Abstract
The Arp2/3 complex is a stable assembly of seven protein subunits including two actin-related proteins (Arp2 and Arp3) and five novel proteins. Previous work showed that this complex binds to the sides of actin filaments and is concentrated at the leading edges of motile cells. Here, we show that Arp2/3 complex purified from Acanthamoeba caps the pointed ends of actin filaments with high affinity. Arp2/3 complex inhibits both monomer addition and dissociation at the pointed ends of actin filaments with apparent nanomolar affinity and increases the critical concentration for polymerization at the pointed end from 0.6 to 1.0 μM. The high affinity of Arp2/3 complex for pointed ends and its abundance in amoebae suggest that in vivo all actin filament pointed ends are capped by Arp2/3 complex. Arp2/3 complex also nucleates formation of actin filaments that elongate only from their barbed ends. From kinetic analysis, the nucleation mechanism appears to involve stabilization of polymerization intermediates (probably actin dimers). In electron micrographs of quick-frozen, deep-etched samples, we see Arp2/3 bound to sides and pointed ends of actin filaments and examples of Arp2/3 complex attaching pointed ends of filaments to sides of other filaments. In these cases, the angle of attachment is a remarkably constant 70 ± 7°. From these in vitro biochemical properties, we propose a model for how Arp2/3 complex controls the assembly of a branching network of actin filaments at the leading edge of motile cells.
Actin polymerization drives forward the leading edge of migrating cells (1, 2). This process depends on elongation of filaments at their fast-growing, barbed ends, organization of filaments into mechanically stable networks, and regulated disassembly of filaments. The mechanisms that initiate barbed end growth and control filament organization in the leading edge are still not known. In vitro, the rate limiting step for filament assembly is nucleation—the formation of actin dimers and trimers (3–5)—and this is the step at which actin assembly is thought to be regulated in vivo. The nucleation of actin filaments in vivo is poorly understood, but one attractive candidate for nucleation factor is the Arp2/3 complex. In addition to five novel polypeptides, the Arp2/3 complex contains two actin-related proteins (Arp2 and Arp3), proposed to form a dimer similar to, but more stable, than an actin dimer (6–8) that might act as a cryptic nucleus for actin filament formation. Models of the atomic structures of Arp2 and Arp3 suggest that they are more likely to initiate growth in a barbed-end than a pointed-end direction (7). Also, Arp2/3 complex localizes to regions of actin assembly such as macropinocytotic cups, the leading edges of motile cells (6–9), the actin comet tails of the intracellular pathogen Listeria monocytogenes (10), and motile actin patches associated with the plasma membrane of budding yeast (11).
Here, we show that purified Arp2/3 complex caps the slow-growing, pointed ends of actin filaments with nanomolar affinity and promotes formation of actin filaments that elongate only at their barbed ends. The abundance of Arp2/3 in Acanthamoeba and its affinity for filament ends are sufficient to cap the pointed ends of all actin filaments in vivo—a previously unsuspected result. Arp2/3 complex organizes actin filaments into a branching network with filament ends attached to the sides of other filaments at a fixed angle of 70°. This capping, nucleating, and fixed-angle branching activity, together with the ability to crosslink actin filaments (12) and induce formation of actin “clouds” around Listeria (10), makes the Arp2/3 complex the best candidate to control actin filament assembly and organization at the leading edge of motile cells and on the surface of intracellular bacteria.
MATERIALS AND METHODS
Protein Purification.
Acanthamoeba actin was purified from DEAE column fractions by polymerization–depolymerization and gel filtration (13). We purified Arp2/3 complex from Acanthamoeba by ion exchange on DEAE, followed by poly-l-proline affinity chromatography (6). Plasma gelsolin was the generous gift of either Bob Robinson or Velia Fowler or was made according to the method of Bryan (14).
Polymerization Assays.
We labeled actin with pyrene iodoacetamide (13) (Molecular Probes) and measured fluorescence with a PTI AlphaScan spectrofluorometer (Photon Technologies International, Princeton). Polymerization medium contained 50 mM KCl, 1.0 mM MgCl2, 1.0 mM EGTA, 0.2 mM ATP, and 10 mM imidazole (pH 7.0). Before use, we converted Ca2+-actin to Mg2+-actin, the physiological form of actin (15).
We formed gelsolin-actin seeds in the presence of 500 μM calcium and 10 mM Tris (pH 8.0) by adding a twofold molar excess of Ca2+ actin to gelsolin and incubating for 2 h at room temperature followed by an overnight incubation at 4°C (14). We warmed these gelsolin–actin dimers to room temperature and added a fivefold excess of actin along with 1 mM EGTA, 1 mM MgCl2, and 50 mM KCl.
Kinetic Modeling.
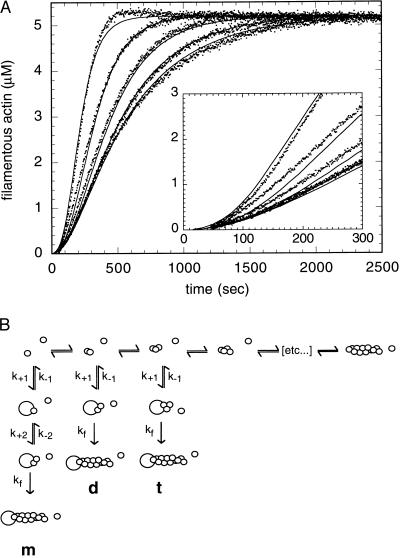
We constructed kinetic models by using kinsim (16) and optimized parameters to fit experimental data with the Marquardt–Levant least-squares optimization algorithm by using the program fitsim (17). The kinetic model for spontaneous actin polymerization consisted of six steps:
 |
The rate constants for the first two steps were varied to obtain a best fit. The third and all subsequent monomers were assumed to add with the rate constants experimentally determined for addition of actin monomers to the barbed end of a stable filament (18). The best fit parameters for this model are given in Table 1. The three models proposed for nucleation by Arp2/3 complex are illustrated in Fig. 2B.
Table 1.
Rate and equilibrium constants for kinetic model of spontaneous actin polymerization
| Step | k+ (μM−1s−1) | k− (s−1) | Kd, M |
|---|---|---|---|
| 1 | 9.2 | 2.9 × 106 | 0.3 |
| 2 | 9.4 | 2.8 × 105 | 0.03 |
| 3 | 10 | 1 | 10−7 |
Figure 2.
Arp2/3 complex nucleates actin filament polymerization. (A) Spontaneous polymerization of 5.3 μM pyrene-labeled (7%) Acanthamoeba actin is accelerated by Arp2/3 complex. Conditions: same as Fig. 1. The concentrations of complex in the samples are, from right to left, 0 μM, 0.037 μM, 0.15 μM, 0.58 μM, and 2.3 μM. Solid lines are kinetic simulations based on binding of Arp2/3 complex to actin dimers and the rate constants in Table 2. (Inset) Early time points of polymerization. Conditions: same as Fig. 1. (B) Models for actin filament nucleation by Arp2/3 complex: Arp2/3 complex interacts sequentially with two actin monomers (m), with an actin dimer (d), or with a trimer (t) to form a nucleus for filament elongation.
Electron Microscopy.
We made gelsolin–actin dimers as described above and then added them to 20 μM actin monomers along with a 0.1 volume of 10x polymerization buffer. These gelsolin-capped filaments were added to either Arp2/3 complex or a buffer blank, incubated for 1–2 min, and then diluted with polymerization buffer to final concentrations of 0.3 μM actin and 0.08 μM Arp2/3 complex and prepared for rapid freezing.
Samples were adsorbed to mica flakes, quick-frozen, and freeze-dried as previously described (19, 20). Electron microscope negatives were placed on a light box and images of selected fields were captured with a video camera and recorded on an optical memory disk recorder; prints of optical memory disk recordings were made with a Sony video printer and were compiled for montages. Montages were scanned with a Microtek ScanMaker III (MicroTek, Hsinchu, Taiwan) and the brightness levels of the scanned images were adjusted for optimum contrast by using Adobe Photoshop version 3.0.5 (Adobe Systems, Mountain View, CA).
RESULTS
Arp2/3 Complex Caps Pointed Ends of Actin Filaments.
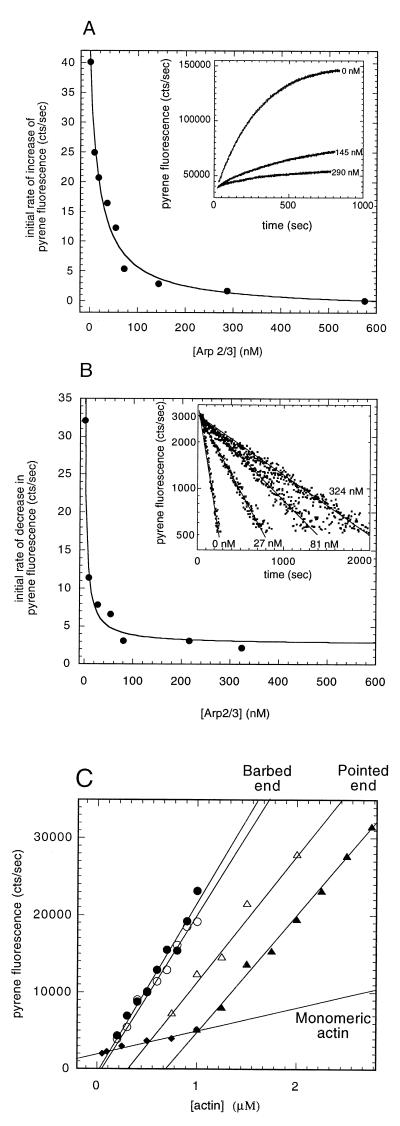
Three lines of evidence show that Arp2/3 complex caps the pointed ends of actin filaments. First, it inhibits pointed-end filament growth from gelsolin-actin dimers with an apparent Kd of 21 ± 3 nM (Fig. 1A). Second, it inhibits dissociation of actin monomers from the pointed ends of gelsolin-capped filaments with an apparent Kd of 4.2 ± 1.0 nM (Fig. 1B). Third, it increases the critical concentration for polymerization at the pointed end of gelsolin-capped filaments from 0.6 to 1.0 μM (Fig. 1C). None of these effects requires Ca2+. These effects of Arp2/3 complex on pointed-end assembly are different from those of the only other known pointed-end capping protein, tropomodulin (21), which requires tropomyosin for high affinity binding.
Figure 1.
Arp2/3 complex caps the pointed ends of actin filaments with nanomolar affinity. (A) Polymerization of 2 μM 7% pyrene-labeled actin monomers was initiated with 3 nM gelsolin-actin dimers in the presence of 0–580 nM Arp2/3 complex as indicated. (Inset) Time courses of pointed-end elongation monitored by pyrene fluorescence. (B) Inhibition of depolymerization of gelsolin-capped actin filaments by Arp2/3 complex. Pyrene-labeled (10%) actin filaments (20 μM), assembled from 0.03 μM gelsolin-actin nuclei, were diluted to 0.2 μM (<pointed end critical concentration) in Arp2/3 concentrations from 0 to 340 nM. (Inset) Pyrene fluorescence traces in the absence (leftmost trace) and presence of increasing concentrations of Arp2/3 complex (progressively slower traces). (C) Effect of Arp2/3 complex on the critical concentration for actin polymerization. A range of concentrations of pyrene-labeled actin (7%) were assembled to steady-state spontaneously (○, •) or from 3 nM gelsolin-actin seeds (▵, ▴) in the absence (open symbols) or presence (closed symbols) of 0.3 μM Arp2/3 complex. The extent of polymerization was measured by pyrene fluorescence after 14 h. We measured critical concentration from the intersections of the filamentous actin fluorescence with actin monomer fluorescence (⧫). Conditions: temperature: 24°C; buffer: 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole (pH 7.0).
Arp2/3 does not affect either the barbed end critical concentration (Fig. 1C) or the rate of barbed-end elongation from preformed actin filaments or spectrin-actin seeds (not shown). Therefore, in contrast to the pointed end, the Arp2/3 complex has no effect on subunit association or dissociation at the barbed end.
Arp2/3 Complex Nucleates Actin Polymerization.
Arp2/3 complex accelerates spontaneous actin polymerization in a concentration-dependent manner but does not abolish the lag phase of polymerization even at high concentration (Fig. 2A). Formally, acceleration of polymerization could be due to one of three mechanisms: (i) Severing of newly formed filaments, which increases the number of growing barbed ends (25); (ii) altering in the kinetics of monomer addition to the barbed end—either by increasing the monomer association rate or decreasing the monomer dissociation rate; or (iii) increasing the rate of nucleation of actin filaments. We rule out the first possibility because, unlike severing proteins, Arp2/3 complex has no effect on elongation from preformed actin filaments (not shown) and does not decrease low shear viscosity of solutions containing actin filaments (12). We rule out the second possibility because the Arp2/3 complex alters neither the critical concentration at the barbed end (Fig. 1C) nor barbed-end elongation from spectrin-actin seeds (not shown) and thus does not affect either association or dissociation of monomers from the barbed end. Therefore, we propose that Arp2/3 complex accelerates polymerization by increasing the rate of nucleation. Barbed end capping factors such as capping protein nucleate filament formation by stabilizing small actin oligomers and have kinetics similar to those of Arp2/3 complex (22, 23). In particular, polymerization accelerates slowly after a lag of ≈1 min even at high concentrations of capping protein. In contrast, preformed actin filaments initiate polymerization at the maximum rate without a lag (24).
We tested several possible mechanisms of nucleation by kinetic simulation. First, we selected rate constants for a standard model (described in Materials and Methods) of nucleation and elongation of actin alone. We performed a global least-squares fit of parameters defined by the model, to full time courses of spontaneous polymerization at four different actin monomer concentrations (3, 5). We then postulated several mechanisms (Fig. 2B) by which the Arp2/3 complex could generate nuclei by forming or stabilizing transient intermediates along the assembly pathway. To distinguish among these models, we searched for the single set of rate constants for each model that gave a best global fit to a set of full time courses of polymerization in the presence of 0.036 to 2.3 μM Arp2/3 complex. The first mechanism is functionally identical to nucleation by the barbed end of an actin filament or, with opposite polarity, by gelsolin. The other nucleation mechanisms are similar to those proposed for capping protein (3, 23).
Dimer- and trimer-binding models fit the data equally well, but the equilibrium binding constants argue for the dimer model (Fig. 2A, solid lines). The association rate for the trimer binding model (Table 2) is more than an order of magnitude higher than the diffusion-limited rate of monomer association to a barbed end, and the equilibrium constant predicted for trimer binding is 10−8 μM. We consider both values implausible. The dimer model predicts a reasonable rate constant for dimer association and a 2-nM affinity of Arp2/3 for actin dimers (Table 2), similar to the Kd we measured for pointed end capping. In contrast, no set of rate constants provides a good fit to the monomer-binding model, particularly at early times.
Table 2.
Rate and equilibrium constants for kinetic models of actin nucleation by Arp2/3 complex
| Monomer | Dimer | Trimer | |
|---|---|---|---|
| k+1 | 27.7 μM−1s−1 | 1.22 μM−1s−1 | 371 μM−1s−1 |
| k−1 | 290 s−1 | 0.002 s−1 | 10−5 s−1 |
| Kd1 | 10.4 μM | 0.0016 μM | 2.7 × 10−8 μM |
| k+2 | 6.4 μM−1s−1 | ||
| k−2 | 650 s−1 | ||
| Kd2 | 101 μM | ||
| kf | 4.2 × 10−5 s−1 | 3.91 × 10−5 s−1 | 8.0 × 10−4 s−1 |
For acceptable fits, all models required a very slow first order reaction between actin binding and formation of a stable filament end. The physical significance of this step is unknown, but it may reflect a two-step binding process or a requirement for nucleotide hydrolysis by Arp2, Arp3, or bound actin. The best-fit forward rate constants for this step are similar for all models, and omission of this step does not change the relative goodness of the fits or calculated affinities of Arp2/3 complex for actin. Because of the slow first order reaction after binding, 49 of 50 actin dimers dissociate from Arp2/3 complex before initiating a new filament with a tightly capped pointed end.
Our kinetic models are meant to help discriminate between the most obvious possible nucleation mechanisms. Direct measurement of interactions between Arp2/3 complex and actin monomers, dimers, and trimers will be required to establish the details of the mechanism. One piece of experimental evidence in support of our model, however, is the effect of profilin on nucleation by Arp2/3 complex. Profilin inhibits spontaneous nucleation of actin filaments by blocking formation of small oligomers but does not alter elongation from preformed filaments. Addition of profilin slows nucleation by Arp2/3 complex (not shown), suggesting that it requires spontaneous formation of oligomers.
Arp2/3 Complex Links the Pointed Ends of Actin Filaments to the Sides of Other Filaments.
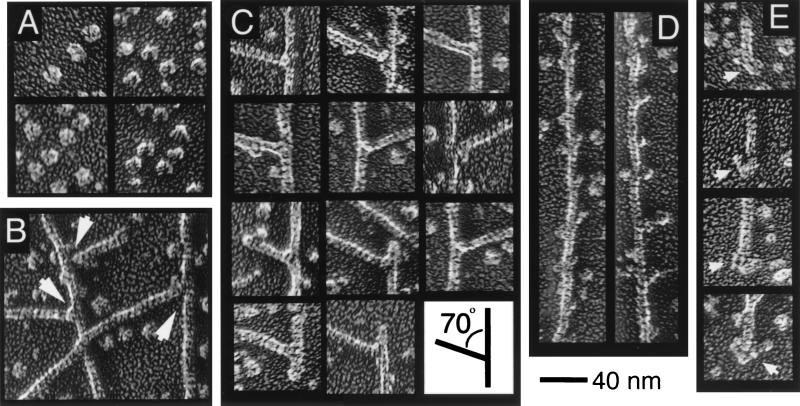
Electron micrographs of rapidly frozen specimens reveal that Arp2/3 complex links actin filaments end-to-side at a fixed angle (Fig. 3 B and C). Purified Arp2/3 (Fig. 3A) is a globular particle with dimensions of ≈10 × 14 nm and in certain orientations appears bilobed or clefted. This structure is similar in most respects to Arp2/3 prepared by glycerol spraying and air drying (8). On filaments partially decorated with Arp2/3 complex, we observed individual Arp2/3 molecules binding to the sides of filaments with a distinct polarity and the expected periodicity of ≈37 nm—the spacing of the crossover of the two-stranded actin helix (Fig. 3D).
Figure 3.
Electron micrographs of quick-frozen, deep-etched, and rotary-shadowed samples of Arp2/3 complex (A) and complex mixed with gelsolin-capped actin filaments (B-D). In the presence of Arp2/3, complex actin filaments form branching arbors with numerous end-to-side connections between filaments (B). The branch points appear to be rigid attachments with a fixed 70° angle between actin filaments (C) and frequently contain a globular mass at the point of attachment (C, left arrow in B). (D) Filaments partially decorated with Arp2/3 complex. (E) Globular masses associated with filament pointed ends in the presence of Arp2/3 complex. Conditions: buffer same as Fig. 1.
When mixed with preformed, gelsolin-capped filaments, the complex induces formation of end-side junctions between filaments (Fig. 3 B and C). The angle between filaments in these junctions is a remarkably consistent 70 ± 7° (n = 48), indicating that the attachment is rigid. In both cases observed, the free ends of branching filaments were capped by a small mass indistinguishable from gelsolin on the barbed end of an actin filament. In three cases in which we observed an end of a filament bearing a branch, the end on the acute side of the branch (70°) appeared to be capped by gelsolin, whereas the end on the obtuse side of the branch (110°) was capped by a larger mass, consistent with Arp2/3 complex (Fig. 3C, image on lower left). A mass is present at many branches, but it is not as distinct as Arp2/3 complex attached to the side of a filament (Fig. 3B).
We identified barbed ends of actin filaments by morphology, comparing images of filament ends with previously obtained (J.A.H., unpublished observations) images of gelsolin bound to actin filaments. Images of filament ends that did not conform to this morphology were of several types but all were capped by large asymmetric masses, some similar in morphology to Arp2/3 complex and others much larger—perhaps fragments of filaments bound to an Arp2/3 molecule (Fig. 3E).
DISCUSSION
The Arp2/3 complex has been proposed to play a role in initiation of actin polymerization (7, 10, 26) and in organization of actin filaments into rigid networks (12) at the leading edges of motile cells. From the data in the present study, we propose that the two activities are coupled tightly and combine to determine the structure of the actin filament network at the leading edge.
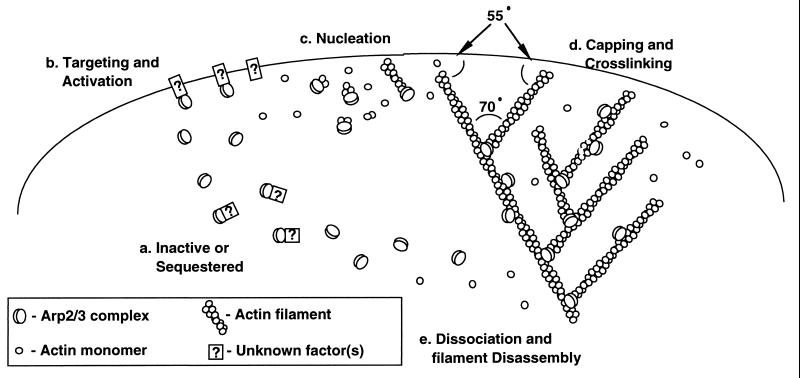
Our nucleation data are consistent with features of a model proposed from structural considerations by Kelleher et al. (7), namely that nucleation by Arp2/3 complex produces filaments that elongate from their barbed-ends. The lack of effect on initial rates of polymerization, however, argues against Arp2 and Arp3 forming a structure that precisely mimics a conventional actin dimer or the barbed end of an actin filament as originally proposed. Welch et al. (10) observed that, in vitro, Arp2/3 complex promotes accumulation of “clouds” of actin filaments around Listeria. Although this assay cannot distinguish between side-binding, end-binding, and nucleation of actin filaments, the authors proposed two nucleation-based models for “cloud formation.” One was essentially the barbed end nucleation model of Kelleher et al. (7), and the other was a nucleation-and-release type mechanism involving transient association of Arp2/3 with filament barbed-ends and, at least initially, elongation of filaments from the pointed end. Our nucleation and pointed end-capping data appear to rule out a nucleation-and-release style mechanism altogether. Instead, we propose a “dendritic nucleation” model (Fig. 4) in which nucleation, pointed-end capping, and filament side binding of Arp2/3 complex combine to produce a distinctive structure.
Figure 4.
Dendritic nucleation model for actin polymerization, capping, and network formation at the leading edge of a motile cell. Inactive or sequestered Arp2/3 complex (a) is recruited to the leading edge of the cell by an unknown mechanism (b). The complex nucleates new actin filaments either while free (c) or while bound to the side of existing filaments (d). The complex anchors the branch at a fixed angle of 70° relative to the barbed ends of the two filaments. Rapid growth of new filaments in the barbed direction, fed by actin subunits from the actin-profilin pool, expands the actin filament network. Subsequent disassembly of the network deep in the cytoplasm regenerates subunits for later growth of the cortical network.
Arp2/3 complex caps filament pointed ends with high-affinity, making it only the second pointed end capping factor discovered and the only one that nucleates actin filaments and binds with high affinity in the absence of tropomyosin. The estimated affinity of Arp2/3 complex for pointed ends (5–20 nM) is 10-fold weaker than the affinities of capping protein (23), villin (27), or gelsolin (28) for barbed ends or of tropomodulin for pointed ends of tropomyosin-bound filaments (21). However, in Acanthamoeba, the micromolar concentration of Arp2/3 complex (7, 29) exceeds the Kd and the concentration of actin filaments (30), so many cellular actin filaments will be capped on their pointed ends by Arp2/3 complex.
Tilney and coworkers (31) argued that the pointed ends of actin filaments in isolated Listeria tails are capped, but most investigators assume that the pointed ends of cellular actin filaments are free and that the primary mode of filament disassembly in vivo is monomer dissociation from the pointed end (32). Capping of a significant fraction of pointed ends in vivo, however, causes current models for treadmilling of actin filaments to fall apart completely (32). Further work is required to understand the disassembly of actin filaments capped at both ends, but disassembly almost certainly involves filament severing or uncapping in addition to monomer dissociation.
Our electron micrographs show that the side and pointed-end binding activities of the Arp2/3 complex are not mutually exclusive but rather combine to form T and Y junctions of actin filaments (Fig. 3 B and C) in which the pointed end of one filament is attached rigidly to the side of another at a fixed angle of 70°. Svitkina et al. (26) recently observed that pointed ends of actin filaments at the leading edge of a fish keratocyte are attached via T junctions to the sides of other filaments and suggested that either ABP-280 or Arp2/3 complex might be responsible. Although they did not measure the branching angles at these junctions, the average angle of the branches presented in their study is 75 ± 10° (n = 17), similar to branches produced by Arp2/3 complex.
The dendritic nucleation activity of the Arp2/3 complex provides an attractive molecular model (Fig. 4) for the assembly of the branching network of actin filaments at the leading edge of a motile cell (26)—one that also may apply to the actin comet tail generated by Listeria (31). Several features of this model require elaboration and verification, particularly the mechanisms of recruitment and activation (Fig. 4b), as well as the order of events, because binding to the sides of filaments could occur before or after nucleation and binding of Arp2/3 complex to the side of a filament could influence its nucleation activity.
Stimulation of Dictyostelium cells with chemoattractant produces an increase in filament number by either nucleation or filament severing (33). In Acanthamoeba, nucleation by Arp2/3 complex may be sufficient for the extension of the leading edge, because the actin cytoskeleton is poised for explosive but transient barbed end growth. An 80-μM pool of actin bound to profilin (V. K. Vinson, E. M. De La Cruz, D. A. Kaiser, H. N. Higgs, and T.D.P., unpublished work; D. A. Kaiser, V. K. Vinson, D. B. Murphy, and T.D.P., unpublished work) can add on to barbed ends (34) and cause newly formed filaments to grow rapidly (≈500 molecules/s). Capping protein will bind to free barbed ends and terminate elongation with a half time of ≈1.5 s (33, 35) to produce an average length for new filaments of 2 μm. The membrane-proximal barbed ends may continue to elongate by selective removal of capping protein by poly-phosphoinositides (35).
An important feature of the dendritic nucleation model is that pointed ends of newly formed filaments are capped and anchored in the existing network, so that elongation of free barbed ends can drive forward the leading edge (36, 37). The fixed branching angle within the filament arbor will tend to create an orthogonal network in which growing barbed ends meet the plasma membrane at an angle of ≈45°, which is mechanically optimal for translating filament elongation into membrane displacement (37).
Acknowledgments
We thank Velia Fowler for providing spectrin actin seeds and gelsolin and Bob Robinson for gelsolin. R.D.M. is indebted to members of the Pollard lab, particularly Joe Kelleher and Mike Ostap, for useful discussions. This work was supported by a National Institutes of Health grant to T.D.P. and a fellowship from the Jane Coffin Childs Fund for Medical Research to R.D.M.
ABBREVIATION
- Arp
actin-related protein
References
- 1.Tilney L G, Bonder E M, Coluccio L M, Mooseker M S. J Cell Biol. 1983;97:112–142. doi: 10.1083/jcb.97.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zigmond S H. Cell Motil Cytoskel. 1993;25:309–316. doi: 10.1002/cm.970250402. [DOI] [PubMed] [Google Scholar]
- 3.Cooper J A, Buhle E L, Jr, Walker S B, Tsong T Y, Pollard T D. Biochemistry. 1983;22:2193–2202. doi: 10.1021/bi00278a021. [DOI] [PubMed] [Google Scholar]
- 4.Tobacman L S, Brenner S L, Korn E D. J Biol Chem. 1983;258:8806–8812. [PubMed] [Google Scholar]
- 5.Frieden C. Proc Natl Acad Sci USA. 1983;80:6513–6517. doi: 10.1073/pnas.80.21.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Machesky L M, Atkinson S J, Pollard T D. J Cell Biol. 1994;127:107–115. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelleher J F, Atkinson S J, Pollard T D. J Cell Biol. 1995;131:385–397. doi: 10.1083/jcb.131.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mullins R D, Stafford W F, Pollard T D. J Cell Biol. 1997;136:331–343. doi: 10.1083/jcb.136.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welch M D, DePace A H, Verma S, Iwamatsu A, Mitchison T J. J Cell Biol. 1997;138:375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welch M D, Iwamatsu A, Mitchison T J. Nature (London) 1997;385:265–269. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- 11.Moreau V, Madania A, Martin R P, Winsor B. J Cell Biol. 1996;134:117–132. doi: 10.1083/jcb.134.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullins R D, Kelleher J F, Xu J, Pollard T D. Mol Biol Cell. 1998;9:841–852. doi: 10.1091/mbc.9.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollard T D. J Cell Biol. 1984;99:1970–1980. doi: 10.1083/jcb.99.6.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryan J. J Cell Biol. 1988;106:1553–1562. doi: 10.1083/jcb.106.5.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young C, Southwick F S, Weber A. Biochemistry. 1990;29:2232–2240. doi: 10.1021/bi00461a005. [DOI] [PubMed] [Google Scholar]
- 16.Barshop A, Wrenn R F, Frieden C. Anal Biochem. 1983;130:134–145. doi: 10.1016/0003-2697(83)90660-7. [DOI] [PubMed] [Google Scholar]
- 17.Zimmerle C T, Frieden C. Biochem J. 1989;258:381–387. doi: 10.1042/bj2580381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollard T D. J Cell Biol. 1986;103:2747–2754. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heuser J E. J Mol Biol. 1983;169:155–195. doi: 10.1016/s0022-2836(83)80179-x. [DOI] [PubMed] [Google Scholar]
- 20.Heuser J E. J Electron Microsc Tech. 1989;13:244–263. doi: 10.1002/jemt.1060130310. [DOI] [PubMed] [Google Scholar]
- 21.Weber A, Pennise C R, Babcock G G, Fowler V M. J Cell Biol. 1994;127:1627–1635. doi: 10.1083/jcb.127.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper J A, Pollard T D. Biochemistry. 1985;24:793–799. doi: 10.1021/bi00324a039. [DOI] [PubMed] [Google Scholar]
- 23.Caldwell J E, Waddle J A, Cooper J A. J Biol Chem. 1989;264:2648–2652. [Google Scholar]
- 24.Pollard T D. Anal Biochem. 1983;134:406–412. doi: 10.1016/0003-2697(83)90316-0. [DOI] [PubMed] [Google Scholar]
- 25.Maciver, S. K., Zot, H. G. & Pollard, T. D. (1991) 115, 1611–1620. [DOI] [PMC free article] [PubMed]
- 26.Svitkina T M, Verkhovsky A B, McQuade K M, Borisy G G. J Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walsh T P, Weber A, Higgins J, Bonder E M, Mooseker M S. Biochemistry. 1984;23:2613–2621. doi: 10.1021/bi00307a012. [DOI] [PubMed] [Google Scholar]
- 28.Selve N, Wegner A. Eur J Biochem. 1986;160:379–387. doi: 10.1111/j.1432-1033.1986.tb09982.x. [DOI] [PubMed] [Google Scholar]
- 29.Kelleher, J. F., Mullins, R. D. & Pollard, T. D. (1998) Methods Enzymol., in press. [DOI] [PubMed]
- 30.Gordon D J, Eisenberg E, Korn E D. J Biol Chem. 1976;251:4778–4786. [PubMed] [Google Scholar]
- 31.Tilney L G, DeRosier D J, Weber A, Tilney M S. J Cell Biol. 1992;118:83–93. doi: 10.1083/jcb.118.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carlier M F, Pantaloni D. J Mol Biol. 1997;269:459–467. doi: 10.1006/jmbi.1997.1062. [DOI] [PubMed] [Google Scholar]
- 33.Eddy R J, Han J, Condeelis J S. J Cell Biol. 1997;139:1243–1253. doi: 10.1083/jcb.139.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollard T P, Cooper J A. Biochemistry. 1984;23:6631–6641. doi: 10.1021/bi00321a054. [DOI] [PubMed] [Google Scholar]
- 35.Schafer D A, Jennings P B, Cooper J A. J Cell Biol. 1996;135:169–179. doi: 10.1083/jcb.135.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forscher P, Smith S J. J Cell Biol. 1988;107:1505–1516. doi: 10.1083/jcb.107.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mogliner A, Oster G. Biophys J. 1996;71:3030–3045. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]