[11, 20]. Another ligand for the family member Tyro3 is the serum protein, Protein S [21]. Both Gas6 and Protein S bind phosphatidylserine on apoptotic cells and it is through this molecular bridging interaction that the TAM family members bind apoptotic cells[22].
In addition, to their role in phagocytosis of apoptotic cells, the TAM family members have also been shown to have a role in the innate immune response. Mice lacking functional Mertk (mertk−/−) are hypersensitive to Gram-negative bacterial LPS and are susceptible to endotoxic shock that is mediated by prolonged NF-κB activity resulting in over production of TNFα[16]. This regulation of NF-κB by Mertk shown in macrophages has been corroborated in DC and the phagocytosis of apoptotic cells by DC appears to be important for self tolerance in a model of diabetes[23] Furthermore, knock-out mice lacking all three TAM family members (TAM−/−) mice also displayed an increase in serum TNFα levels following LPS injection[18] and DC from TAM−/− mice have been shown to have hyper-immune responses to TLR ligands[24]. Interestingly Axl and Interferon receptor appear to cooperate to modulate STAT inhibiting the innate immune response to TLR mediated activation by the induction of SOCS1[24]. However in these studies, the response to whole bacteria and importantly, phagocytosis of bacteria was not thoroughly examined.
Previous studies that have investigated the involvement of TAM receptors in phagocytosis of bacteria have provided conflicting results[10, 18]. Macrophages isolated from mertk−/− mice were shown to phagocytize the Gram-positive bacterium L. monocytogenes similarly to wild-type[10]. However, the TAM−/− macrophages displayed an increased ability to phagocytize Gram-negative E. coli[18].
Based on the importance of TLRs for phagocytosis, the ability of the Mertk to regulate LPS signaling, and the increase in phagocytosis of E. coli by the TAM−/−macrophages, we sought to determine whether Mertk regulated phagocytosis of the Gram-negative bacterium, E. coli. In this study we demonstrate, by multiple methods, that individually or collectively the TAM family members are not essential for phagocytosis of E. coli by thioglycollate-elicited macrophages. In addition, we show that individual TAM receptors are also not important for phagocytosis of bacteria by resident and bone marrow-derived macrophages. Complementary to the lack of a role in phagocytosis, we show that Mertk is not essential for killing of E. coli or modulation of growth of the intracellular pathogen F. tularensis.
Methods
Animals
Mice were housed in a specific pathogen-free facility and maintained according to the UNC-Chapel Hill Institutional Animal Use and Care Committee guidelines. mertk−/− mice were developed here at UNC. mertk−/− is the updated nomenclature which was described previously as merkd or mertkkd[10, 11]. Although the cytoplasmic kinase domain was targeted, the lack of protein expression makes these mice null for Mertk. axl−/−, and tyro3−/− mice were kindly provided by Dr. Stephen P. Goff (Columbia University, New York) and Dr. Greg Lemke (Salk Institute for Biological Studies, San Diego) respectively. TAM−/−mice were generated by breeding of backcrossed single knock-outs. All genotypes were backcrossed at least 6 generations to the C57BL/6J background. Male mice 8–12 weeks old were used in all studies. Wild-type mice used were strain C57BL/6J. Macrophages
Thioglycollate-elicited macrophages were obtained as previously described[10]. Briefly, 3 mL of 3% thioglycollate was injected intraperitoneally (I.P.) 3 days prior to harvest by peritoneal wash in PBS. Cells were washed 3 times in PBS and allowed to adhere for 3 hours in media (RPMI 1640 (Gibco) supplemented with 5% fetal bovine serum, 1 mM sodium pyruvate, 2 × 10−5 2-ME, 10 mM HEPES, 50 units penicillin G and 50 µg/mL streptomycin sulphate). Nonadherent cells were washed off and fresh media was added. In order to eliminate activity due to stimulation by thioglycollate, macrophages were allowed to rest at 37°C 5% CO2 for seven days prior to experimental use.
Resident peritoneal macrophages were obtained by 60 seconds of gentle peritoneal lavage in Versene. Cells were removed, washed and allowed to adhere onto glass coverslips in 24 well plates or plastic tissue culture dishes. Non-adherent cells were washed off which resulted in a highly enriched population of resident macrophages.
Bone marrow-derived macrophages were obtained by harvesting bone marrow from mouse femurs. Cells were flushed from femurs, washed and incubated overnight in DMEM-H (Gibco) supplemented with 50 units penicillin G, 50 µg/mL streptomycin sulphate and 10 mM HEPES. Non-adherent cells were removed and plated in media containing 10 ng/mL recombinant mouse M-CSF (Peprotech). Fresh media was added on day 4. Macrophages were harvested on day 8 and re-plated at appropriate density for experimental use on day 9. Bone marrow-derived macrophages were confirmed to be 100% CD11b–positive and 100% CD11c–negative by flow cytometry (data not shown).
Phagocytosis Assays
Phagocytosis assays were preformed using a MOI of 10 E. coli 0111:B4 (ATCC) transformed with green fluorescent protein plasmid (pEGFP; Clontech). Green fluorescent protein (GFP)-containing E. coli (E. coli O111:B4-GFP) were added to antibiotic-free, serum-free media and aliquoted into wells containing macrophages on glass coverslips. Macrophages were allowed to phagocytize bacteria for the indicated time periods. Macrophages were then washed 5 times in PBS and 0.2% trypan blue was added to wells to quench fluorescence from extracellular bacteria. Cells were then fixed in 2% paraformaldehyde and coverslips were mounted on slides with Vectamount mounting media for fluorescence (Vector). Phagocytosis was determined by overlays of bright field and GFP images captured using an Olympus fluorescent microscope equipped with a DP70 digital camera and Image Pro Plus software (Opelco). At least 100 macrophages were counted per time point with triplicate samples per genotype.
Fluorimetric phagocytosis assays were also preformed. Briefly, cells were plated in 96 well plates and a MOI of 10 bacteria in serum-free and antibiotic-free media was added to each well. After indicated time periods, cells were washed as described above and trypan blue was added to quench any extracellular fluorescence. Plates were then read on a Fluoroskan Ascent FL fluorimeter (Thermo Labsystems) according to manufacturers instructions. E. coli and S. aureus Bioparticles used for fluorimetric assays were obtained from Molecular Probes. Normalized fluorescent intensity was calculated by obtaining a ratio of fluorescent cell tracker green-labeled wild-type macrophages to mertk−/−macrophages (measured in separate wells) and multiplying this ratio to the ingested bacterial bioparticle fluorescence.
In addition, phagocytosis assays were performed and analyzed by flow cytometry. For these experiments, cells were plated in 60 mM dishes and a MOI of 10 bacteria in serum-free, antibiotic-free media was added for indicated time periods. Cells were then washed and trypan blue was added as previously described. Cells were then fixed and analyzed using a BD FACScan flow cytometer.
Apoptotic Cells
Apoptotic cells were harvested as previously described [11]. Briefly, thymi from 4–6 week old wild-type mice were harvested and tissues were mechanically dissociated using forceps. Cells were then washed and incubated in media containing 2 µM Dexamethasone. After 5 hours, Cell tracker Orange (Molecular Probes) was added to thymocytes for 30 minutes according to manufacturer’s instructions. Cells were then washed in 1x PBS and then allowed to incubate in media for 30 minutes prior to experimentation.
Bactericidal Assays
Thioglycollate-elicited macrophages were plated and rested as described above. Cells were washed 3 times with PBS to remove media containing antibiotics. MOI of 10 E. coli O111:B4-GFP was added to wells. After 30 minutes 5 µg/mL gentamicin was added to each well to kill extracellular bacteria. After the allotted times, cells were harvested by scraping in PBS and plated in serial dilutions on LB agar containing ampicillin to select only E. coli O111:B4-GFP.
Gentamicin protection assays
All Francisella tularensis Live Vaccine Strain (LVS) gentamicin protection assays were performed under appropriate bio safety precautions. Approximately 105 bone marrow-derived macrophages were plated per well in a 96-well tissue culture dish. Bacteria were added at a multiplicity of infection of 10:1 or 100:1 in a volume of 100 µL per well. Supernatants were aspirated and replaced with media containing gentamicin (25 µg/mL) to kill extracellular bacteria at 4 hours post-inoculation. Six and 24 hours post-inoculation, cells were rinsed with 1x PBS and scraped from the wells with sterile wooden applicator sticks, vortexed, diluted, and plated onto chocolate agar for bacterial recovery. All gentamicin protection experiments were performed in triplicate and the mean and standard deviations were calculated.
Statistics
Statistics were performed using Graph Pad Prism Software. Where appropriate students t-test or 2-way ANOVA with Bonferroni’s post-tests were performed. A p value of less than 0.05 was considered significant.
Results
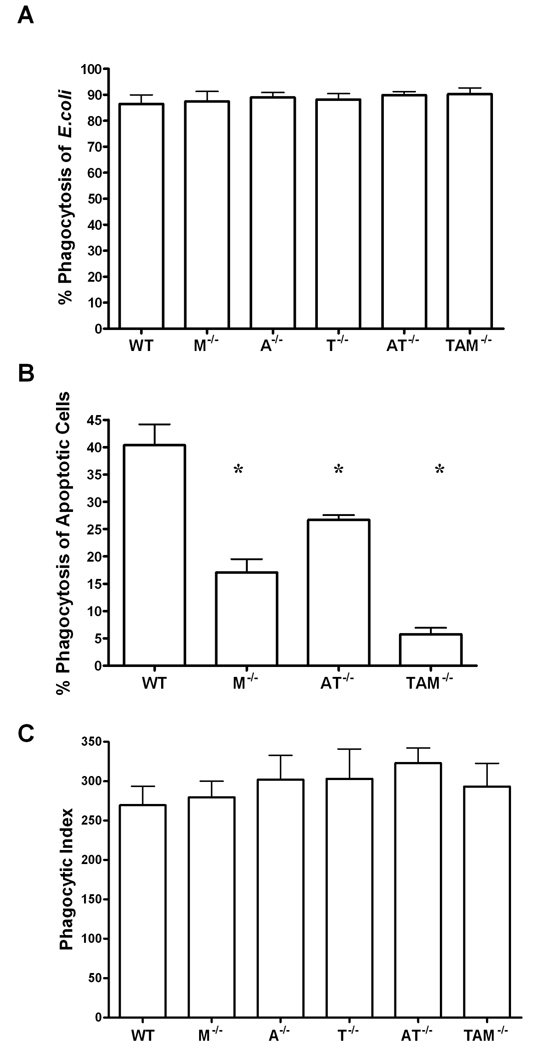
While a role for the TAM receptors in phagocytosis of apoptotic cells has been well documented[10, 11], their participation in phagocytosis of bacteria remains controversial. To determine if the TAM receptors play a role in the phagocytosis of bacteria, thioglycollate-elicited macrophages were harvested from wild-type, mertk−/−, axl−/−, tyro3−/−, axl−/−/tyro3−/− double knock-outs and TAM−/−triple knock-out mice and exposed to E. coli O111:B4-GFP for one hour. No significant differences were found in terms of the number of cells that had phagocytized at least one bacterium (percent phagocytosis, Figure 1A). In contrast, macrophages from mertk−/− and axl−/−/tyro3−/− mice harvested at the same time were deficient in phagocytosis of apoptotic cells similar to previous publications, and documented for the first time, this deficiency was most apparent when all three receptors were lacking (Figure 1B). To determine whether one strain was phagocytizing more bacteria on a per cell basis, we also performed a phagocytic index. The phagocytic index was calculated as the total number of bacteria ingested by 100 cells. In corroboration of the percent phagocytosis results, the total number of bacteria ingested was similar between strains tested across the time course (Figure 1C). Thus, despite the importance of Mertk, and to a lesser extent Axl and Tyro3, in the phagocytosis of apoptotic cells, neither Mertk nor the other family members appear to play a prominent role in bacterial engulfment.
Figure 1. TAM receptors are not required for the phagocytosis of E. coli O111:B4 GFP by macrophages.
A, Phagocytosis assay using MOI of 10 E. coli O111:B4 GFP by thioglycollate-elicited macrophages from wild-type (WT), mertk−/−(M−/−), axl−/−(A−/−), and tyro3−/−(T−/−), axl−/−/tyro3−/−(AT−/−), and TAM−/− mice. B, In vitro phagocytosis assay of apoptotic cells 10:1 to thioglycollate-elicited macrophages from wild-type (WT), mertk−/−(M−/−),axl−/−/tyro3−/−(AT−/−), and TAM−/−mice. C, Phagocytic index calculated as number of bacteria phagocytized by 100 cells from samples in 1A. Figures are mean and standard error, n=3 and representative of at least 2 separate experiments. * p<0.01 by students t test vs. wild-type. A and C are not statistically significant by one-way ANOVA or post test.
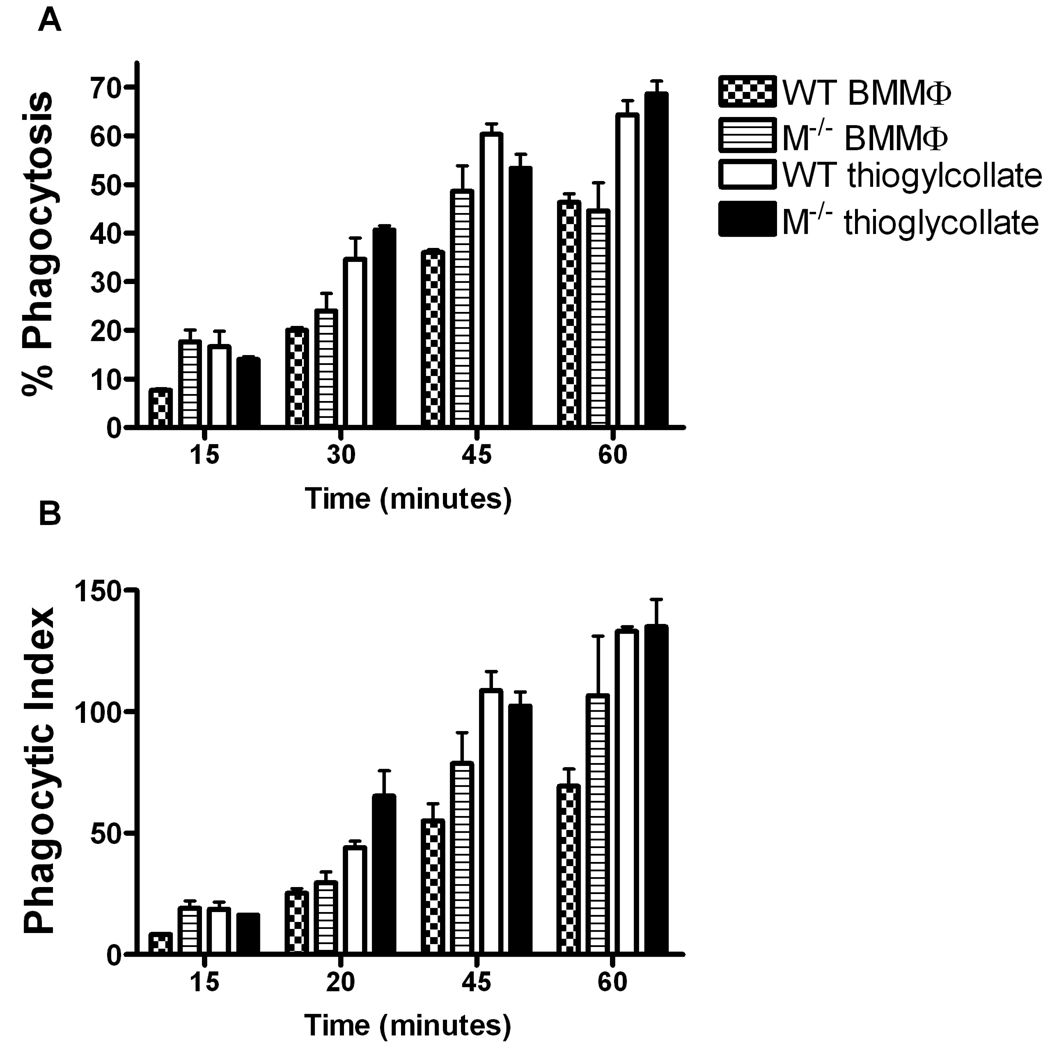
To determine whether Mertk could affect phagocytosis of E. coli at earlier time points, we preformed a similar experiment using wild-type and mertk−/− thioglycollate-elicited macrophages and bone marrow-derived macrophages (BMMΦ) from 15 minutes to 1 hour. While phagocytosis proceeded in a time dependent manner for both types of macrophages, there were no statistically significant differences between wild-type and mertk−/− strains in percent phagocytosis (Figure 2A) or phagocytic index (Figure 2B). Interestingly, though independent of genotype, the thioglycollate-elicited macrophages were found to be significantly more active in phagocytosis of bacteria compared to BMMΦ by 2-way ANOVA in percent phagocytosis and phagocytic index (Figure 2Aand Figure 2B).
Figure 2. Phagocytosis of E coli O 111:B4 is time dependent and Mertk independent in thioglycollate-elicited and bone marrow-derived macrophages.
A, In vitro phagocytosis assay using a MOI of 10 E. coli O111:B4 GFP by thioglycollate and BMMΦ from wild-type and mertk−/− mice, over a time course of 15 to 60 minutes. B, Phagocytic index was calculated as number of bacteria phagocytized by 100 cells from samples in A. Figures are mean and standard error, n=3 and representative of at least 2 separate experiments. No statistically significant differences were found between genotypes of either cell type by 2 way ANOVA for both panel A and panel B. Statistically significant differences were found between BMMΦ and thioglycollate-elicited macrophages by 2 way ANOVA in both genotypes p<0.0001 for Figure 2A. Bonferroni’s post tests also show significance between wild type BMMΦ and thioglycollate-elicited macrophages at 30, 45, and 60 minutes p<0.01 for figure 2A. Statistical significance was also found by Bonferroni’s post test between mertk−/− BMMΦ and thioglycollate-elicited macrophages at 30 and 60 mins p<0.05 and 0.001 respectively for Figure 2A. For Figure 2B statistically significant differences were found between BMMΦ and thioglycollate-elicited macrophages by 2-way ANOVA in wild-type and mertk−/−p<0.0001, p<0.05 respectively. Post tests indicate significance at the 45 and 60 minute time points for wild-type, p<0.001.
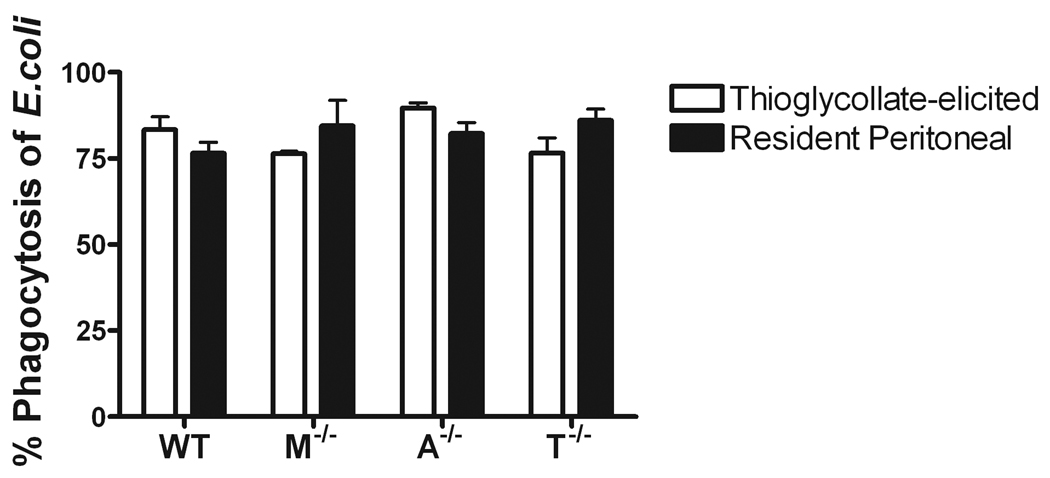
Since the macrophages used in the previous study were either thioglycollate-elicited or derived in vitro with cytokines, we wanted to confirm our results using resident peritoneal macrophages. Unlike BMMΦ, resident peritoneal macrophages phagocytize similarly to thioglycollate-elicited macrophages. However, there still remained no differences in phagocytosis of E. coli between wild-type and mertk−/− macrophages. In addition to mertk−/−, macrophages from axl−/− and tyro3−/− single knock-out mice also showed no statistical differences in percent phagocytosis when compared to wild-type resident macrophages (Figure 3).
Figure 3. Phagocytosis of E. coli O 111:B4 is similar in thioglycollate and resident peritoneal macrophages.
In vitro phagocytosis assay of MOI of 10 E. coli O111:B4 GFP and thioglycollate (□) and resident peritoneal (▪) macrophages from wild-type (WT), mertk−/−(M−/−), axl−/−(A−/−), and tyro3−/−(T−/−). Figures are mean and standard error, n=3 and representative of at least 2 separate experiments. No statistically significant differences found between genotypes or cell types by one-way ANOVA.
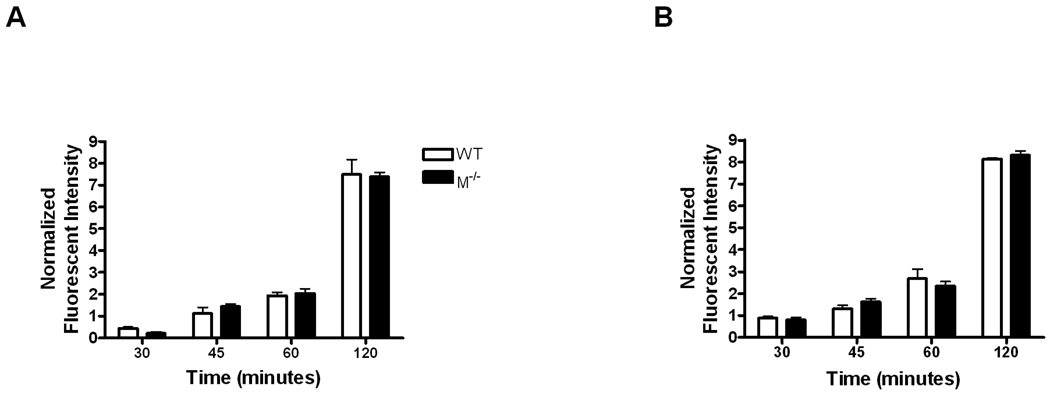
To confirm that phagocytosis was indeed similar between wild-type and mertk−/− macrophages, we also assessed the phagocytosis of E. coli O111:B4 GFP by thioglycollate-elicited macrophages using a flow cytometry-based approach. This approach allows for a large scale analysis of the amount of phagocytosis that is taking place. As the cells phagocytize bacteria, they accumulate GFP, become fluorescent and are therefore detectable by flow cytometry. As shown in Figure 4, the number of fluorescent thioglycollate-elicited macrophages increased rapidly with time; however, similar to our previous findings, wild-type, mertk−/−, and TAM−/− macrophages showed similar numbers of cells containing fluorescence at all time points tested. In addition, even at 120 minutes a few cells did not become fluorescent indicating that not all macrophages will phagocytize a bacterium (Figure 4).
Figure 4. In vitro phagocytosis assay by flow cytometry.
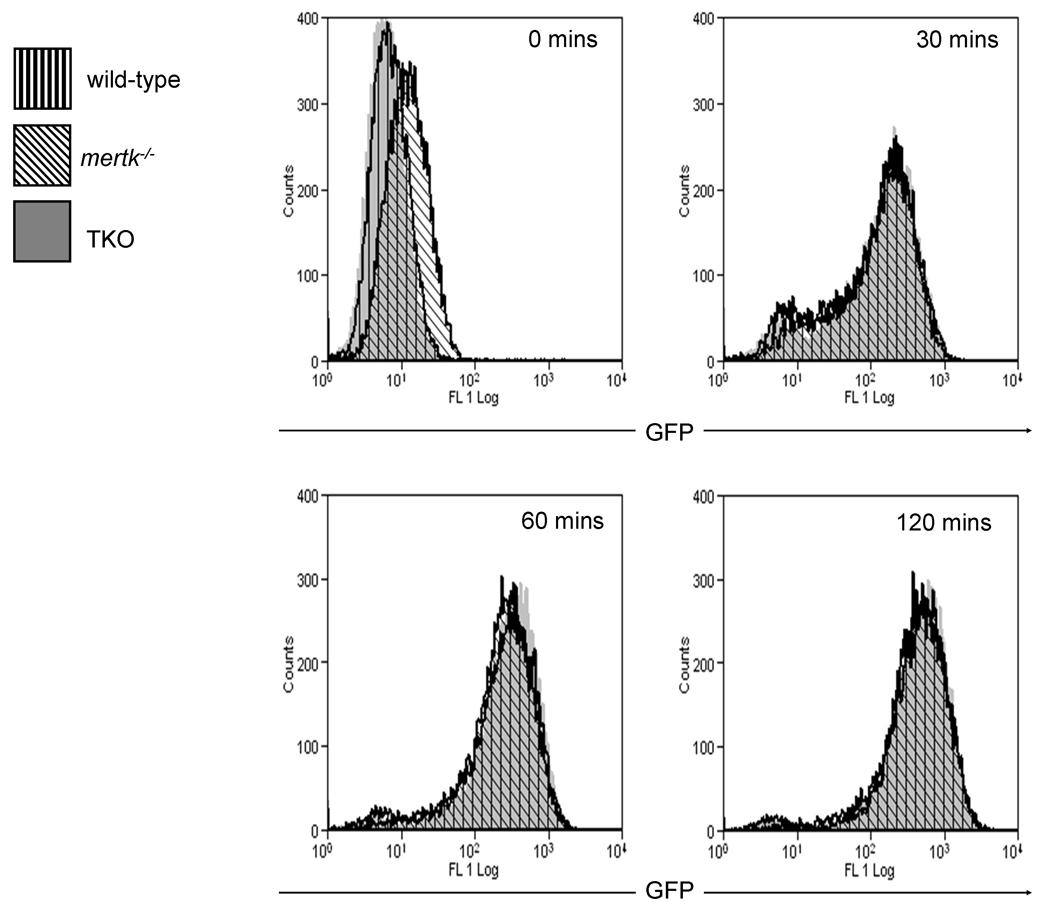
A time course of 30 to 120 minutes was used to examine the phagocytosis of MOI of 10 E. coli O111:B4 GFP by wild-type, mertk−/−, and TAM−/− thioglycollate-elicited macrophages. Data is representative of 2 separate experiments
E. coli O111:B4 is known to be an extracellular bacteria that deliberately prevents phagocytosis and forms pedestals on the gut epithelial cells which it normally encounters[25]. To rule out a role for this process and lack of an effect by Mertk on phagocytosis of bacteria, we performed a phagocytosis assay using E. coli bioparticles. Bioparticles are chemically or heat-killed bacteria that are fluorescently labeled and thus easily detected by a fluorimeter. E. coli bioparticles were phagocytized similarly by both wild-type and mertk−/−thioglycollate-elicited macrophages (Figure 5A). Thus, regardless of whether the E. coli were alive or dead; their phagocytosis was unimpaired in wild-type or mertk−/− macrophages. Since components of Gram-positive and Gram-negative bacteria are recognized by different TLRs, we wanted to demonstrate that our results were not specific to Gram-negative bacteria. Therefore, we performed the same phagocytosis assay using Gram-positive S. aureus bioparticles. As with the E. coli, wild-type and mertk−/− macrophages phagocytized S. aureus bioparticles equally (Figure 5B). Therefore, Mertk is not essential for phagocytosis of bacterial bioparticles regardless of Gram status.
Figure 5. Lack of affect in mertk −/− macrophages is not dependent on Gram status.
In vitro phagocytosis assay of E. coli (A) and S. aureus (B) bioparticles at a ratio of 10:1 by wild-type (□) and mertk−/− (▪) thioglycollate-elicited macrophages. Bioparticles were incubated over a time course of 15–120 minutes. Groups at each time point are mean and standard error of n=4, and is representative of 3 experiments. No statistically significant differences were found between genotypes by 2-way ANOVA.
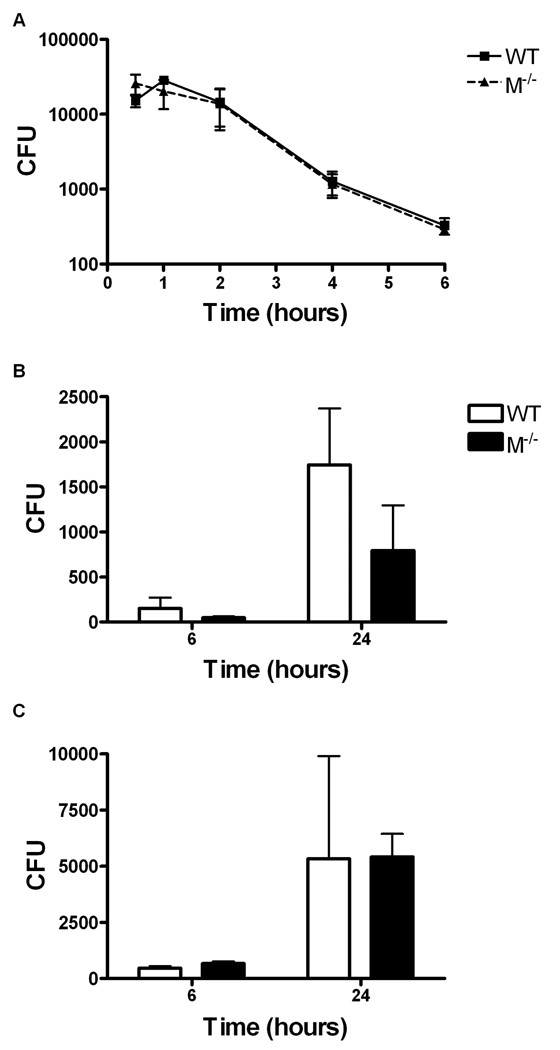
Previous reports indicate mertk−/− macrophages produce greater amounts of TNFα in response to E. coli LPS[16]. Thus, we hypothesized that after phagocytosis these hyper-activated macrophages may be more efficient at bacterial killing than wild-type. To determine if the lack of Mertk affected the ability of the macrophages to kill bacteria, we performed a bactericidal assay using the naturally extracellular E. coli O111:B4 GFP used in previous experiments. As shown in Figure 6A, mertk−/− macrophages ingested and killed the bacteria equally to wild-type macrophages over a 6 hour period. Thus, Mertk does not inhibit or enhance the ability of the macrophage to kill this strain of bacteria.
Figure 6. Killing and replication of bacteria are not Mertk dependent.
A, In vitro bactericidal assay of E. coli O111:B4 GFP by thioglycollate-elicited wild-type (▪) and mertk−/− (▲) macrophages was conducted over a 6 hour time course and at a MOI of 10:1. B and C, In vitro gentamicin protection assay was used to examine the killing of the intracellular pathogen F. tularensis at a MOI of 10 (B) or MOI of 100 (C) by wild-type (□) and mertk−/− (▪) BMMΦ. Each bar is the mean and standard deviation of triplicate wells, n=3 and is representative of 3 separate experiments. No statistically significant differences were between genotypes found by 2-way ANOVA.
We further examined whether the presence or absence of Mertk would affect the killing of intracellular pathogens that replicate in the cytoplasm. Francisella tularensis is a Gram-negative facultative bacterium that is able to escape the phagolysosome and replicates in the cytosol of the host cells such as macrophages[26]. F. tularensis is able to down-regulate phagocytosis and TLR signaling and inhibits apoptosis of macrophages which appears to provide an advantage in propagation of the bacterium. As shown in Figure 6B and C, F. tularensis Live Vaccine Strain (LVS) had similar replication kinetics in mertk−/− and wild-type macrophages at two different MOIs. These data suggest that Mertk does not affect the ability of the macrophage to slow intracellular replication or kill bacteria.
Discussion
Many receptors including CD14, scavenger receptor, integrins, complement receptors and Fc receptors recognize and participate in the phagocytosis of apoptotic cells as well as bacteria[2, 9, 27]. The Tyro3, Axl, and Mertk (TAM) family of receptors has been shown to be important for the phagocytosis of apoptotic cells[10, 11]. Previous studies have found conflicting evidence for the role of the TAM receptors in the phagocytosis of bacteria[10, 18]. Our past findings suggested that the Mertk receptor did not play a critical role in the phagocytosis of the Gram-positive bacteria L. monocytogenes by macrophages[10]. This study used live L. monocytogenes and this intracellular bacteria can invade host cells independent of phagocytosis[28]. As such, it is possible that the results from this study were merely a consequence of the bacteria being equally able to invade cells in the presence or absence Mertk. A second study used only the TAM triple knock-out macrophages and examined phagocytosis of the Gram-negative bacteria E. coli [18]. In this study, the investigators found that phagocytosis of bacteria was increased over 3-fold in the TAM−/− cells; however, these cells were derived from mice with a heterogeneous background of founder genes that may have influenced the phenotype.
In this report, we find that macrophages from mertk−/−, axl−/−, tyro3−/−, axl−/− /tyro3−/− double knock-outs and TAM−/− triple knock-out mice on the C57BL/6 background ingest E. coli bacteria equally to wild-type macrophages (Figure 1, Figure 2, Figure 3, Figure 4). Our current findings that TAM family members do not play a role in phagocytosis of E. coli is in contrast to a previously published report[18]. Furthermore, we examined the phagocytosis of S. Aureus and found Mertk does not affect the engulfment of Gram-positive bacteria, corroborating our past finding with L. monocytogenes[10]. Interestingly prior exposure of macrophages to apoptotic cells significantly reduced the phagocytosis of bacteria; however this process too was independent of the TAM family members as knock out macrophages also could not ingest bacteria after treatment with apoptotic cells (data not shown). It is plausible that apoptotic cells bind multiple receptors that share recognition of bacterial components such as complement receptors, Scavenger Receptor or CD14 and these could be acting as competitive ligands that prevent macrophages from recognizing the bacteria. Nonetheless, our results strongly suggest that Mertk and the family members, Axl and Tyro3 are not essential for the engulfment of Gram-positive or Gram-negative bacteria. It is likely that other receptors present on macrophages that recognize microbial molecular components may compensate for the lack of TAM receptors.
We also examined the role of Mertk on three populations of macrophages in the event that different populations might use the TAM receptors preferentially. We show fewer bone marrow-derived macrophages (BMMΦ) were phagocytic for bacteria compared to thioglycollate-elicited macrophages (Figure 2). Even though the thioglycollate-elicited macrophages were rested prior to phagocytosis for 7 days, the data suggests that they phagocytize in greater numbers than BMMΦ. For example, at the one hour time point, about 65% of wild-type thioglycollate-elicited macrophages had engulfed at least one bacterium as compared to only about 45% of the wild-type BMMΦ (Figure 2). Interestingly resident unstimulated macrophages ingest E. coli similar to thioglycollate macrophages (Figure 3). These results point to the possibility that different macrophage inducing agents produce different phagocytic activities among macrophages. Those collected after thioglycollate exposure tend to be more phagocytic whereas BMMΦ may be more differentiated and less phagocytic.
Since mertk−/− macrophages produce elevated TNFα in response to LPS [14], we hypothesized that macrophages from these mice may be able to kill bacteria more efficiently. As we showed in Figure 6A, the lack of Mertk had no effect on the ability of the macrophage to kill the naturally extracellular pathogen E. coli. In addition, we examined a similar scenario using an intracellular pathogen, F. tularensis LVS. F. tularensis is Gram-negative bacterium that is able to escape the phagolysosome and replicate in the cytosol of macrophages [26]. We examined the ability of the macrophages lacking Mertk to inhibit this replication. As with the E. coli, we saw no difference in the replication of F. tularensis LVS between the mertk−/− and wild-type macrophages (Figure 6B and Figure 6C). Thus, Mertk also does not play an essential role in inhibition of replication or killing of bacteria.
In summary, while previously published data document a role for TAM receptors in response to bacterial by products, we demonstrate that Mertk, Axl, and Tyro3 appear to have no essential role in the phagocytosis of Gram-negative or Gram-positive bacteria or in the killing of extracellular or intracellular bacteria. This is in contrast to our previous understanding of TAM receptors in bacterial clearance.
Acknowledgements
This work was funded in part by National Institute for Allergy and Infectious Disease Grant 5-31702
The authors would like to thank Stephen Goff for the axl−/− and Greg Lemke for the tyro3−/− mice. We are also grateful to Heather Seitz and Paul Gholke for technical assistance and Andrea Portbury, Lorelei Taylor, and Kasturi Puranam for critical review of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4(7):499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 2.Aderem A, Underhill DM. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 3.Blander JM, Medzhitov R. Regulation of phagosome maturation by signals from toll-like receptors. Science. 2004;304(5673):1014–1018. doi: 10.1126/science.1096158. [DOI] [PubMed] [Google Scholar]
- 4.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420(6916):629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 5.Mejia E, Bliska JB, Viboud GI. Yersinia controls type III effector delivery into host cells by modulating Rho activity. PLoS Pathog. 2008;4(1):e3. doi: 10.1371/journal.ppat.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groves E, et al. Molecular mechanisms of phagocytic uptake in mammalian cells. Cell Mol Life Sci. 2008;65(13):1957–1976. doi: 10.1007/s00018-008-7578-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watson RW, et al. Neutrophils undergo apoptosis following ingestion of Escherichia coli. J Immunol. 1996;156(10):3986–3992. [PubMed] [Google Scholar]
- 8.Stuart LM, Ezekowitz RA. Phagocytosis: elegant complexity. Immunity. 2005;22(5):539–550. doi: 10.1016/j.immuni.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Platt N, et al. Role for the class A macrophage scavenger receptor in the phagocytosis of apoptotic thymocytes in vitro. Proc Natl Acad Sci U S A. 1996;93(22):12456–12460. doi: 10.1073/pnas.93.22.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott RS, et al. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature. 2001;411(6834):207–211. doi: 10.1038/35075603. [DOI] [PubMed] [Google Scholar]
- 11.Seitz HM, et al. Macrophages and dendritic cells use different Axl/Mertk/Tyro3 receptors in clearance of apoptotic cells. J Immunol. 2007;178(9):5635–5642. doi: 10.4049/jimmunol.178.9.5635. [DOI] [PubMed] [Google Scholar]
- 12.Graham DK, et al. Cloning and developmental expression analysis of the murine c-mer tyrosine kinase. Oncogene. 1995;10(12):2349–2359. [PubMed] [Google Scholar]
- 13.Caraux A, et al. Natural killer cell differentiation driven by Tyro3 receptor tyrosine kinases. Nat Immunol. 2006;7(7):747–754. doi: 10.1038/ni1353. [DOI] [PubMed] [Google Scholar]
- 14.Graham DK, et al. Cloning and mRNA expression analysis of a novel human protooncogene, c-mer. Cell Growth Differ. 1994;5(6):647–657. [PubMed] [Google Scholar]
- 15.Cohen PL, et al. Delayed apoptotic cell clearance and lupus-like autoimmunity in mice lacking the c-mer membrane tyrosine kinase. J Exp Med. 2002;196(1):135–140. doi: 10.1084/jem.20012094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camenisch TD, et al. A novel receptor tyrosine kinase, Mer, inhibits TNF-alpha production and lipopolysaccharide-induced endotoxic shock. J Immunol. 1999;162(6):3498–3503. [PubMed] [Google Scholar]
- 17.Lu Q, et al. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature. 1999;398(6729):723–728. doi: 10.1038/19554. [DOI] [PubMed] [Google Scholar]
- 18.Lu Q, Lemke G. Homeostatic regulation of the immune system by receptor tyrosine kinases of the Tyro 3 family. Science. 2001;293(5528):306–311. doi: 10.1126/science.1061663. [DOI] [PubMed] [Google Scholar]
- 19.Nagata K, et al. Identification of the product of growth arrest-specific gene 6 as a common ligand for Axl, Sky, and Mer receptor tyrosine kinases. J Biol Chem. 1996;271(47):30022–30027. doi: 10.1074/jbc.271.47.30022. [DOI] [PubMed] [Google Scholar]
- 20.Wallet MA, et al. MerTK is required for apoptotic cell-induced T cell tolerance. J Exp Med. 2008;205(1):219–232. doi: 10.1084/jem.20062293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stitt TN, et al. The anticoagulation factor protein S and its relative, Gas6, are ligands for the Tyro 3/Axl family of receptor tyrosine kinases. Cell. 1995;80(4):661–670. doi: 10.1016/0092-8674(95)90520-0. [DOI] [PubMed] [Google Scholar]
- 22.Wu Y, Tibrewal N, Birge RB. Phosphatidylserine recognition by phagocytes: a view to a kill. Trends Cell Biol. 2006;16(4):189–197. doi: 10.1016/j.tcb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Sen P, et al. Apoptotic cells induce Mer tyrosine kinase-dependent blockade of NF-kappaB activation in dendritic cells. Blood. 2007;109(2):653–660. doi: 10.1182/blood-2006-04-017368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rothlin CV, et al. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell. 2007;131(6):1124–1136. doi: 10.1016/j.cell.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 25.Vallance BA, Finlay BB. Exploitation of host cells by enteropathogenic Escherichia coli. Proc Natl Acad Sci U S A. 2000;97(16):8799–8806. doi: 10.1073/pnas.97.16.8799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clemens DL, Lee BY, Horwitz MA. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect Immun. 2004;72(6):3204–3217. doi: 10.1128/IAI.72.6.3204-3217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Areschoug T, Gordon S. Scavenger receptors: role in innate immunity and microbial pathogenesis. Cell Microbiol. 2009 doi: 10.1111/j.1462-5822.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- 28.Vazquez-Boland JA, et al. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14(3):584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]