Abstract
Genetic alteration in oral premalignant lesions (OPLs), the precursors of oral squamous cell carcinomas (OSCCs), may represent key changes in disease initiation and development. We ask if DNA amplification occurs at this early stage of cancer development and which oncogenic pathways are disrupted in OPLs. Here we evaluated 50 high-grade dysplasias and low-grade dysplasias that later progressed to cancer for gene dosage aberrations using tiling-path DNA microarrays. Early occurrences of DNA amplification and homozygous deletion were frequently detected, with 40% (20/50) of these early lesions exhibiting such features. Expression for 88 genes in seven recurrent amplicons were evaluated in five independent head and neck cancer datasets, with 40 candidates found to be overexpressed relative to normal tissues. These genes were significantly enriched in the canonical ERK/MAPK, FGF, p53, PTEN, and PI3K/AKT signaling pathways (P = 8.95x10-3--3.18×10-2). These identified pathways share interactions in one signaling network, and amplification-mediated deregulation of this network was found in 30.0% of these preinvasive lesions. No such alterations were found in 14 low-grade dysplasias that did not progress, while 43.5% (10/23) of OSCCs were found to have altered genes within the pathways with DNA amplification. Multi-target FISH showed that amplification of EGFR and CCND1 can co-exist in single cells of an oral dysplasia, suggesting the dependence on multiple oncogenes for OPL progression. Taken together, these findings identify a critical biological network that is frequently disrupted in high-risk OPLs, with different specific genes disrupted in different individuals.
Keywords: oral premalignant lesion, gene amplification, homozygous deletion, DNA microarray, signaling pathway
Introduction
Oral squamous cell carcinoma (OSCC) is one of the most common head and neck neoplasm, with more than 30,000 cases identified in the United States each year.1 Despite advances in treatment, the 5-year survival rate of all stages and advanced stage (stage III and IV) remains at less than 50% and 25%, respectively, for the past three decades.2 The poor survival rate is mainly because most patients were diagnosed at the advanced stages of the disease. Early detection and treatment at the premalignant stages with intensified follow-up would improve patient survival.2, 3
Oral cancer is believed to progress from hyperplasia, through various extent of dysplastic changes, carcinoma in situ (CIS), and finally breaking through the basement membrane at the invasive SCC stage.4 In oral premalignant lesions (OPLs), the presence and degree of epithelial dysplasia is used to assess the risk for progression to malignancy. By definition, dysplasia is characterized by cellular atypia and loss of normal maturation and stratification and has no evidence of invasion.5 High-grade preinvasive lesion, including severe dysplasia and CIS, is associated with the strongest risk for malignant transformation.3, 6-9 Thus, patients with these lesions are treated in British Columbia to prevent further development into invasive SCC.3 However, the majority of the low-grade dysplasia, mild and/or moderate dysplasia, will not progress to cancer. Previous studies have started integrating imaging and molecular analysis with histopathologic evaluation for improving the ability to predict progression risk in OPLs.10-15 An understanding of the molecular mechanisms that govern the promotion of OPLs into cancer would be very relevant for clinical practice in the identification of genes suitable for therapeutic targeting, prognostic and risk predictive markers.
In oral cancer, the accumulation of genetic alterations is associated with the progression of OPL to invasiveness.16, 17 Molecular analysis of OPLs is necessary to identify key changes in disease initiation and progression. However, studies of OPLs are rare and have not been attempted on a genome-wide scale, owing mostly to the minute amount of DNA obtainable from primary OPLs.16 Changes in gene dosage occur frequently in cancer genomes. Gains and losses of genomic regions may contain proto-oncogenes and tumor suppressor genes, which may lead to aberrant expression useful for malignant transformation. Low-level copy number changes involving large regions with many genes have been frequently observed in OSCCs, but their effect on gene expression remains ambiguous.18-21 High-level copy number changes, DNA amplification or homozygous deletion, encompass focal changes and often lead to the discovery of cancer-causing genes. Amplicons are defined as DNA segments less than 20 megabase pairs (Mbps) of which at least five copies exist in a single cell.22 They are useful for oncogene discovery because these unstable regions are under relentless selection and thus harbor genes advantageous for tumor growth.23-25 Amplified oncogenes are also clinically useful for therapeutic development.22, 23 Furthermore, amplification of EGFR with more than 12 copies per cell in head and neck SCC (HNSCC) is associated with poor survival.26 On the other hand, biallelic loss such as homozygous deletion contributes to functional inactivation, facilitating the localization of tumor suppressors.27, 28 High-resolution whole genome DNA microarrays have been useful in exploring genetic alterations in formalin-fixed paraffin-embedded (FFPE) specimens without the need for sample amplification.18, 29 Indeed this technology has a functional resolution of 50 kbp, improving the localization of regions with gene amplification and homozygous deletion.29, 30
Oral high-grade dysplasias are known to have a high likelihood of cancer progression, whereas low-grade lesions have a low probability of progression and can even regress.14 We evaluated genome wide gene dosage alteration in 50 manually microdissected FFPE high-risk OPLs, which included 43 high-grade dysplasias and seven low-grade dysplasias that are known to have later progressed to cancer. We focused on the identification of high-level DNA amplification and homozygous deletions instead of low-level copy number change. This type of analysis has never been undertaken for such early stage lesions. We also compared these findings to 23 OSCC and 14 low-grade dysplasias that never progressed. Expression of gene candidates within recurrent amplicons in OPLs were analyzed in public datasets with 188 HNSCCs and further confirmed in 61 oral cancers. Taken together, our analysis suggests that a common signaling network involving the ERK/MAPK, FGF, p53, PTEN, and PI3K/AKT signaling pathways is frequently deregulated in high-risk OPLs.
Materials and Methods
Tissue Samples
This study involves 87 (64 dysplasias and 23 OSCCs) archival FFPE specimens obtained from the British Columbia Oral Biopsy Service (Supp. Table S1). The group of “high-risk OPLs” includes 43 high-grade preinvasive lesions (22 severe dysplasia and 21 CIS) and seven low-grade lesions (one hyperplasia and six mild and/or moderate dysplasias) that later progressed to high-grade dysplasias or OSCCs. These lesions represent 86 patients, all with no prior history of cancer. Samples from one patient with a severe dysplasia on the lower lip (sample Oral7) and an OSCC on the tongue (sample Oral80) are both included in the study as they are from distinct anatomical sites. For low-grade lesions, patients were followed up with a median duration of eight years in a longitudinal study established at the BC Oral Cancer Prevention Program. Low-grade lesions that did not progress between 1985 and 2009 consisted of one hyperplasia and 13 mild and moderate dysplasias. All diagnoses were confirmed by the study pathologist (LZ) using criteria established by the World Health Organization (WHO).5 Areas of dysplasia were identified using hematoxylin and eosin (H&E) stained sections cut from FFPE tissues. Epithelial cells in these areas were meticulously dissected from adjacent non-epithelium tissue under an inverted microscope using a 23G needle. DNA was extracted as previously described.18
Whole genome DNA microarray analysis
Tiling-path genomic arrays (SMRT v.1 and v.2) were obtained from the BC Cancer Research Centre Array Laboratory.29 The whole genome is represented as 26,819 overlapping bacterial artificial chromosome (BAC) clones spotted in duplicate with complete coverage of the human genome, allowing breakpoint detection at a resolution of 50 kbp.29, 30 Briefly, each sample DNA and normal reference pooled male genomic DNA (Novagen, Mississauga, ON, Canada) (250 ng each) were random prime labeled with cyanine-3 and cyanine-5 dCTP, respectively, mixed with 100 μg of human Cot-1 DNA, purified and hybridized to the array at 45°C for 36 hours before washing. Hybridized arrays were scanned as previously described.31, 32
Data analysis
Array images were analyzed using SoftWoRx Tracker Spot Analysis software (Applied Precision, Issaquah, WA). A three-step normalization procedure, including LOWESS fitting, spatial, and median normalization, was used to remove systematic biases.33 SeeGH software was used to display log2 signal intensity ratios in relation to genomic locations in the hg17 assembly (NCBI Build 35).34 Data points with standard deviation >0.075 and signal to noise ratio <3 in either channel were removed. None of the 87 genomic profiles contain technical artefacts of wavy pattern which are often observed from FFPE samples.35 All profiles has been deposited to Gene Expression Omnibus (GEO) database at NCBI, series accession number GSE9193.36
High-level DNA amplifications and presumptive homozygous deletions were identified by a moving-average based algorithm as previously described.31 The threshold for high copy number was set to log2 signal intensity ratio > 0.8 for amplification or < -0.8 for homozygous deletions. Only those alterations containing ≥ 3 overlapping clones were identified in order to avoid false-positives due to hybridization artifacts. Recurrent minimal altered regions of amplification were identified by the presence of a given amplicon in at least two high-risk OPLs. Genes within such altered regions were mapped according to the RefSeq Genes track release 25. As copy number variation-associated BAC clones are often associated with amplification and DNA rearrangement, such BACs are included in the analysis.31, 37
Transcript expression analysis
Independent transcript analyses of genes mapped within minimal altered regions of amplification were performed using the Oncomine database.38 Five studies within Oncomine analyzed expression patterns between head and neck tumors (N = 188) and normal tissues (N = 38) (Supp. Table S2).39-43 In Oncomine, Student's t test was performed to reflect the significance of differential expression observed in tumors compared to normal tissue in each study. Furthermore, two public GEO datasets (GSE10121 and GSE9844) containing 61 oral tumors and 18 normal samples were downloaded.41, 43, 44 The normalized data from each dataset were extracted. Two-sided student's t-test along with a Benjamini-Hochberg multiple testing correction was performed comparing the oral tumors with the normal samples per study. The significance threshold was set at P<0.05.
Real-time polymerase chain reaction
Total RNA from eight OSCCs and nine normal oral mucosal tissue from different healthy individuals were extracted using TRIzol (Invitrogen, Carlsbad, CA). Five hundred nanograms of total RNA from each sample were converted to cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) in a total volume of 20 μL. Real-time PCR using TaqMan Universal PCR master mix was performed to analyze TLN1 and CREB3 expression levels with Applied Biosystems Fast Real-Time PCR System. TaqMan gene expression assays (Assay ID) of TLN1 (Hs00196775_m1), CREB3 (Hs00197255_m1), and 18S rRNA (Hs99999901_s1) were purchased from Applied Biosystems. Reactions were performed in triplicate and according to manufacturer's protocol. The 2-ΔΔCt method was used to calculate relative expression values using the average of cycle thresholds of target genes and 18S rRNA, and the value of 1 was arbitrarily assigned to one normal sample. Expression levels between OSCCs and normal samples were compared by a two-sided Wilcoxon-rank sum test.
Biological functions and pathway analysis
To define the biological functions of all the genes mapped within recurrent amplicons in OPLs, we interrogated the Biological Function Analysis in Ingenuity Pathway Analysis (IPA) (version 7.0) (Ingenuity® Systems, www.ingenuity.com). Biological Function Analysis identifies biological functions and diseases that are significantly enriched in the data relative to chance alone by Fisher's exact test. In addition, genes in recurrent amplicons in high-risk OPLs and associated with transcript overexpression in at least one HNSCC dataset were further explored using the Canonical Pathways Analysis. Canonical Pathways Analysis explores 48 well-characterized metabolic and signaling pathways for the significant enrichment of the dataset in these pathways, again using Fisher's exact test to calculate the probability that the association between genes in the dataset and the canonical pathway is explained by chance alone.
Fluorescence in situ hybridization (FISH)
FISH assays were performed as described in Romeo et al.,45 except with the modifications of using a lower concentration of pepsin (0.032%) and longer digestion time (80-90 minutes). Two sets of dual colored probe (Vysis, Downers Grove, IL) were sequentially performed on the same tissue section according to manufacturer's instruction, which included the pair of CEP11 (centromere 11p11.11-q11, SpectrumGreen)/ CCND1 (11q13, SpectrumOrange), and CEP7 (7p11.1-q11.1, SpectrumGreen)/ EGFR (7p12, SpectrumOrange). Signals were captured and imaged using Olympus BX61 and ImagePro Plus 5.1.
Results
Early occurrence of DNA amplification and homozygous deletion in OPLs
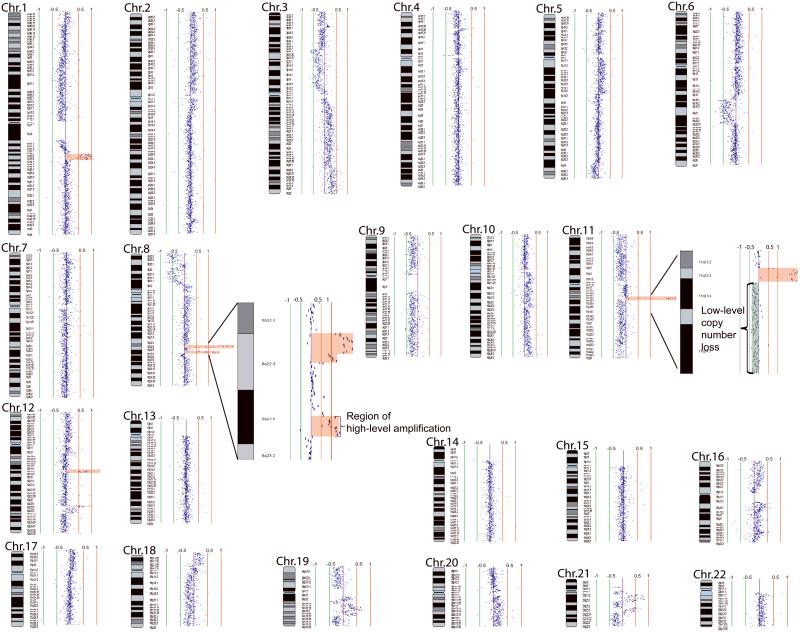
It is evident that copy number alterations are more frequent among OSCCs relative to high-grade dysplasias across the whole genome (Supp. Figure S1). High-level copy number alteration, including DNA amplification and homozygous deletion, is a frequent event in high-risk OPLs. In total, 40% of high-risk OPLs (19/43 high-grade dysplasias and 1/7 progressing low-grade dysplasias) exhibited at least one region of high-level copy number change. A frequency of 65.2% (15/23) was found in OSCCs. No such changes were detected in low-grade lesions that did not progress. A whole genome karyogram of one high-grade dysplasia Oral42 with seven regions of gene amplification is illustrated in Figure 1.
Figure 1.
Whole genome tiling-path array profile of a carcinoma in situ (CIS) Oral42. Each data point represents one BAC-derived segment on the array. The log2 signal intensity ratios of a competitive hybridization with pooled male genomic DNA are plotted by SeeGH software.34 The red and green bar lines are positive and negative log2 signal intensity ratio lines scaled by an increment of 0.5. Data points to the left and right of the centre line represent DNA copy number losses and gains, respectively. Specifically, seven regions of gene amplification on 1q23.2-q23.3, 1q23.3-q24.1, 8q22.2-q22.3, 8q23.1, 11q13.3-q13.4, 12q14.3, and 12q23.2-q23.3 are shaded red. Magnified views of the two amplicons on chromosome 8, and a region of low-level copy number loss and a region of gene amplification on chromosome 11 are shown in two insets.
Recurrent amplicons and rare regions of homozygous deletion harbor known and novel candidate cancer genes
To distinguish genetic events at the premalignant stage from late-stage events, high-level copy number changes were identified separately in 50 high-risk OPLs and 23 OSCCs. In the 20/50 high-risk OPLs with high-level copy number alteration, 43 incidents of gene amplification and six regions of homozygous deletion were detected. At a similar level, 46 occurrences of amplification and two regions of homozygous deletion were identified in 15/23 OSCCs (Supp. Table S3). Many of these lesions have at least two regions of high-level copy number alteration, including 11/20 of the high-risk OPLs and 9/15 of the OSCCs. In addition, many of the detected amplicons overlap, suggesting that these regions do not occur by chance.
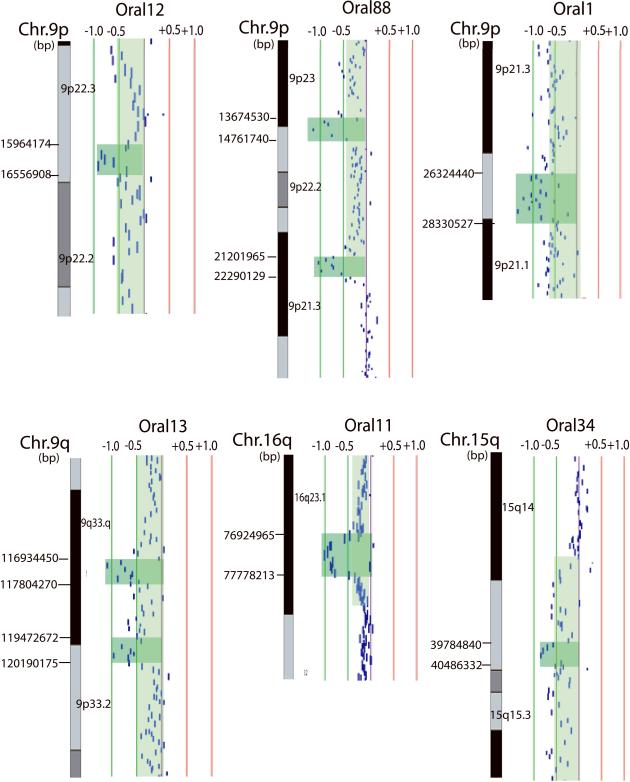
Homozygous deletions were seen less frequently than DNA amplification, occurring in five high-grade dysplasias and one OSCC (Fig. 2). The eight identified regions of homozygous deletion do not overlap (Table 1), but two regions on 9p22.3 (in samples Oral12 and Oral88) are separated by 1.2 Mbp. In total, 44 genes were identified as homozygously deleted, including known tumor suppressors CDKN2A (9p21.3), CDKN2B (9p21.3), MTAP (9p21.3), and WWOX (16q23.1). Genes bounded by the novel regions of homozygous deletion that we identified (9p21.1-p21.2, 9p22.3, 9p22.3-p23, 9q33.1, 9q33.1-q33.2, and 15q15.1) may represent tumor suppressors driving oral carcinogenesis (Supp. Table S4).
Figure 2.
Graphical representations of region of homozygous deletion in oral lesions. Each data point represents one BAC-derived segment on the array. The detected region of homozygous deletion in each sample is shaded in dark green, whereas single copy loss is shaded in pale green on the corresponding data points. The red and green lines are positive and negative ratio lines scaled by an increment of log2 signal ratios of 0.5.
Table 1.
Regions of homozygous deletion.
| Sample ID | Chromosomal band | Proximal flanking clone* | Start bp coordinate | Distal flanking clone* | End bp coordinate | Size (Mbp) | Number of genes | Known tumor suppressors |
|---|---|---|---|---|---|---|---|---|
| Oral88 | 9p22.3-p23 | 692G11 | 13674530 | 55P10 | 14761740 | 1.09 | 4 | - |
| Oral12 | 9p22.3 | 97O17 | 15964174 | 554H2 | 16556908 | 0.59 | 1 | - |
| Oral88 | 9p21.3 | 380P16 | 21201965 | 145H12 | 22290129 | 1.09 | 14 | CDKN2A, CDN2B, MTAP |
| Oral1 | 9p21.1-p21.2 | 802E2 | 26324440 | 133E6 | 28330527 | 2.01 | 10 | - |
| Oral13 | 9q33.1 | 235P13 | 116934450 | 121P18 | 117804270 | 0.87 | 2 | - |
| Oral13 | 9q33.1-q33.2 | 57K1 | 119472672 | 374B16 | 120190175 | 0.72 | 0 | - |
| Oral34 | 15q15.1 | 723A20 | 39784840 | 468N2 | 40486332 | 0.70 | 13 | - |
| Oral11 | 16q23.1 | 730M21 | 76924965 | 556H2 | 77778213 | 0.85 | 1 | WWOX |
All the listed human BAC clones were selected from the RPCI-11 library.
DNA amplifications occurring in OPLs may activate genes that facilitate the development of oral cancer. Seven recurrent amplicons, ranging from 0.45 Mbp to 2.26 Mbp in size, were identified in our dataset of 50 high-risk OPLs (Table 2). Minimal altered regions of these recurrent amplicons contain 88 unique genes, 15 of which are known to be involved in cancer as determined by IPA Functional Analysis (P = 1.31 × 10-5—1.41 × 10-2) (Table 3, Supp. Table S5). Except for chromosomal loci at 2q11.2 and 8q22.3, all amplicons harbor at least one cancer-related gene according to the IPA knowledgebase (though literature searches for genes in the outstanding regions identified potential cancer genes) (Table 2). This further substantiates the need to investigate the genes within these recurrent amplicons in OPLs.
Table 2.
Recurrent regions of gene amplification in OPLs. Minimal altered regions recurrent in at least two high-risk OPLs are listed.
| Chromosome | Proximal flanking clone* | Start (bp) | Distal flanking clone* | End (bp) | Size (Mbp) | Frequency of amplification in OPLs (N = 50) | Frequency of amplification in OSCCs (N = 23) | Candidate Genes |
|---|---|---|---|---|---|---|---|---|
| 2q11.2 | 793A22 | 96361719 | 61O17 | 97032268 | 0.67 | 4% | 0 | CIAO1 |
| 4q12 | 273B19 | 55487333 | 345F18 | 56688451 | 1.20 | 4% | 0 | KDR |
| 7p11.2 | 164O17 | 54583596 | 708P5 | 55194026 | 0.61 | 8% | 23.1% | EGFR |
| 8q11.21 | 770E5 | 49653988 | 259D18 | 50105411 | 0.45 | 4% | 0 | SNAI2 |
| 8q22.3 | 302J23 | 102005665 | 375I14 | 102513409 | 0.51 | 6% | 4.35% | YWHAZ |
| 9p13.3 | 121D5 | 33990614 | 312A20 | 36034564 | 2.04 | 4% | 0 | CCL19, CCL21, CCL27, DCTN3, OPRS1, TLN1, CREB3 |
| 11q13.2-q13.4 | 715N9 | 67947069 | CTD-2011L1 3 | 70204378 | 2.26 | 14% | 26.1% | CCND1, FGF3, FGF19, GAL, FGF4 |
Unless otherwise stated, all the listed human BAC clones were selected from the RPCI-11 library.
Table 3.
Cancer-related genes mapped within recurrent regions of amplicon in high-risk OPLs.
| Gene Name | EntrezGene ID for human | Chromosome band |
|---|---|---|
| RNF103 | 7844 | 2p11.2 |
| KDR | 3791 | 4q11-q12 |
| EGFR | 1956 | 7p12 |
| TACC1 | 6867 | 8p11 |
| LSM1 | 27257 | 8p11.2 |
| STAR | 6770 | 8p11.2 |
| WHSC1L1 | 54904 | 8p11.2 |
| ADAM9 | 8754 | 8p11.23 |
| FGFR1 | 2260 | 8p11.2-p11.1 |
| EIF4EBP1 | 1978 | 8p12 |
| SNAI2 | 6591 | 8q11 |
| CCL19 | 6363 | 9p13 |
| CCL21 | 6366 | 9p13 |
| CCL27 | 10850 | 9p13 |
| CREB3 | 10488 | 9p13.3 |
| OPRS1 | 10280 | 9p13.3 |
| CA9 | 768 | 9p13-p12 |
| RECK | 8434 | 9p13-p12 |
| CCND1 | 595 | 11q13 |
| FGF3 | 2248 | 11q13 |
| FGF19 | 9965 | 11q13.1 |
| GAL | 51083 | 11q13.2-q13.4 |
| FGF4 | 2249 | 11q13.3 |
Transcript analysis of independent HNSCC datasets
Candidate oncogenes within amplicons are likely to have increased mRNA expression in cancer specimens. Thus, we evaluated transcript levels of genes in the recurrent amplicons to refine the gene list using five independent studies of HNSCC.39-43 Of the 88 genes identified within recurrent amplicons, 40 were overexpressed in at least one dataset of HNSCC relative to normal tissues. This validation at the transcript level in independent datasets suggests the potential importance of the 40 candidate genes in cancer development.
Frequent oncogenic activation of a common signaling network in OPLs
We hypothesize that genes with elevated copy number in recurrent OPLs and increased mRNA levels in HNSCC are important for oral cancer development. To understand the signaling defects in OPLs, we interrogated 48 well-characterized signaling pathways in the IPA canonical pathway database to examine which pathways are significantly enriched with the candidate genes. The top five deregulated canonical pathways include the ERK/MAPK, FGF, p53, PTEN, and PI3K/AKT signaling pathways (Table 4, P = 8.95×10-3, 1.63×10-2, 1.96×10-2, 1.96×10-2, and 3.18×10-2 respectively). Graphical representation of each canonical pathway is provided in Supp. Figures S2, S3, S4, S5, S6.
Table 4.
Disruption of canonical signaling pathways in oral premalignant lesions.
| Canonical pathways identified | Overexpressed genes in head and neck SCC | p-values |
|---|---|---|
| ERK/MAPK Signaling | CREB3, TLN1, YWHAZ | 8.95E-03 |
| FGF Signaling | CREB3, FGF3 | 1.63E-02 |
| p53 Signaling | CCND1, SNAI2 | 1.96E-02 |
| PTEN Signaling | CCND1, EGFR | 1.96E-02 |
| PI3K/AKT pathway | CCND1, YWHAZ | 3.18E-02 |
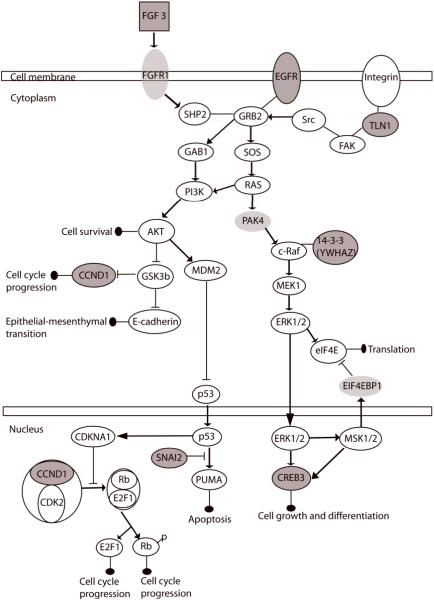
It is important to note that several genes within recurrent amplicons, including the CREB3, CCND1, and YWHAZ, participate in multiple canonical pathways. In addition, amplified genes including the FGFR and EGFR could both activate the ERK/MAPK and the PI3K/AKT pathways. Furthermore, 14 amplified genes share direct and indirect relationships in one network (Supp. Fig. S7). As multiple pathways could contribute to cancer development, the interactions among the top significantly over-represented canonical pathways were considered as a network (Fig. 3). Seven candidate genes (FGF3, EGFR, TLN1, YWHAZ, CCND1, CREB3 and SNAI2) within recurrent amplicons in high-risk OPLs participate in this signaling network. Of these seven candidate genes, FGF3, EGFR, and CCND1 have been frequently associated with gene amplifications in OSCCs,46-48 and protein expression of YWHAZ has been detected in oral dysplasias.49; while SNAI2 is often described as an important regulator for epithelialmesenchymal transition.50 We validated expression levels of TLN1 and CREB3 by quantitative PCR in eight OSCCs and nine normal oral mucosa tissues. Expression of TLN1 and CREB3 both showed increased expression in the OSCC group relative to normal tissues (p = 4.53×10-3, 8.14×10-3, respectively, Wilcoxon rank-sum test) (Supp. Fig. S8). Genes within this signaling network not identified in recurrent amplicons include PAK4, FGFR1, and EIF4EBP1. PAK4 is amplified in one high-grade dysplasia and one OSCC, while FGFR1 and EIF4EBP1were amplified in one high-grade dysplasia. Taken together, gene amplification of at least one of these 10 genes within this signaling network was found in 30.0% (15/50) of high-risk OPLs and 43.5% (10/23) of OSCCs.
Figure 3.
Multiple disruptions in a single network driven by the mechanism of gene amplification. The top significantly deregulated canonical pathways in oral dysplasias are the ERK/MAPK, FGF, p53, PTEN, and PI3K/AKT signaling pathways, which share common nodes and interplay as a single network. Genes colored in grey with outlined circle were recurrently deregulated by gene amplification in oral dysplasias and significantly overexpressed in independent head and neck cancer datasets, whereas those colored in light grey were found to be amplified only in one preinvasive lesion. Altogether, 25 oral lesions (one progressing low-grade lesions, 14 high-grade dysplasias, and ten OSCCs) exhibited high-level gene amplification of different genes inside this network, contributing to a disruption of 34.2% of all the progressing low-grade dysplasias, high-grade dysplasias, and OSCCs (N = 73).
To further substantiate the importance of this signaling network, we investigated two genetic features in all 87 oral lesions: 1) the presence of multiple amplicons harboring genes of this signaling network within a single specimen, and 2) the presence of overexpression in members of this signaling network that are not deregulated by gene amplification. We found that three high-grade dysplasias and one OSCC maintained multiple regions of gene amplification of different genes of this network. For example, one high-grade dysplasia (Oral22), exhibited three regions of high-level amplification on 7p11 (EGFR), 11q13 (CCND1, FGF3), and 19q13.2 (PAK4). The presence of multiple amplicons targeting a common network illustrates the potential importance for the disruption of multiple components within single samples. Next, we evaluated mRNA overexpression among 47 genes of this network not having DNA amplification in five independent HNSCC datasets. Among them, AKT1, AKT3, NRAS, PIK3CA, PIK3CB, PIK3CG, PRKACB, PRKAR1A, PRKCA, PRKCB1, PRKCE, PRKCI, RRAS, and RRAS2 were significantly overexpressed in at least two of the five datasets. This further demonstrates the frequent deregulation of this signaling network in the development of HNSCC.
Validation of mRNA levels in OSCCs
It has been previously suggested that different anatomical sites of cancers could affect mRNA expression profiles. However, the availability of OSCC expression datasets deposited in GEO database is limited, thus hindering sub-group analysis from different sites in the oral cavity or the head and neck region. Focusing on oral cancer, we analyzed expression levels of two oral cancer-specific studies (N = 61) to validate the importance of the 88 genes in recurrent amplicons for oral carcinogenesis.41, 43, 44 Of the 88 genes, 46 genes were overexpressed in at least one OSCC dataset, including 28 of the 40 overexpressed genes in HNSCC datasets (Supp. Table S6). Components of the signaling network, including the FGF3, FGF4, FGF19, EGFR, CCND1, CREB3, and YWHAZ, were also found to be significantly overexpressed in OSCC datasets. Thus, the expression pattern identified from oral cancer datasets was found to be similar to that of HNSCC.
Single cells of oral dysplasia exhibit co-amplification of EGFR and CCND1
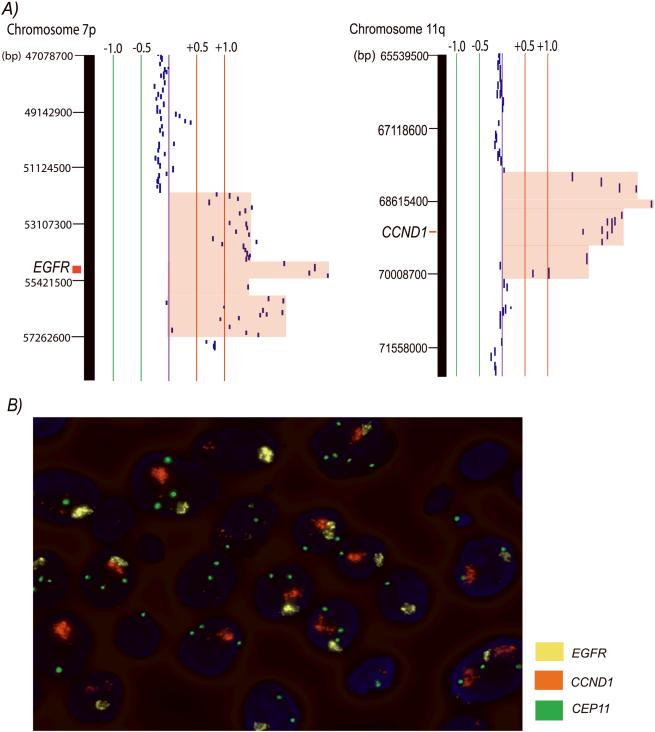
Co-amplification of at least two regions of the genome exists in 11 high-risk OPLs and 8 invasive SCC, constituting 55.9% (19/34) of samples with DNA amplification. We ask if the observed co-amplification is a manifestation of intra-lesion heterogeneity or if synchronous gene amplifications exist in single cells of the lesion. We performed FISH using probes spanning genomic regions of EGFR, CCND1, and the centromere of chromosome 11 on high-grade dysplasia Oral22 to examine this question (Fig. 4). Gene amplifications of EGFR and CCND1 were observed to co-exist in single cells of high-grade dysplasia, demonstrating that both of these changes can occur synchronously at the premalignant stage during oral cancer development.
Figure 4.
Co-amplification of EGFR and CCND1 in high-grade dysplasia Oral22. A) The left SeeGH profile represents amplification at 7p11.2 (EGFR locus), whereas the right profile represents amplification at 11q13.3 (CCND1 locus) of sample Oral22. The red and green lines are positive and negative ratio lines scaled by an increment of log2 signal ratios of 0.5. Amplified region was shaded in red. B) FISH visualization of co-amplification of EGFR and CCND1 in single dysplastic cells of Oral22. Sequential FISH was performed with probes mapping to CCND1, EGFR, and the centromeric region of chromosome 11, respectively displayed as orange, yellow, and green signal (original magnification, 1000x). Note the large clusters of amplification signals of CCND1 and EGFR relative to the centromeric region of chromosomes 11 in the same nucleus.
Discussion
Genetic alterations of DNA amplification and homozygous deletion have long been recognized as chromosomal regions that contain genes important for cancer development.23, 31, 51-57 As genetic alterations accumulate to produce a neoplastic phenotype, we focus on preinvasive stages of high-grade and low-grade lesions that are known to further develop into cancer, aiming to identify early genetic events during cancer development. Amplicons recurrently present in OPLs were identified, and genes that are overexpressed in HNSCC datasets were further distinguished. The purpose of this study includes: identifying early genetic events in cancer development, understanding the underlying pathways governing the progression of OPLs, and discovering gene candidates that might have therapeutic value for the prevention and treatment of oral cancer patients.
Amplifier phenotype in oral high-grade dysplasias
Gene amplification is a major mechanism of oncogene activation and has been associated as poor prognostic indicator in human cancers.22 Here, we detected frequent events of gene amplification in OPLs. Previous studies have reported gene amplifications in OSCC and oral cancer cell lines, with some studies evaluating known oncogenes or using interval-marker CGH.18, 20, 21, 58-62 Here, using an unbiased genome-wide approach we show that the early stages of oral lesions already suffer from increased genomic complexity. Seven amplicons were found to be recurrently present in oral dysplasias, ranging in size from 0.45 Mbp to 2.26 Mbp. Previously, Snijders et al. examined 89 oral invasive tumors by interval-marker array and identified nine amplicons smaller than 3 Mbp.21 The similar number of amplicons detected in our set of 50 high-risk OPLs was interesting, as it is believed that genetic alterations accumulate as oral dysplasias progress to invasiveness. Although tiling-path array provided increased genomic coverage compared with interval-marker, parallel analysis of our dataset of 23 OSCCs also detected similar level of genomic complexities between high-risk OPLs and invasive carcinomas. In addition, none of the 14 non-progressing low-grade lesions harbor region of DNA amplification. All the identified amplicons in oral dysplasias contain at least one cancer-related gene, supporting the concept that DNA amplification is likely to activate oncogene and contributes to OPL progression. Taken together, these results show that an amplifier phenotype exists in OPLs, and these amplicons might directly contribute to oral carcinogenesis. As gene amplification is readily detectable in clinical specimen, and our data showed that amplification frequently exists in high-risk OPLs, it might be plausible for gene amplification to serve as marker in OPLs predictive for aggressive progression to malignancy. However, a larger sample size of low-grade lesions with clinical outcome would be required to address this hypothesis.
Disruption of multiple components of a signaling network in oral dysplasias
It is crucial to understand the deregulated molecular pathways that govern the progression of oral premalignancy. By evaluating OPLs for gene amplification and examining genes with transcript overexpression in HNSCC and OSCC datasets, we identified genes that are involved in the ERK/MAPK, FGF, p53, PTEN, and PI3K/AKT signaling pathways. The FGF signaling pathway regulates developmental processes and angiogenesis, and has been an important therapeutic target in human cancers.63, 64 FGF signaling can activate the PI3K/AKT signaling cascade, leading to an induction of epithelial-mesenchymal transition and cell migration;65 while the FGF-stimulated ERK/MAPK signaling pathway is implicated in cell differentiation, proliferation, and survival.66 Activation of Akt by phosphorylation has been shown as an early event in oral preneoplastic lesions, and its expression is correlated with poor outcome in oral cancer patients.67 By examining the molecular interactions among the most significantly deregulated pathways as a single network (Fig. 3), we demonstrated the diverse mechanisms for the activation of this network, emphasizing the need for molecular targeted therapies of multiple signaling pathways.
Our study identified the genes that are amplified early during carcinogenesis. These genes were targeted towards one signaling network, and gene amplification frequently disrupted this signaling network as early as the low-grade dysplasia. Moreover, mRNA overexpression was frequently found in members of this network not activated by gene amplification in HNSCC datasets. The HNSCC datasets were chosen because of their large sample size, public availability, and could potentially broaden the scope of this oral cancer specific study to other HNSCCs. Nevertheless, expression analysis of oral cancer samples was also performed using two public datasets and similar results were found. In conclusion, our data highlights the early and frequent activation of one signaling network in OPLs. This also gives important insights into therapeutic strategies as different genes are mechanistically altered in different individuals.
Pathway addiction in oral dysplasias
The term “oncogene addiction” was first coined to describe the physiological dependence of cancer cells on a single activated oncogene for the maintenance of their malignant phenotype.68 In contrast to this phenomenon, a majority of our samples with amplification harbor more than one amplicon, suggesting the dependence on multiple oncogenes for OPL progression. As amplicons are known to be unstable regions, the maintenance of multiple amplicons must allow them to gain a selective advantage for clonal growth.23, 25 Interestingly, four oral lesions, including three high-grade dysplasias and one invasive carcinoma, exhibited multiple amplicons harboring genes from the same signaling pathways, suggesting the phenomenon of “pathway addiction” in these early stage lesions.69 Although intra-lesional heterogeneity might exist in dysplastic cells, which could contribute to the detection of various amplicons in different clonal populations, we observed co-amplification of two genes in this signaling network, EGFR and CCND1, could occur in single cells of a preinvasive lesion. This demonstrates the occurrence of an amplifier phenotype in single cells of oral dysplasia, and that multiple oncogenes are potentially important for overgrowth of such cells to form cancer. Whether the FGF signaling network is causative in oral cancer development needs to be investigated in further studies.
The potential “pathway addiction” to this signaling network yields important insight for drug development. Targeting a receptor tyrosine kinase might not be sufficient if downstream targets are also disrupted. In addition, genetic testing of multiple components of this network, possibly by FISH assays to detect amplification hotspots, might enhance therapeutic efficacy.70,71
Summary
This is the first comprehensive examination of DNA amplification and homozygous deletion in OPLs. We identified one signaling network that is frequently deregulated at different components in oral dysplasias, reflecting diverse mechanisms but common underlying biology governing the progression from oral premalignancy into invasiveness. This study suggests the importance of multiple signaling pathways in the early stage of oral carcinogenesis. Combined targeting of these oncogenic pathways might be effective for treatment of oral cancer patients since several genes of this network could be activated in a single specimen. Furthermore, since different targets are altered in different specimens, it is important to identify multiple markers to stratify patients that may benefit from personalized therapies for oral cancer.71
Supplementary Material
Acknowledgements
Grant support: NIDCR grants R01DE15965 and R01DE13124; Genome Canada; Canadian Institutes of Health Research (CIHR); Scholarships to IFLT and CFP from CIHR and Michael Smith Foundation for Health Research.
We thank Yuqi Zhu, Raj Chari, Bradley Coe, William Lockwood, Timon Buys, and Paul Boutros for advice and discussion.
Abbreviations
- OPL
oral premalignant lesion
- OSCC
oral squamous cell carcinoma
- HNSCC
head and neck squamous cell carcinoma
- CIS
carcinoma in situ
- FFPE
formalin-fixed paraffin-embedded
- BAC
bacterial artificial chromosome
- IPA
Ingenuity Pathway Analysis
- FISH
fluorescence in situ hybridization
References
- 1.Cancer Facts & Figures 2007. American Cancer Society; Atlanta: 2007. [Google Scholar]
- 2.Epstein JB, Gorsky M, Cabay RJ, Day T, Gonsalves W. Screening for and diagnosis of oral premalignant lesions and oropharyngeal squamous cell carcinoma: role of primary care physicians. Can Fam Physician. 2008;54:870–5. [PMC free article] [PubMed] [Google Scholar]
- 3.Poh CF, Ng S, Berean KW, Williams PM, Rosin MP, Zhang L. Biopsy and histopathologic diagnosis of oral premalignant and malignant lesions. J Can Dent Assoc. 2008;74:283–8. [PubMed] [Google Scholar]
- 4.Warnakulasuriya S, Reibel J, Bouquot J, Dabelsteen E. Oral epithelial dysplasia classification systems: predictive value, utility, weaknesses and scope for improvement. J Oral Pathol Med. 2008;37:127–33. doi: 10.1111/j.1600-0714.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 5.Definition of leukoplakia and related lesions: An aid to studies on oral precancer. Oral Surgery, Oral Medicine, Oral Pathology. 1978;46:518–39. [PubMed] [Google Scholar]
- 6.Crissman JD, Zarbo RJ. Dysplasia, in situ carcinoma, and progression to invasive squamous cell carcinoma of the upper aerodigestive tract. Am J Surg Pathol. 1989;13(Suppl 1):5–16. [PubMed] [Google Scholar]
- 7.Fresko D, Lazarus SS. Oral carcinoma in situ. Its progression to squamous, basosquamous, and basal-cell carcinoma. Arch Pathol Lab Med. 1981;105:15–9. [PubMed] [Google Scholar]
- 8.Hayward JR, Regezi JA. Oral dysplasia and in situ carcinoma: clinicopathologic correlations of eight patients. J Oral Surg. 1977;35:756–62. [PubMed] [Google Scholar]
- 9.Summerlin DJ. Precancerous and cancerous lesions of the oral cavity. Dermatol Clin. 1996;14:205–23. doi: 10.1016/s0733-8635(05)70351-x. [DOI] [PubMed] [Google Scholar]
- 10.Hall GL, Shaw RJ, Field EA, Rogers SN, Sutton DN, Woolgar JA, Lowe D, Liloglou T, Field JK, Risk JM. p16 Promoter methylation is a potential predictor of malignant transformation in oral epithelial dysplasia. Cancer Epidemiol Biomarkers Prev. 2008;17:2174–9. doi: 10.1158/1055-9965.EPI-07-2867. [DOI] [PubMed] [Google Scholar]
- 11.Guillaud M, Zhang L, Poh C, Rosin MP, MacAulay C. Potential use of quantitative tissue phenotype to predict malignant risk for oral premalignant lesions. Cancer Res. 2008;68:3099–107. doi: 10.1158/0008-5472.CAN-07-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao L, Lee JS, Fan YH, Ro JY, Batsakis JG, Lippman S, Hittelman W, Hong WK. Frequent microsatellite alterations at chromosomes 9p21 and 3p14 in oral premalignant lesions and their value in cancer risk assessment. Nat Med. 1996;2:682–5. doi: 10.1038/nm0696-682. [DOI] [PubMed] [Google Scholar]
- 13.Partridge M, Pateromichelakis S, Phillips E, Emilion GG, A'Hern RP, Langdon JD. A case-control study confirms that microsatellite assay can identify patients at risk of developing oral squamous cell carcinoma within a field of cancerization. Cancer Res. 2000;60:3893–8. [PubMed] [Google Scholar]
- 14.Rosin MP, Cheng X, Poh C, Lam WL, Huang Y, Lovas J, Berean K, Epstein JB, Priddy R, Le ND, Zhang L. Use of allelic loss to predict malignant risk for low-grade oral epithelial dysplasia. Clin Cancer Res. 2000;6:357–62. [PubMed] [Google Scholar]
- 15.Rosin MP, Lam WL, Poh C, Le ND, Li RJ, Zeng T, Priddy R, Zhang L. 3p14 and 9p21 loss is a simple tool for predicting second oral malignancy at previously treated oral cancer sites. Cancer Res. 2002;62:6447–50. [PubMed] [Google Scholar]
- 16.Garnis C, Buys TP, Lam WL. Genetic alteration and gene expression modulation during cancer progression. Mol Cancer. 2004;3:9. doi: 10.1186/1476-4598-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao L, Hong WK, Papadimitrakopoulou VA. Focus on head and neck cancer. Cancer Cell. 2004;5:311–6. doi: 10.1016/s1535-6108(04)00090-x. [DOI] [PubMed] [Google Scholar]
- 18.Baldwin C, Garnis C, Zhang L, Rosin MP, Lam WL. Multiple microalterations detected at high frequency in oral cancer. Cancer Res. 2005;65:7561–7. doi: 10.1158/0008-5472.CAN-05-1513. [DOI] [PubMed] [Google Scholar]
- 19.Noutomi Y, Oga A, Uchida K, Okafuji M, Ita M, Kawauchi S, Furuya T, Ueyama Y, Sasaki K. Comparative genomic hybridization reveals genetic progression of oral squamous cell carcinoma from dysplasia via two different tumourigenic pathways. J Pathol. 2006;210:67–74. doi: 10.1002/path.2015. [DOI] [PubMed] [Google Scholar]
- 20.Smeets SJ, Braakhuis BJ, Abbas S, Snijders PJ, Ylstra B, van de Wiel MA, Meijer GA, Leemans CR, Brakenhoff RH. Genome-wide DNA copy number alterations in head and neck squamous cell carcinomas with or without oncogene-expressing human papillomavirus. Oncogene. 2006;25:2558–64. doi: 10.1038/sj.onc.1209275. [DOI] [PubMed] [Google Scholar]
- 21.Snijders AM, Schmidt BL, Fridlyand J, Dekker N, Pinkel D, Jordan RC, Albertson DG. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene. 2005;24:4232–42. doi: 10.1038/sj.onc.1208601. [DOI] [PubMed] [Google Scholar]
- 22.Myllykangas S, Bohling T, Knuutila S. Specificity, selection and significance of gene amplifications in cancer. Semin Cancer Biol. 2007;17:42–55. doi: 10.1016/j.semcancer.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22:447–55. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 24.Myllykangas S, Tikka J, Bohling T, Knuutila S, Hollmen J. Classification of human cancers based on DNA copy number amplification modeling. BMC Med Genomics. 2008;1:15. doi: 10.1186/1755-8794-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miele M, Bonatti S, Menichini P, Ottaggio L, Abbondandolo A. The presence of amplified regions affects the stability of chromosomes in drug-resistant Chinese hamster cells. Mutat Res. 1989;219:171–8. doi: 10.1016/0921-8734(89)90012-x. [DOI] [PubMed] [Google Scholar]
- 26.Temam S, Kawaguchi H, El-Naggar AK, Jelinek J, Tang H, Liu DD, Lang W, Issa JP, Lee JJ, Mao L. Epidermal growth factor receptor copy number alterations correlate with poor clinical outcome in patients with head and neck squamous cancer. J Clin Oncol. 2007;25:2164–70. doi: 10.1200/JCO.2006.06.6605. [DOI] [PubMed] [Google Scholar]
- 27.Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–3. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 28.Kikuchi R, Tsuda H, Kozaki K, Kanai Y, Kasamatsu T, Sengoku K, Hirohashi S, Inazawa J, Imoto I. Frequent inactivation of a putative tumor suppressor, angiopoietin-like protein 2, in ovarian cancer. Cancer Res. 2008;68:5067–75. doi: 10.1158/0008-5472.CAN-08-0062. [DOI] [PubMed] [Google Scholar]
- 29.Ishkanian AS, Malloff CA, Watson SK, DeLeeuw RJ, Chi B, Coe BP, Snijders A, Albertson DG, Pinkel D, Marra MA, Ling V, MacAulay C, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 30.Coe BP, Ylstra B, Carvalho B, Meijer GA, Macaulay C, Lam WL. Resolving the resolution of array CGH. Genomics. 2007;89:647–53. doi: 10.1016/j.ygeno.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 31.Lockwood WW, Chari R, Coe BP, Girard L, Macaulay C, Lam S, Gazdar AF, Minna JD, Lam WL. DNA amplification is a ubiquitous mechanism of oncogene activation in lung and other cancers. Oncogene. 2008 doi: 10.1038/onc.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coe BP, Lockwood WW, Girard L, Chari R, Macaulay C, Lam S, Gazdar AF, Minna JD, Lam WL. Differential disruption of cell cycle pathways in small cell and non-small cell lung cancer. Br J Cancer. 2006;94:1927–35. doi: 10.1038/sj.bjc.6603167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khojasteh M, Lam WL, Ward RK, MacAulay C. A stepwise framework for the normalization of array CGH data. BMC Bioinformatics. 2005;6:274. doi: 10.1186/1471-2105-6-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chi B, deLeeuw RJ, Coe BP, Ng RT, MacAulay C, Lam WL. MD-SeeGH: a platform for integrative analysis of multi-dimensional genomic data. BMC Bioinformatics. 2008;9:243. doi: 10.1186/1471-2105-9-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van de Wiel MA, Brosens R, Eilers PH, Kumps C, Meijer GA, Menten B, Sistermans E, Speleman F, Timmerman ME, Ylstra B. Smoothing waves in array CGH tumor profiles. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp132. [DOI] [PubMed] [Google Scholar]
- 36.Tsui IF, Rosin MP, Zhang L, Ng RT, Lam WL. Multiple aberrations of chromosome 3p detected in oral premalignant lesions. Cancer Prev Res (Phila Pa) 2008;1:424–9. doi: 10.1158/1940-6207.CAPR-08-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong KK, deLeeuw RJ, Dosanjh NS, Kimm LR, Cheng Z, Horsman DE, MacAulay C, Ng RT, Brown CJ, Eichler EE, Lam WL. A comprehensive analysis of common copy-number variations in the human genome. Am J Hum Genet. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cromer A, Carles A, Millon R, Ganguli G, Chalmel F, Lemaire F, Young J, Dembele D, Thibault C, Muller D, Poch O, Abecassis J, et al. Identification of genes associated with tumorigenesis and metastatic potential of hypopharyngeal cancer by microarray analysis. Oncogene. 2004;23:2484–98. doi: 10.1038/sj.onc.1207345. [DOI] [PubMed] [Google Scholar]
- 40.Ginos MA, Page GP, Michalowicz BS, Patel KJ, Volker SE, Pambuccian SE, Ondrey FG, Adams GL, Gaffney PM. Identification of a gene expression signature associated with recurrent disease in squamous cell carcinoma of the head and neck. Cancer Res. 2004;64:55–63. doi: 10.1158/0008-5472.can-03-2144. [DOI] [PubMed] [Google Scholar]
- 41.Pyeon D, Newton MA, Lambert PF, den Boon JA, Sengupta S, Marsit CJ, Woodworth CD, Connor JP, Haugen TH, Smith EM, Kelsey KT, Turek LP, et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67:4605–19. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chung CH, Parker JS, Karaca G, Wu J, Funkhouser WK, Moore D, Butterfoss D, Xiang D, Zanation A, Yin X, Shockley WW, Weissler MC, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5:489–500. doi: 10.1016/s1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- 43.Toruner GA, Ulger C, Alkan M, Galante AT, Rinaggio J, Wilk R, Tian B, Soteropoulos P, Hameed MR, Schwalb MN, Dermody JJ. Association between gene expression profile and tumor invasion in oral squamous cell carcinoma. Cancer Genet Cytogenet. 2004;154:27–35. doi: 10.1016/j.cancergencyto.2004.01.026. [DOI] [PubMed] [Google Scholar]
- 44.Ye H, Yu T, Temam S, Ziober BL, Wang J, Schwartz JL, Mao L, Wong DT, Zhou X. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;9:69. doi: 10.1186/1471-2164-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romeo MS, Sokolova IA, Morrison LE, Zeng C, Baron AE, Hirsch FR, Miller YE, Franklin WA, Varella-Garcia M. Chromosomal abnormalities in non-small cell lung carcinomas and in bronchial epithelia of high-risk smokers detected by multi-target interphase fluorescence in situ hybridization. J Mol Diagn. 2003;5:103–12. doi: 10.1016/s1525-1578(10)60459-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheu JJ, Hua CH, Wan L, Lin YJ, Lai MT, Tseng HC, Jinawath N, Tsai MH, Chang NW, Lin CF, Lin CC, Hsieh LJ, et al. Functional genomic analysis identified epidermal growth factor receptor activation as the most common genetic event in oral squamous cell carcinoma. Cancer Res. 2009;69:2568–76. doi: 10.1158/0008-5472.CAN-08-3199. [DOI] [PubMed] [Google Scholar]
- 47.Freier K, Sticht C, Hofele C, Flechtenmacher C, Stange D, Puccio L, Toedt G, Radlwimmer B, Lichter P, Joos S. Recurrent coamplification of cytoskeleton-associated genes EMS1 and SHANK2 with CCND1 in oral squamous cell carcinoma. Genes Chromosomes Cancer. 2006;45:118–25. doi: 10.1002/gcc.20270. [DOI] [PubMed] [Google Scholar]
- 48.Lese CM, Rossie KM, Appel BN, Reddy JK, Johnson JT, Myers EN, Gollin SM. Visualization of INT2 and HST1 amplification in oral squamous cell carcinomas. Genes Chromosomes Cancer. 1995;12:288–95. doi: 10.1002/gcc.2870120409. [DOI] [PubMed] [Google Scholar]
- 49.Ralhan R, Desouza LV, Matta A, Chandra Tripathi S, Ghanny S, Dattagupta S, Thakar A, Chauhan SS, Siu KW. iTRAQ-multidimensional liquid chromatography and tandem mass spectrometry-based identification of potential biomarkers of oral epithelial dysplasia and novel networks between inflammation and premalignancy. J Proteome Res. 2009;8:300–9. doi: 10.1021/pr800501j. [DOI] [PubMed] [Google Scholar]
- 50.Cobaleda C, Perez-Caro M, Vicente-Duenas C, Sanchez-Garcia I. Function of the zinc-finger transcription factor SNAI2 in cancer and development. Annu Rev Genet. 2007;41:41–61. doi: 10.1146/annurev.genet.41.110306.130146. [DOI] [PubMed] [Google Scholar]
- 51.Johnson BE, Ihde DC, Makuch RW, Gazdar AF, Carney DN, Oie H, Russell E, Nau MM, Minna JD. myc family oncogene amplification in tumor cell lines established from small cell lung cancer patients and its relationship to clinical status and course. J Clin Invest. 1987;79:1629–34. doi: 10.1172/JCI112999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kubo T, Yamamoto H, Lockwood WW, Valencia I, Soh J, Peyton M, Jida M, Otani H, Fujii T, Ouchida M, Takigawa N, Kiura K, et al. MET gene amplification or EGFR mutation activate MET in lung cancers untreated with EGFR tyrosine kinase inhibitors. Int J Cancer. 2008 doi: 10.1002/ijc.24150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Myllykangas S, Himberg J, Bohling T, Nagy B, Hollmen J, Knuutila S. DNA copy number amplification profiling of human neoplasms. Oncogene. 2006;25:7324–32. doi: 10.1038/sj.onc.1209717. [DOI] [PubMed] [Google Scholar]
- 54.Weir B, Zhao X, Meyerson M. Somatic alterations in the human cancer genome. Cancer Cell. 2004;6:433–8. doi: 10.1016/j.ccr.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 55.Cairns P, Polascik TJ, Eby Y, Tokino K, Califano J, Merlo A, Mao L, Herath J, Jenkins R, Westra W, et al. Frequency of homozygous deletion at p16/CDKN2 in primary human tumours. Nat Genet. 1995;11:210–2. doi: 10.1038/ng1095-210. [DOI] [PubMed] [Google Scholar]
- 56.Lerman MI, Minna JD. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. The International Lung Cancer Chromosome 3p21.3 Tumor Suppressor Gene Consortium. Cancer Res. 2000;60:6116–33. [PubMed] [Google Scholar]
- 57.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 58.Freier K, Schwaenen C, Sticht C, Flechtenmacher C, Muhling J, Hofele C, Radlwimmer B, Lichter P, Joos S. Recurrent FGFR1 amplification and high FGFR1 protein expression in oral squamous cell carcinoma (OSCC) Oral Oncol. 2007;43:60–6. doi: 10.1016/j.oraloncology.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 59.Reshmi SC, Huang X, Schoppy DW, Black RC, Saunders WS, Smith DI, Gollin SM. Relationship between FRA11F and 11q13 gene amplification in oral cancer. Genes Chromosomes Cancer. 2007;46:143–54. doi: 10.1002/gcc.20394. [DOI] [PubMed] [Google Scholar]
- 60.Xia J, Chen Q, Li B, Zeng X. Amplifications of TAOS1 and EMS1 genes in oral carcinogenesis: association with clinicopathological features. Oral Oncol. 2007;43:508–14. doi: 10.1016/j.oraloncology.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 61.Garnis C, Baldwin C, Zhang L, Rosin MP, Lam WL. Use of complete coverage array comparative genomic hybridization to define copy number alterations on chromosome 3p in oral squamous cell carcinomas. Cancer Res. 2003;63:8582–5. [PubMed] [Google Scholar]
- 62.Hsu LC, Huang X, Seasholtz S, Potter DM, Gollin SM. Gene amplification and overexpression of protein phosphatase 1alpha in oral squamous cell carcinoma cell lines. Oncogene. 2006;25:5517–26. doi: 10.1038/sj.onc.1209563. [DOI] [PubMed] [Google Scholar]
- 63.Katoh M, Katoh M. Cross-talk of WNT and FGF signaling pathways at GSK3beta to regulate beta-catenin and SNAIL signaling cascades. Cancer Biol Ther. 2006;5:1059–64. doi: 10.4161/cbt.5.9.3151. [DOI] [PubMed] [Google Scholar]
- 64.Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-3-reviews3005. REVIEWS3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 66.McKay MM, Morrison DK. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26:3113–21. doi: 10.1038/sj.onc.1210394. [DOI] [PubMed] [Google Scholar]
- 67.Massarelli E, Liu DD, Lee JJ, El-Naggar AK, Lo Muzio L, Staibano S, De Placido S, Myers JN, Papadimitrakopoulou VA. Akt activation correlates with adverse outcome in tongue cancer. Cancer. 2005;104:2430–6. doi: 10.1002/cncr.21476. [DOI] [PubMed] [Google Scholar]
- 68.Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297:63–4. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 69.Molinolo AA, Amornphimoltham P, Squarize CH, Castilho RM, Patel V, Gutkind JS. Dysregulated molecular networks in head and neck carcinogenesis. Oral Oncol. 2008 doi: 10.1016/j.oraloncology.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F, Gambacorta M, Siena S, Bardelli A. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279–86. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 71.Yamatodani T, Ekblad L, Kjellen E, Johnsson A, Mineta H, Wennerberg J. Epidermal growth factor receptor status and persistent activation of Akt and p44/42 MAPK pathways correlate with the effect of cetuximab in head and neck and colon cancer cell lines. J Cancer Res Clin Oncol. 2008 doi: 10.1007/s00432-008-0475-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.