Abstract
In this paper, we propose a novel method for the registration of volumetric images of the brain that optimizes the alignment of both cortical and subcortical structures. In order to achieve this, relevant geometrical information is extracted from a surface-based morph and diffused into the volume using the Navier operator of elasticity, resulting in a volumetric warp that aligns cortical folding patterns. This warp field is then refined with an intensity driven optical flow procedure that registers noncortical regions, while preserving the cortical alignment. The result is a combined surface and volume morph (CVS) that accurately registers both cortical and subcortical regions, establishing a single coordinate system suitable for the entire brain.
Keywords: Cortical and subcortical alignment, elastic warp, magnetic resonance imaging (MRI), volumetric registration
I. Introdution
PAIR-WISE brain image registration is an active area of research in the medical imaging community. Various algorithms have addressed the generic problem of registering information from two brain scans using an array of different techniques that can be broadly classified as either volume based or surface based. Volumetric registration (see [1] for a thorough survey) seeks a 3-D deformation field that is driven by either raw intensity information or features derived from image intensities. A different approach is to extract geometric features from surface models of anatomical structures such as the neocortex, and to reformulate the complex correspondence problem in a surface matching framework.
Both of these approaches have advantages and weaknesses. Surface-based methods [2]–[5] have been shown to accurately align the highly complex folding pattern of the human cerebral cortex, and to result in increased statistical power for averaging of functional data across subjects, presumably due to their alignment of functionally homologous regions across subjects [3]. This accuracy stems from the direct use of geometric information that is generally unavailable to volumetric methods, and the relatively close relationship between folding patterns and functional properties of the neocortex. Conversely, volumetric methods [6]–[10] provide a correspondence field across the entire brain, including subcortical and ventricular regions that are not in the domain of the surface-based alignment procedures, but typically fail to align corresponding cortical folds across subjects.
The failure of nonlinear volumetric procedures to align cortical folding patterns stems from the highly nonconvex nature of the driving energy functionals when initialized with affine transforms. Fundamentally, a 12 parameter affine transform, which is typically used as an initial estimate of the displacement fields, does not bring many homologous folds into close enough correspondence to allow nonlinear techniques to align them. In essence, if the initialization of a volumetric nonlinear warp does not bring a fold within one half the interfold spacing of the homologous fold then intensity-based algorithms will converge to the incorrect solution.
In this paper, we propose a method that combines surface-based and volumetric approaches, preserving the strengths of each. This is accomplished by integrating surface-based information into a volumetric registration procedure through the use of a biomechanical deformation model. The result is a 3-D displacement field that aligns both cortical folding patterns and subcortical structures across subjects. While the idea of using surface-based registration to drive volumetric registration is not new [11]–[13], here we focus on the automated and accurate registration of both cortical folding patterns and noncortical regions in 3-D space.
Regularizing registration procedures by employing biophysical models of the underlying deformable objects is an attractive approach in the field of medical image registration as it brings to bear branches of mathematics that allow the generation of correspondence fields with specified properties (e.g., smoothness and invertibility). One of the first such presentations of this type of registration was that of Bajcsy et al. [6], who built on the work of Broit [14], where images were considered to be solid linear elastic bodies constrained at certain position by applied external forces. Extensions of this early work have been since proposed by Davatzikos [7], Ferrant [12], and Toga [13], who use sparse displacement fields extracted from curves or surfaces to produce a sparse displacement field on the boundary of the cortex, which is then diffused using a volumetric elastic model. A drawback of these methods is that they are accurate close to the boundary driving the deformation, but they fail to properly align structures distant from it.
Collins [15] was the first to propose a method that addresses this problem. He used surface information through manually extracted sulcal traces together with intensity information to obtain a deformation field that was accurate both in the cortical areas as well as in regions distant from the cortex.
Other hybrid methods, such as HAMMER [16] implicitly incorporate surface as well as volume information in the alignment through the use of feature vectors that represent both intensity as well as geometric information in the registration process. More recent work extending HAMMER [17] uses the result of HAMMER as an initialization to further improve cortical surface alignment between the two subjects being registered.
Our current approach [18] is similar to that of Joshi et al. [19], [20]. These two methods have the same basic ingredients. First, a surface-based registration algorithm is applied and is used to derive a volumetric displacement field. Subsequently, an intensity-based registration is computed to match regions distant from the surfaces. It is worth pointing out that the surface registration used in the two approaches differ significantly in that the surface-based registration method employed by Joshi et al. is not automated, requiring a trained neuroanatomist to specify a set of corresponding landmarks across subjects. The current algorithm extends our preliminary work [18], and uses an automated algorithm for surface-based registration [21] to obtain an initial estimate of a displacement field that aligns two individual brain images. The resulting alignment is based on a biophysical model of the brain, thus yielding a smooth, invertible deformation field. Using this deformation as an initialization, we then apply an intensity-based registration [9] and [22].
Initializing the deformation field by aligning cortical folds provides an extremely well-constrained alignment of noncortical regions, which can then be brought into register using a nonlinear intensity-based volumetric deformation.
The remainder of this paper is organized as follows. In Section II we give an overview of the preprocessing steps that lead to accurate and topologically correct cortical surface models given an MRI image, as well as of the surface-based registration that brings the geometric features of these surfaces into register across subjects. The extraction and registration of the cortical surfaces are performed automatically using previously described techniques and freely available software (http://surfer.nmr.mgh.harvard.edu/fswiki), hence, we only describe them briefly. In Section III we provide details about the diffusion of the surface-based registration results into the volume, using the Navier operator derived from the theory of linear elasticity. We detail the discretization of the Navier operator, the numerical scheme used in the implementation, and the technical aspects of working with tetrahedral meshes. In Section IV, we briefly explain the intensity-based registration algorithm that was previously presented in [9]. Finally in Section V we present our results on a varied set of real medical data sets, showing the accuracy of the resulting registration procedure both cortically and throughout the brain.
II. Surface-Based Registration
We present an algorithm for registering two structural brain scans, arbitrarily denoted target and moving. This registration involves three main steps. First, cortical folding patterns are registered using information derived from cortical surface models. Then this warp is projected back to the volumetric domain and diffused using linear elasticity constraints. Finally an optical flow intensity-based registration algorithm is applied, using the output of the second step as an initialization. This framework results in a nonlinear displacement field which simultaneously registers cortical and subcortical structures.
In the preprocessing step, the input brain scans are independently processed to obtain accurate, topologically correct reconstructions of the cortical surfaces (see [21], [23]–[26] for details). As a result we obtain four detailed cortical surfaces for each brain scan. For each hemisphere, one of the surfaces is extracted from the boundary between the gray and white matter (denoted the white surface), while the other surface is automatically positioned at the pia between cortical gray matter and cerebrospinal fluid (CSF) (denoted the pial surface).
The reconstruction of the cortical surfaces is a complex procedure [24] that is broken into a number of subtasks. First, intensity variations due to magnetic field inhomogeneities are corrected [27] and a normalized intensity image is created from a T1-weighted anatomical 3-D MRI data set [9]. Next, extracerebral voxels are removed, using a “skull-stripping” procedure [23]. The intensity normalized, skull-stripped image is segmented based on the geometric structure of the gray-white interface. Cutting planes are then computed that separate the cerebral hemispheres and disconnect subcortical structures from the cortical component [24]. This generates a preliminary segmentation that is partitioned using a connected components algorithm. Any interior holes in the white matter components are filled, resulting in a single filled volume for each cortical hemisphere. Finally, the resulting volume is covered with a triangular tessellation and deformed to produce an accurate and smooth representation of the gray-white interface as well as the pial surface. This surface departs from a simple spherical topology due to the presence of subcortical gray matter as well as various midbrain structures. These topological “defects” are automatically removed, resulting in a surface that is both geometrically accurate and topologically correct [23], [25], [28].
Each of the corresponding surfaces of the target and moving scans are then registered in the spherical space [3], [29]. The algorithm minimizes the following energy functional:
| (1) |
where Jp measures the alignment, based on cortical depth and curvature information, while the other two terms act as regularizers. JA is a topology-preservation term, while Jd controls the amount of metric distortion allowed
| (2) |
| (3) |
In (2) and (3), superscripts denote integration time with 0 being the starting point, T and V are the number of triangles and vertices in the tessellation, is the position of vertex i at iteration n, N(i) is the set of neighbors of the ith vertex and Ai denotes the oriented area of triangle i. The result of the surface registration is a 1-to-1 mapping that transports all surfaces of the moving scan to the counterpart surfaces in the target image , where S can be any of the left/right pial/white surfaces of the brain.
An example of the representation of the sulcal patterns being registered in spherical space is displayed in Fig. 1. On the left-hand side it is the subject to be registered, in the middle the already registered subject and on the right-hand side the target that is displayed. The colormap we use represents average convexity information, or the signed distance a point has to move to reach the inflated surface [21].
Fig. 1.
Example of the sulcal pattern rendered on the sphere showing the effects of spherical registration. From left to right: initial pose of subject, registered subject, target sulcal pattern.
III. Volumetric Registration of the Cortical Structures
When the surface registration is completed, we obtain a displacement vector field that provides a 1-to-1 mapping between the hemisphere surfaces of the target and moving brain scans in Euclidean space. In the following we show how we diffuse this vector field from the cortical surfaces to the rest of the volume.
Let be the target image domain. We define an arbitrary transformation of the target image as: ϕ(x) = x + u(x) where denotes the displacement field. The goal is to find a function ϕ such that ϕ(x) = Freg(x), for any (one of the left/right pial/white surfaces of brain ), where Freg is the specified displacement through the surface registration procedure. Since the surfaces represent a space of co-dimension 1, in order for this problem to be well-posed, we impose an additional regularity constraint. We require the displacement field to be an elastic deformation, i.e., a smooth, orientation-preserving deformation which satisfies the equations of static equilibrium in elastic materials. This means that we (additionally) require u to satisfy
| (4) |
where is an operator we define below.
The choice of the operator and the discretization method to numerically extrapolate the displacement field has numerous solutions that have been proposed in the registration literature. Some of the widely used ones are the thin plate splines, proposed by Bookstein [30], and the B-splines-based free-form deformations proposed by Rueckert et al. [31]. We have chosen to use the Navier operator from the linear elasticity theory together with the finite element method. This choice was motivated by the high level of flexibility needed in order to satisfy the constraints imposed by the displacement fields on all four surface models of the brain (left and right hemisphere white matter and pial surfaces).
We consider this component of the registration framework to be the most technically significant. The remainder of this section is divided as follows. We start with a short presentation of the partial differential equation of static elasticity. Next, we derive the external forces directly from the output of the surface registration algorithm, as a set of prescribed displacements. Finally, technical aspects regarding our discretization method are presented.
A. Elastic Operator
In order to solve the problem stated above, we use the equilibrium equation for elastic materials. This states that at equilibrium, the elastic energy equals the external forces applied to the body [32]
| (5) |
Here, ∇u = ((∂u)/(∂x), (∂u)/(∂y), (∂u)/(∂z)) denotes the gradient and the second Piola-Kirchoff stress tensor is defined as
and
is the Green-St. Venant strain tensor [32]. Here λ and μ are the Lamé elastic constants that characterize the elastic properties of an isotropic material. The linear approximation to the above operator uses the Fréchet derivative of
| (6) |
since no deformation occurs in the absence of external forces. Finally, is computed by dropping the nonlinear terms in , which results in
| (7) |
where S = λtr(E)I + 2μE is the linearized stress tensor and E = (1)/(2)(∇u + ∇uT) is the linearized strain tensor. Hence, the linear approximation of (4) can be written as
| (8) |
The main drawback of (8) is that it is only valid for small-magnitude deformations. To overcome this restriction, we implement an extension to the linear model, as presented in [11]. Specifically, given external forces that describe large displacements, one can iteratively solve for small linear increments using the linearized Navier equation (8)
| (9) |
or, by neglecting the last term
| (10) |
Using this iterative process, the solution of (10) converges to the solution of (4) (see [32] for the proof).
The Lamé constants λ and μ are specified as functions of Young's modulus of elasticity ε and the Poisson ratio ν: λ = (εν)/((1 + ν)(1 − 2 ν)) and μ = (ε)/(2(1 + ν)). In all of our experiments we used ε = 1 and ν = 0.3.
B. Finite Element Method
The finite element method (FEM) is a discretization technique aimed at obtaining approximate solutions to various partial differential equations. As opposed to finite differences that discretize the differential operators on a structured grid, the FEM approach discretizes the solution space of possible functions. This involves the recasting of one or more equations to hold in some average sense over subdomains of simple geometry.
Using FEM, the given continuous domain is subdivided into a set of M elements, which constitute a mesh. In our case, these elements are tetrahedra, which means that the approximate solution is linear in the interior of each tetrahedron. The displacement field is built as linear combinations of the displacements observed at the nodes of the mesh.
More precisely, the deformation within one element of the mesh is approximated through barycentric interpolation of the shape functions with the k-element (k = 4, in our case) nodal points as coefficients:
| (11) |
The shape function of node i of element el is defined as
| (12) |
where Vel is the volume of element el and coefficients , , etc., are computed such that , for xj the position of node j = 1 ··· 4 of element el and δij being the Kronecker symbol.
The linearized elasticity (10) can be approximated using a variational form, which aims at minimizing the elastic potential energy
| (13) |
where . Here, Kel represents the element stiffness matrix, which can be written as [33]
D in the above is the symmetric elasticity matrix, defined as
where
is obtained by applying the linearized Navier operator to the shape function Nel:
and
An important aspect regarding the FEM is the assembly step, in which all the nodal matrices are inserted into a compact representation of the global elastic potential energy
where r is a re-sizing function which keeps track of individual nodes. Similarly, the displacement at any given point inside the mesh domain can be represented by linear interpolation of the displacement at nodal points, which is encoded by a vector U. There is a clear correspondence between the order of the elements of the vector U and the rows and columns of the global elasticity matrix K. Thus, components (3i + j) with i = 0 ··· Nnodes – 1, j = 0 ··· 2 represent component j of the nodal displacement i. Then (13) becomes
| (14) |
Finally, it should be noticed that individual displacements at an arbitrary point x ∈ Ω can be written as u(x) = N(x)U, which uses the same resizing function r and transcribes (11). This means, that a point x will be inside exactly one tetrahedron, hence its displacement can be expressed using the shape functions inside that tetrahedron as a function of the nodal displacements.
C. External Forces
The theory presented above resolves the discretization of the elastic potential energy. One important issue that remains to be addressed is how the point displacements resulting from the spherical registration are implemented in (10).
One method that implements point displacements is described in [11]. It allows constraining the displacements of certain mesh nodes by explicitly modifying the rows and the columns of the unknowns in the displacement vector U. However, our numerical experiments indicate that this method of constraining the linear elastic system is too restrictive in the sense that it can lead to undesirable warps in regions where anatomical differences exist between the two individual anatomies being registered (such as a split or extra fold).
An alternative method of introducing surface information is via the relaxation of the constraints imposed above. We reformulate the problem of finding a mapping ϕ(x) = x + u(x) such that and (with Si the surfaces of target brain ) as an energy minimization problem. Assuming that the points of the target surfaces are sampled as x1 ··· xV, we solve for a function u that minimizes
| (15) |
where yi = Freg(xi) – xi, i = 1 ··· V are the values of the displacement field obtained from the surface registration in the spherical space.
This relaxation of hard constraints addresses the concern of handling qualitative anatomical differences (such as a split sulcus) between the target and the moving anatomy. When α is large, the constraints are almost perfectly satisfied at the expense of an under-regularized warp. On the other hand, when α is small, the elastic energy term dominates, so that the overall strain obtained is small. In our experiments, we typically used α = 100.
As shown before, u(xi) can be expressed in terms of the unknowns U as u(xi) = N(xi)U through barycentric interpolation. Since all the terms in (15) are strictly convex functions, the overall energy is strictly convex. Hence, the minimum is attained when the gradient is null. By differentiation, we obtain the linear system
| (16) |
This formulation is well suited for a fast iterative linear solver, such as the conjugate gradient-based optimizer.
D. Mesh Construction and Surgery
One difficult algorithmic issue that needs to be addressed is the tetrahedralization of the domain. Beyond the requirements that the tetrahedra (elements) must create a partition of the domain of interest, they must also meet certain quality constraints.
The quality of the elements in the mesh directly impacts the speed and accuracy of the approximate solutions obtained when solving a partial differential equation. One common measure is the aspect ratio of the elements, which is the ratio of the maximum side length to the shortest edge. For a high-quality mesh, this value should be as small as possible.
In addition, there are second-order requirements. Since the basic underlying assumption when meshing a domain is that the approximate solution being sought is parameterized by simple functions inside each element, we would like our mesh to place more elements in regions where the solution varies more rapidly.
In our implementation, we used a flip-based algorithm for generating constrained Delaunay tetrahedralizations [34]. We used the implementation provided by TetGen [35]–[37] to build a Delaunay tetrahedral mesh that is adapted to the input surfaces. We included constraints on the quality of the generated tetrahedra (which was 1.414) and an upper bound on the volume of each tetrahedron (which was set to 3 mm3). Additionally, to meet the conditions stated above, we also required the mesh to be finer near the input surfaces, in order to give maximum flexibility to the resulting solution. An example of a mesh used at one iteration of the algorithm is presented in Fig. 2.
Fig. 2.
Example of tetrahedral mesh used for one iteration of the elastic solver. Notice how the mesh is denser near the input surfaces.
1) Fixing Topology Defects
After solving the linear elastic system of (10) using the numerical scheme in (16), the components of the solution vector U are distributed as displacements of the mesh nodes. The resulting displacement field must meet the usual registration constraint of invertibility. Through the local inversion theorem, this is equivalent to having a nonnegative Jacobian for each tetrahedron.
In rare cases, the resulting tetrahedra can have negative Jacobians. This is due to the fact that some displacements are too large and break the implicit assumptions that allowed us to use the linearized model and then to approximate the solution with a piecewise linear function.
A few methods have been published that can be used to resolve these situations. Conceptually, they follow one or a combination of these two strategies: the first strategy is to apply smoothing to the mesh nodal positions, while the second general technique is more local, aiming to fix tangles locally through an energy minimization approach [38]. Ideally, these techniques should provide some guarantee that the resulting mesh is not inverted. While such proofs exist for 2-D structured meshes, the same has not been currently shown for the 3-D case.
In the present case, our goal is to remove the tangles in the deformed mesh (i.e., after applying the displacement field obtained through the linear elastic equation) while modifying the nodal displacements as little as possible. This task is facilitated by the fact that, in the absence of displacements, the initial mesh is guaranteed to be topologically correct.
Our solution is based on local smoothing. After detecting the topological defects in the deformed configuration of the mesh, a neighborhood of each defect is considered in the mesh topology and we perform mesh smoothing inside this cropped region. See Fig. 3 for an intuitive example of a cropped mesh. To determine the neighborhood of the tangled tetrahedron, the dual mesh is considered. Then, the local neighborhood where local smoothing is applied is determined by breadth-first-search around the problematic node. Typically, we explore a neighborhood of radius 3. The differential operator used for smoothing is again the linear elastic operator. The challenge when operating with only a local submesh and changing node positions is not to create other topological defects. We minimize this potential problem by not allowing nodes with neighbors in both the cropped mesh and the rest of the original mesh to move. With these constraints implemented as external forces, we solve a linear elastic problem inside the cropped mesh.
Fig. 3.
Intuitive representation of a topology defect in 2-D—the blue triangle has a negative Jacobian; in this case, a neighborhood of size 3 around the triangle is selected. The graph on which this neighborhood is selected is formed by considering the graph dual to the mesh (namely, elements are nodes and nodes become the edges). The circled nodes in the figure represent the target points, which are meant to preserve consistency with the rest of the mesh.
We iteratively perform this procedure until all the inverted tetrahedra have been removed. Although there is no formal proof, in our experience with dozens of brains this algorithm has resolved all topological problems to date.
E. Implementation
The algorithm effectively used for the diffusion of the surface registration from the cortical surfaces to the rest of the volume is now reviewed for clarity. Before proceeding, a few precisions are necessary: first, in the attempt to equally divide the displacement being covered at each iteration, we divide the remaining distance to the target in equal sized steps. We can also specify that there will be N + 2 iterations and divide the displacements in N equal parts [see the coefficient in (7)]. Then, as shown in equation (10), through an iterative solution of small linear increments, we can cope with large displacements. However, for efficiency reasons, it seems reasonable to initialize the diffusion in a pose that affinely registers the subject to the target. We do that by minimizing the square distance of a subset of the surface landmarks to be registered. For simplicity, we used a Powell optimizer.
The iterative process described above results in the following.
Using the results of the surface-based spherical registration for all of the surfaces of the target and moving brain scans, recover a sparse displacement field in the Euclidean space .
Regress out the affine component R0 from the sparse displacement field obtained above; this results in the updated sparse displacement field .
- Apply the linear incremental model in N steps; i.e., loop j = 1 ··· or until the mean square error of the prescribed sparse displacements does not decrease anymore.
- Get current morphed positions and create sparse displacement field
(17) - Create a tetrahedral mesh based on the current surface positions and initialize the stiffness matrix and the external forces.
- Solve the linear system.
- Resolve potential topology defects.
In the steps above, sN)j) = (1)/(max(1, N – j + 1)), whose role is to equally split the distance covered in each iteration. If we choose to perform 1 or more iterations past N, this will usually be done in the attempt to get closer to the target points (hence the presence of the max operator).
Also, as mentioned above, prior to the elastic iterative solver, we eliminate the linear component R0, as this greatly reduces the computational burden. We do this by finding the matrix which minimizes the expression described in (15)
Our algorithm has been implemented in C++. We used TetGen [35]–[37] for the tetrahedral mesh generation, and PETSc [39], [40] for the solving of the linear system through the Conjugated Gradient method. To speed up computations, we used MPI to distribute the computations between different processes.
IV. Intensity-Based Registration
After completing the diffusion of the surface displacements, the obtained registration yields a good correspondence of the cortical sheet, while remaining moderately successful for subcortical structures. To further improve the alignment throughout the brain, we complete the proposed pipeline by applying an optical flow intensity-based registration with the initial field being given by the result of the elastic registration.
We emphasize the fact that this particular registration is used as an example, but that the key is the initialization of any reasonable nonlinear volumetric registration using surface alignment. In our experience this type of initialization results in an alignment that is close to the true minimum and well into the correct basin of attraction.
Nonlinear optical flow intensity based registration algorithms have received significant attention in the past two decades in the medical imaging community [41]. We use the algorithm first proposed in [9], which formulates the registration problem as an energy minimization. The energy being minimized is constructed in a manner similar to [3] and contains four terms: one encouraging smooth deformation fields, one maximizing the log likelihood of the similarity at each atlas location, one ensuring invertibility and one minimizing metric distortions
| (18) |
where GI is an intensity term, GT is a topology constraining one, GM ensures the metric preservation properties, and GS is a smoothness preservation term.
Assuming the atlas is an array of V nodes at locations ri and we are trying to bring a subject into register with it, the image likelihood can be written as
| (19) |
where is the warping function we are seeking. The topology term encodes the desire to avoid functions with negative Jacobians
| (20) |
where Di is the Jacobian of the current mapping. The coefficient k modulates the negative force associated with regions that are either noninvertible or almost.
The metric distortion of the morph can be computed in a straightforward manner as the mean squared difference between the distance of the nodes at time t denoted dt and their original distance d0. In a similar manner, the smoothness of the deformation can be quantified as the difference between the vector at location i and the mean of its neighbors. Denoting the closest six neighbors of each node i by N(i), these two terms are then given by
| (21) |
| (22) |
V. Results
We apply the pipeline detailed above to three intrasubject and intersubject registration problems. We start with a challenging intrasubject registration experiment, after which inter-subject registration is evaluated using two independent data sets. The first cross-subject evaluation is based on the work of Fischl et al. [42]. It has the advantage of containing detailed manual segmentation labels for both cortical as well as subcortical regions. The second data set was obtained from the publicly available IBSR database of the Center for Morphometric Analysis at the Massachusetts General Hospital (http://www.cma.mgh.harvard.edu/ibsr/). It contains manually labeled brain acquisitions where the available labels are mostly subcortical. In both sets of intersubject registration experiments, we use the segmentation labels to quantitatively characterize the performance of our algorithm.
In order to demonstrate the performance of our registration framework, we compare it to two standard algorithms from the literature: an affine registration algorithm, implemented in FLIRT [43] and the publicly available version of HAMMER [16]1. In all of our experiments, we first arbitrarily select one of the input brains as a target and then register the rest of the data set to it. To quantify the alignment accuracy of each method, we compute the Jaccard Coefficient metric [45], which is often referred to as Tanimoto Coefficient in the literature and is closely related to the Dice overlap measure. It describes the similarity or overlap between the labels of registered images (V and T) as
| (23) |
Additionally, in order to assess overall accuracy, we also compute an extended version of this overlap measure. If S is a set of labels for which we want to compute the overlap, given two volumes V and T, this measure is defined as
| (24) |
where [V ∈ S] is by definition an abbreviated way of writing {i ∈ I : V(i) ∈ S}. In (24) it is implicitly assumed, by a slight abuse of notation, that V(i) represents the morphed subject labels in the target frame. (All the volumes are sampled in the same frame of reference and I is the set of coordinate voxels). This generalized version of the overlap measure is similar to the generalized pair-wise multilabel Tanimoto Coefficient defined by Crum et al. for fuzzy labels [46].
A. Deformation Fields and Convergence
Before describing the specifics of our three sets of different registration experiments, we show an example deformation field and a graph of the residual distance (in the elastic phase) from one of our experiments. These figures provide a visual intuition of the deformation characteristics of CVS and they can be taken as a good representation of the remainder of our results.
We start by showing an example deformation field obtained by forward morphing a surface in the midcoronal plane section of the target space in Fig. 4. As expected, the surface-driven elastic displacement field smoothly diffuses from the cortical surfaces, and it is reasonably smooth in the ventricular areas. On the contrary, the intensity-driven morph mostly deforms the ventricular areas, with almost no contribution in the cortical areas. For all of our warps we constrained the Jacobian at the mesh node locations to be positive definite, therefore prohibiting self-intersection. As the alignment of folding patterns in the spherical registration step requires a weakly regularized warp, it is somewhat sensitive to noise and thus the 2-D slices displayed in Fig. 4 has regions with apparently degenerate morph components. We emphasize, however, that the warp is constrained to be diffeomorphic and these regions are not discontinuities.
Fig. 4.
Visualization of the resulting warp. From left to right, the first image represents a sample plane mesh to be warped. The plane is chosen in the midcoronal section of the target brain. The pial surface of the brain is indicated in yellow, while the gray/white surface is in red. The second image renders the same mesh, warped using the surface-driven elastic warp—the change of frame is due to the affine alignment performed. The last image displays, in addition to the previous displacement field, the result of the intensity-based registration. The contour lines of the intersection of the target brain cortical surfaces are also morphed along with the surface.
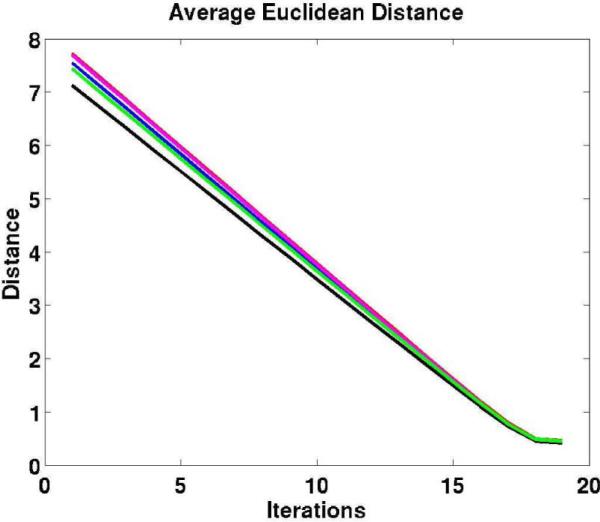
In Fig. 5, we include five plots of average Euclidean distances between the current position of the warped subject and the target positions (based on information provided by the surface registration) computed after each iteration of the linear solver corresponding to five random subjects. These traces empirically show that our numerical method converges. The number of iterations of the linear solver used in these cases is N = 18 (the number of total steps, as presented in Section III-E).
Fig. 5.
The plots display five traces of the average Euclidean distance (the mean average distance) between the current position of the warped subject and the target positions after each iteration of the linear elastic solver for five random subjects.
And finally we display a plot of Euclidean distances computed on the left and right hemispheres between the target and subject surfaces after the spherical registration, elastic morph and the full CVS registration pipeline in Fig. 6. The distances computed for 35 subjects confirm that the elastic warp manages to keep corresponding surfaces at a sufficiently close distance (≈1 mm on average). Even though the intensity-based volu-metric step worsens somewhat that accuracy (< 2 mm on average), as we show in the below discussion, the label overlap accuracy still remains almost unaffected. That observation is important, as in our registration framework we do not fix the solution resulting from the surface-based registration step, so it could potentially be possible for the subsequent operations to decrease the already achieved alignment accuracy in the cortex. As our results show, however, that does not happen.
Fig. 6.
Euclidean distances computed on the left and right hemispheres between the target and subject surfaces after the spherical registration, elastic morph and the full CVS registration pipeline.
B. Intrasubject Registration of Ex Vivo and In Vivo Scans
In the case of our intrasubject registration experiments, we use the first two stages of the CVS framework to align an ex vivo scan with the same hemisphere scanned in vivo. The imaging protocol for the ex vivo tissue is different from that of the in vivo. The former images are acquired using a multiecho flash protocol due to the reduced T1 contrast observed postmortem. This makes the preprocessing step required to obtain the surfaces for the ex vivo images more challenging, however, it does not affect the surfaces being used in the registration process.
We emphasize that registering these two modalities is a challenging problem given the differences between the intensities exhibited by the ex vivo and in vivo scans, together with the large scale deformations induced in the ex vivo hemisphere during the autopsy process, as well as the fact that we are registering a single hemisphere image to one with an entire head. We are currently implementing a cross-contrast intensity procedure that could complement the current intensity-based morph component of our CVS framework. However, as the results show, the elastic volume morph initialized by the surface-based registration already produces highly promising results.
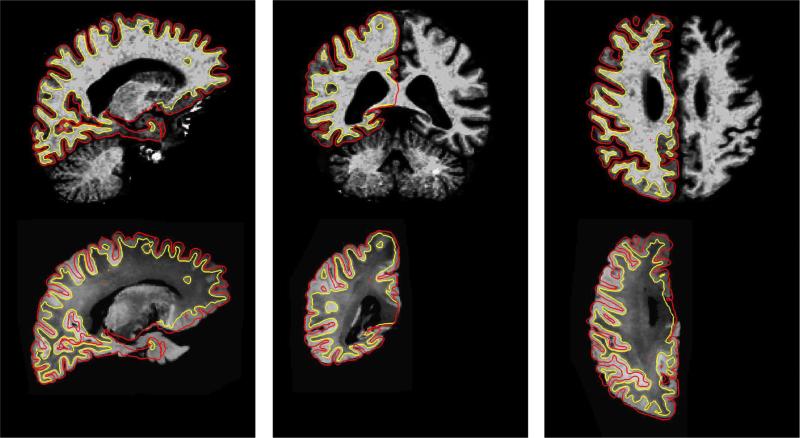
In Fig. 7 we present the results of the surface-driven elastic morph applied to the ex vivo image so that it matches the in vivo one. The resulting correspondence is almost perfect, since the underlying anatomy is the same and the deformation is truly a biomechanical one. However, we remind the reader that the correspondence does contain some inaccuracies near the lateral ventricles. This is because none of the cortical surfaces used to initialize the deformation field are near this region.
Fig. 7.
Results of the surface-driven elastic morph between an ex vivo hemisphere and an in vivo scan of the same subject. The top row shows the in vivo brain, while the bottom row images are snapshots of the registered ex vivo hemisphere. Surfaces are from the in vivo data (pial surface in red and gray/white surface in yellow).
C. Intersubject Registration Experiment 1
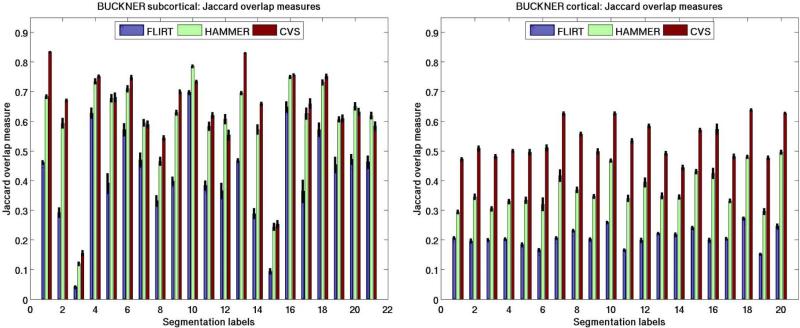
In our first set of inter-subject registration experiments, we used a data set of 36 brains from the work of Fischl et al. [42]. All the input data were individually morphed to a randomly chosen individual considered to be the target. In order to evaluate the quality of the registration, we sampled the set of corresponding cortical segmentation labels into the volume and merged them with the subcortical ones. We used 20 cortical2 and 21 subcortical labels. The numbers shown in Fig. 8 present the mean extended overlap measures (with standard error as vertical black lines) for the labels divided into two sets: cortical and subcortical labels. Here, besides the three registration algorithms to be compared (FLIRT, HAMMER, and CVS), we also include results for the elastic registration without using intensity-based volumetric alignment. In the case of both cortical and subcortical labels, it is the affine method that performs most poorly, followed by HAMMER. CVS clearly outperforms the simply elastic method in the case of subcortical labels and retains the same high level of accuracy for the cortical labels. We emphasize that outperforming HAMMER by such a margin with both our elastic and our combined registration frameworks, especially in the case of the subcortical segmentation labels, has great significance. That is because HAMMER, a volumetric registration tool, should intuitively align subcortical structure more accurately than a method relying on a surface-based component. We note that although the relationship between the folds and subcortical structures is poorly understood, given the massive connectivity with cortex, their position is unlikely to be independent of cortex [47]. Therefore, we believe that the surface-based registration establishes a robust initialization for the elastic (and intensity-based registration) steps and thus helps avoiding local optima. We leave the study of the relationship between the folds and subcortical structures to future work.
Fig. 8.
Extended Jaccard coefficient (24) measures computed for Experiment 1 using 20 cortical and 21 subcortical labels listed in Tables I and II, respectively. The vertical lines represent the standard error of the mean of the measurement.
The poor accuracy of the affine alignment of FLIRT (which, we believe, would be true of any 12 parameter transform, in general) is indicative of the difficulty encountered by nonlinear volumetric procedures initialized with affine transforms. Essentially, the affine initialization is poor over much of the cortex so the nonlinear energy functionals are not in the basin of attraction containing the true minima, and hence do not converge to the correct solution. Initializing HAMMER in a more robust way, we believe, could remove most of its performance differences with our methods.
In Table I, we include the mean and standard error results (indicated in parenthesis) of the Jaccard Coefficient overlap metric computed for all the selected labels individually over all subjects. The labels are superiorfrontal, inferiorparietal, rostralmiddlefrontal, precentral, middletemporal, lateraloccipital, superiorparietal, superiortemporal, postcentral, inferiortemporal, fusiform, and precuneus. The three columns of the table correspond to alignment achieved with FLIRT (Affine), HAMMER, and CVS. Statistically significant (p > 0.05) differences between HAMMER and CVS are indicated by bold typesetting. Similarly, the mean and standard error overlap measures for the 21 subcortical labels are included in Table II. We point out that in the case of cortical labels, all the selected labels perform statistically significantly better than HAMMER and in the case of subcortical labels, out of 12 cases where statistically significant differences are established between HAMMER and CVS, the latter is superior on nine occasions. Identical information, the Jaccard overlap metric computed for individual subcortical and cortical manual segmentation labels is displayed in the bar plots of Fig. 9.
TABLE I.
Mean Jaccard Coefficients and Standard Error Computed for 20 Cortical Labels Across Subjects in Intersubject Registration Experiment 1. We Include the Results of FLIRT (Affine), HAMMER, and CVS. Manual Segmentation Labels for the Left (L) and Right (R) Hemispheres Are: Superiorfrontal (SupFr), Inferiorparietal (InfPar), Rostralmiddlefrontal (RostMiFr), Precentral (PreCnt), Middletemporal (MiTemp), Lateraloccipital (LatOcc), Superiorparietal (SupPar), Superiortemporal (SupTemp), Postcentral (PostCnt), Inferiortemporal (InfTemp), Fusiform, and Precuneus. Statistically Significant Differences Between HAMMER and CVS Are Indicated by Bold Typesetting
| Label |
Affine (SE) |
HAMMER (SE) |
CVS (SE) |
|---|---|---|---|
| L-Fusifm | 0.2073(0.01) | 0.2941(0.01) | 0.4720(0.01) |
| L-InfPar | 0.1964(0.01) | 0.3457(0.01) | 0.5084(0.01) |
| L-InfTemp | 0.1999(0.01) | 0.3052(0.01) | 0.4819(0.01) |
| L-LatOcc | 0.2032(0.00) | 0.3295(0.01) | 0.5002(0.01) |
| L-MiTemp | 0.1849(0.01) | 0.3336(0.01) | 0.4962(0.01) |
| L-PostCnt | 0.1665(0.01) | 0.3201(0.02) | 0.5111(0.01) |
| L-PreCnt | 0.2073(0.01) | 0.4165(0.02) | 0.6264(0.01) |
| L-Precun | 0.2318(0.00) | 0.3686(0.01) | 0.5572(0.01) |
| L-RostMiFr | 0.2029(0.01) | 0.3465(0.01) | 0.4989(0.01) |
| L-SupFr | 0.2599(0.01) | 0.4673(0.01) | 0.6263(0.01) |
| L-SupPar | 0.1671(0.00) | 0.3401(0.01) | 0.5340(0.01) |
| L-SupTemp | 0.1986(0.01) | 0.3941(0.02) | 0.5840(0.01) |
| R-InfPar | 0.2221(0.00) | 0.3483(0.01) | 0.4919(0.01) |
| R-RatOcc | 0.2181(0.01) | 0.3444(0.01) | 0.4435(0.01) |
| R-MiTemp | 0.2411(0.01) | 0.4303(0.01) | 0.5701(0.01) |
| R-PreCnt | 0.1997(0.01) | 0.4246(0.02) | 0.5737(0.02) |
| R-RostMiFr | 0.2045(0.01) | 0.3318(0.01) | 0.4813(0.01) |
| R-SupFr | 0.2735(0.01) | 0.4807(0.01) | 0.6382(0.01) |
| R-SupPar | 0.1525(0.00) | 0.2957(0.01) | 0.4762(0.01) |
| R-SupTemp | 0.2473(0.01) | 0.4959(0.01) | 0.6269(0.01) |
TABLE II.
Mean Jaccard Coefficient and Standard Error Computed for 21 Subcortical Labels Across Subjects in Intersubject Registration Experiment 1. We Include the Results of FLIRT (Affine), HAMMER, and CVS. Manual Segmentation Labels Are: Brain Stem (BrStem), Left and Right (L/R) Cerebral White Matter (CrWM), Cerebral Cortex (CrCtx), Lateral Ventricle (LtVent), Thalamus Proper (Thal), Caudate (Caud), Putamen (Put), Pallidum (Pall), Hippocampus (Hipp), Amygdala (Amyg). Statistically Significant Differences Between HAMMER and CVS Are Indicated by Bold Typesetting
| Label |
Affine (SE) |
HAMMER (SE) |
CVS (SE) |
|---|---|---|---|
| L-CrWM | 0.4622 (0.01) | 0.6830(0.01) | 0.8335 (0.00) |
| L-LtVent | 0.2927 (0.02) | 0.5936(0.02) | 0.6705 (0.01) |
| L-InfLtVent | 0.0417 (0.01) | 0.1194(0.01) | 0.1553 (0.01) |
| L-Thal | 0.6282 (0.02) | 0.7348(0.01) | 0.7514 (0.01) |
| L-Caud | 0.3916 (0.03) | 0.6777(0.01) | 0.6806 (0.02) |
| L-Put | 0.5716 (0.02) | 0.7095(0.01) | 0.7466 (0.01) |
| L-Pall | 0.4703 (0.02) | 0.5947(0.01) | 0.5894 (0.01) |
| ThVent | 0.3320 (0.02) | 0.4650(0.02) | 0.5441 (0.01) |
| FoVent | 0.3987 (0.02) | 0.6292(0.01) | 0.6993 (0.01) |
| BrStem | 0.6979 (0.01) | 0.7858(0.01) | 0.7338 (0.01) |
| L-Hipp | 0.3841 (0.02) | 0.5833(0.02) | 0.6201 (0.01) |
| L-Amyg | 0.3662 (0.03) | 0.6072(0.02) | 0.5537 (0.02) |
| R-CrWM | 0.4678 (0.01) | 0.6954(0.01) | 0.8301 (0.00) |
| R-LtVent | 0.2888 (0.02) | 0.5728(0.02) | 0.6589 (0.01) |
| R-InfLtVent | 0.0943 (0.01) | 0.2439(0.01) | 0.2532 (0.01) |
| R-Thal | 0.6492 (0.02) | 0.7496(0.01) | 0.7563 (0.01) |
| R-Caud | 0.3653 (0.04) | 0.6263(0.02) | 0.6595 (0.02) |
| R-Put | 0.5720 (0.02) | 0.7311(0.01) | 0.7515 (0.01) |
| R-Pall | 0.4541 (0.03) | 0.6062(0.01) | 0.6097 (0.01) |
| R-Hipp | 0.4724 (0.02) | 0.6505(0.01) | 0.6316 (0.01) |
| R-Amyg | 0.4627 (0.02) | 0.6194(0.01) | 0.5843 (0.02) |
Fig. 9.
Experiment 1: Mean Jaccard Coefficient measures computed for individual labels (left: subcortical; right: cortical). The vertical lines represent the standard error for the measurement.
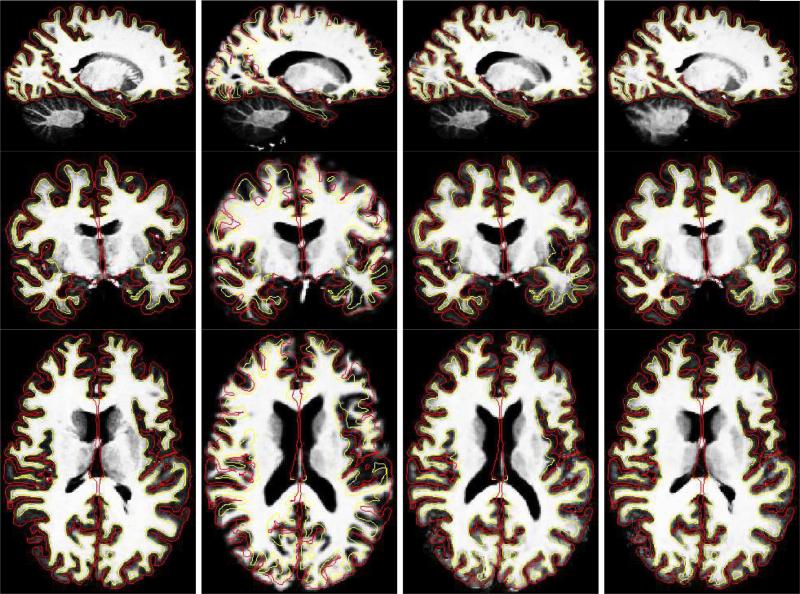
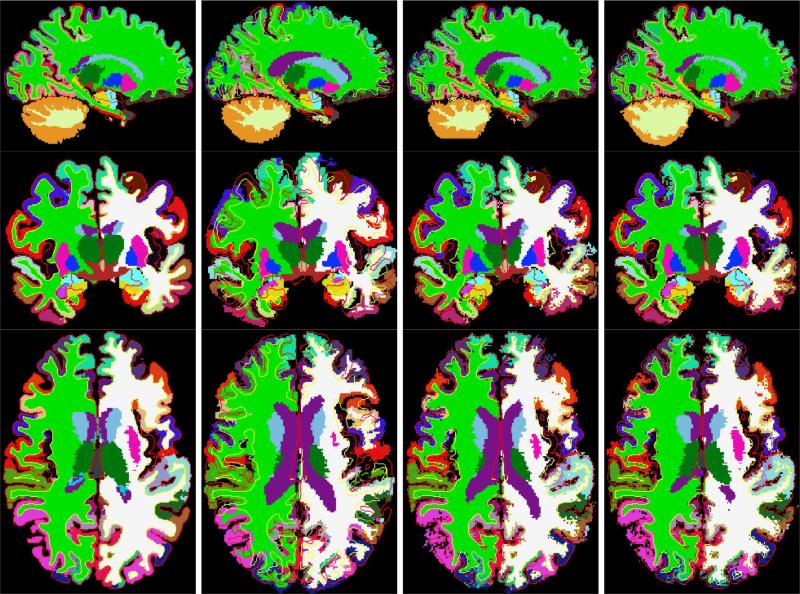
We show registration results for these intersubject registration experiments in two figures. Fig. 10 displays deformed intensity images of a subject and the target and Fig. 11 demonstrates the corresponding deformed manual segmentation volumes (same slice as before). The first column, on both of the figures, displays the target; the second represents the subject after affine registration (using FLIRT); the third displays the subject after the surface-driven elastic registration; and the fourth column shows the results of our combined surface-based and intensity-based registration. In all the images, the surfaces shown are those of the target (pial surfaces in red and gray/white surfaces in yellow). The intensity-based morph mostly affects areas away from the cortical ribbon, such as the ventricles.
Fig. 10.
Intersubject registration results of Experiment 1 applied to intensity images. The first column displays the target, the second the subject after affine registration (using FLIRT), the third the subject after the surface-driven elastic registration and the fourth the results of the complete CVS registration pipeline. In all the images, the surfaces shown are those of the target (pial surfaces in red and gray/white surfaces in yellow). The intensity-based morph mostly affects areas away from the cortical ribbon, such as the ventricles.
Fig. 11.
Intersubject registration results of Experiment 1 applied to manual segmentations. The first column displays the target, the second the subject after affine registration (using FLIRT), the third the subject after the surface-driven elastic registration, and the fourth the results of the complete CVS registration pipeline. In all the images, the surfaces shown are those of the target (pial surfaces in red and gray/white surfaces in yellow). The intensity-based morph mostly affects areas away from the cortical ribbon, such as the ventricles.
D. Intersubject Registration Experiment 2
In our second set of intersubject registration experiments, we used MRI acquisitions from eleven subjects of the IBSR data set. This data set was collected by the Center for Morphometric Analysis, MGH and is publicly available (http://www.cma.mgh.harvard.edu/ibsr/). It includes manual segmentation results for 19 principal gray and white matter structures: brain stem, left and right cerebral white matter, cerebral cortex, lateral ventricle, thalamus proper, caudate, putamen, pallidum, hippocampus, and amygdala. We note that this data set is not ideal to convey the improved performance of our algorithm both in the cortical and subcortical domains, given that most of the available labels identify subcortical areas. Nonetheless, as a recently published registration method similar to our work [19] relied on these acquisitions for validation, we decided to include it as well for easier comparison.
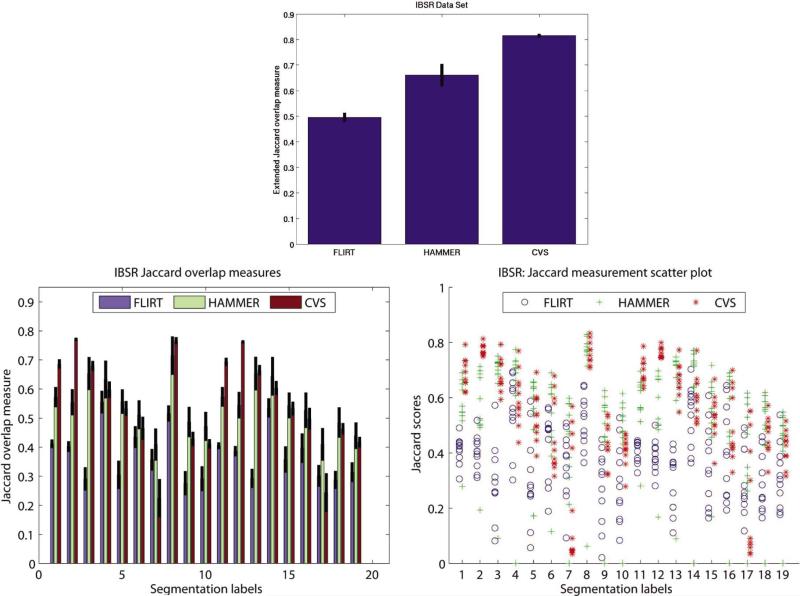
The overlap measures in Table III are computed for each of the available segmentation labels. Again, we evaluated the mean Jaccard Coefficient for the three different alignment algorithms: affine alignment with FLIRT (Affine), HAMMER, and CVS. The standard error (SE) of the measurements is indicated in parenthesis. The same information in bar plot format is presented in Fig. 12. The plot on the top displays the extended Jaccard overlap metric, while the plots on the bottom display the overlap measures computed for each label individually in a bar- and a scatter-plot format, respectively. The latter aids the reader to more easily asses significance of individual measures.
TABLE III.
Mean Jaccard Coefficient and Standard Error Computed Across Subjects for Experiment 2. We Include the Results of FLIRT (Affine), HAMMER, and CVS. Manual Segmentation Labels Are: Brain Stem (BrStem), Left and Right (L/R) Cerebral White Matter (CrWM), Cerebral Cortex (CrCtx), Lateral Ventricle (LtVent), Thalamus Proper (Thal), Caudate (Caud), Putamen (Put), Pallidum (Pall), Hippocampus (Hipp), Amygdala (Amyg). Statistically Significant Differences Between HAMMER and CVS Are Indicated by Bold Typesetting
| Label |
Affine (SE) |
HAMMER (SE) |
CVS (SE) |
|---|---|---|---|
| L-CrWM | 0.4115 (0.02) | 0.5722 (0.04) | 0.6860 (0.02) |
| L-CrCtx | 0.4024 (0.02) | 0.5544 (0.05) | 0.7706 (0.01) |
| L-LtVent | 0.2911 (0.04) | 0.6533 (0.06) | 0.6792 (0.02) |
| L-Thal | 0.5554 (0.04) | 0.6328 (0.07) | 0.5976 (0.03) |
| L-Caud | 0.3049 (0.05) | 0.5566 (0.04) | 0.5331 (0.03) |
| L-Put | 0.4346 (0.04) | 0.5114 (0.05) | 0.4648 (0.04) |
| L-Pall | 0.3566 (0.04) | 0.4091 (0.06) | 0.2245 (0.07) |
| BrStem | 0.5158 (0.03) | 0.7150 (0.07) | 0.7658 (0.01) |
| L-Hipp | 0.2760 (0.04) | 0.4866 (0.05) | 0.4282 (0.02) |
| L-Amyg | 0.2918 (0.04) | 0.4716 (0.05) | 0.4113 (0.02) |
| R-CrWM | 0.4050 (0.01) | 0.5731 (0.03) | 0.6937 (0.01) |
| R-CrCtx | 0.3867 (0.02) | 0.5444 (0.05) | 0.7620 (0.01) |
| R-LtVent | 0.2926 (0.03) | 0.6530 (0.06) | 0.6648 (0.02) |
| R-Thal | 0.5349 (0.03) | 0.6438 (0.07) | 0.6036 (0.02) |
| R-Caud | 0.3576 (0.05) | 0.5436 (0.04) | 0.5327 (0.02) |
| R-Put | 0.3966 (0.05) | 0.5269 (0.06) | 0.4976 (0.04) |
| R-Pall | 0.3016 (0.04) | 0.4095 (0.06) | 0.2437 (0.07) |
| R-Hipp | 0.2875 (0.03) | 0.4847 (0.05) | 0.4630 (0.02) |
| R-Amyg | 0.3142 (0.03) | 0.4395 (0.05) | 0.4171 (0.02) |
Fig. 12.
Experiment 2: Top: Mean extended Jaccard coefficient measures. Bottom: Mean Jaccard coefficient measures computed for individual labels. The vertical lines represent the standard error for the measurement.
As in the previous experiments, for most of the labels, our method outperforms the affine and HAMMER algorithm and also has a reduced standard error, indicating the stability of the registration. In six cases, the difference between the performance of HAMMER and CVS are also statistically significant (as indicated by the bold typesetting). In the case of four of those CVS outperforms HAMMER. Again, as the majority of the IBSR labels are subcortical, we cannot demonstrate on this data set the accuracy of the cortical alignment.
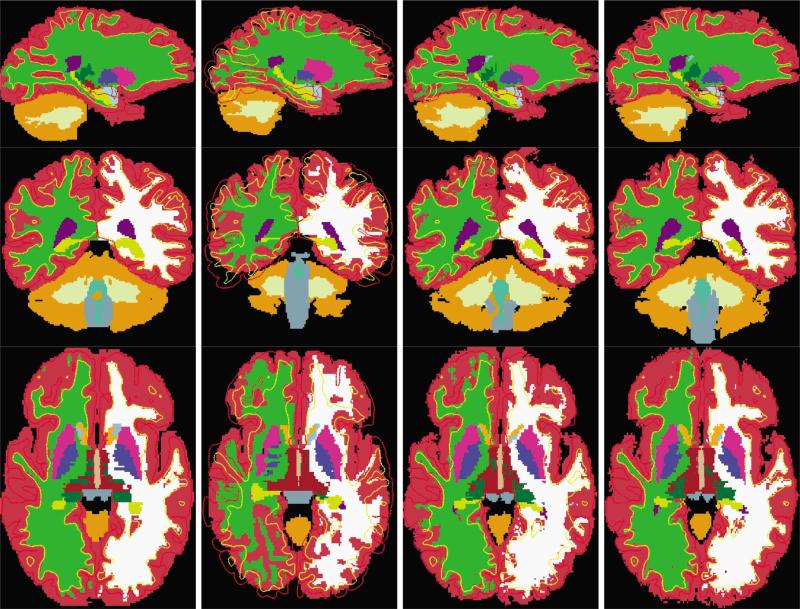
We show registration results for these intersubject registration experiments as well. Fig. 13 displays the segmentation volume of the target and the deformed segmentation volumes of a subject using the three different registration algorithms. The first column displays the target segmentation and the second, third, and fourth columns represent the deformed segmentation volumes resulting from FLIRT (affine), HAMMER, and our new CVS registration. In all the images, the surfaces shown are those of the target (pial surfaces in red and gray/white surfaces in yellow).
Fig. 13.
Examples of results obtained in intersubject Experiment 2. The first column displays the target segmentation and the second, third, and fourth columns represent the deformed segmentation volumes resulting from FLIRT (affine), HAMMER, and our new combined surface-based and intensity-based registration. In all the images, the surfaces shown are those of the target (pial surfaces in red and gray/white surfaces in yellow).
VI. Conclusion
In this paper, we have presented a novel method for the registration of volumetric images of the brain that optimizes the alignment of both cortical and subcortical structures. Given two brain scans, the method consists of two steps. First, a surface-based registration algorithm computes correspondences between the input surfaces. These correspondences are then translated into a sparse displacement field in Euclidean space, and diffused into the volume using the Navier operator of elasticity. In a second step, an optical flow registration algorithm refines the alignment obtained after the first step-resulting in the alignment of noncortical structures, such as the ventricles, thalamus, basal ganglia, etc., which are not near the surfaces used in the first step.
To assess the accuracy of our method, we performed both intrasubject and intersubject registration experiments. First, we used the surface correspondence to solve the challenging problem of accurately aligning in vivo and ex vivo acquisitions of the same brain. The challenge here was to recover the large deformation during the extraction the brain from the skull and the subsequent fixation. Our results demonstrate that the surface-based component of our newly introduced registration framework is independent of the intensities of the underlying images, as it only relies on the geometrical features extracted through the surface registration.
Second, we used two different data sets with manually segmented labels to quantify the accuracy of intersubject registration. We provided quantitative performance measures of our method against two other well-known registration methods, FLIRT and HAMMER. In general, we conclude that our registration framework outperformed these methods both in accuracy and robustness.
The importance of the type of automated procedure presented in this paper is that it allows the establishment of a coordinate system across the entire brain that is accurate for both cortical and noncortical regions. This result should facilitate the analysis of structural and functional imaging data in a unified fashion, and make the techniques more easily used by the broader neuroimaging community.
We are in the process of making this software freely available as part of the FreeSurfer software suite (http://surfer.nmr. mgh.harvard.edu/). That release, we believe, will prove the robustness and reproducibility of our registration approach with respect to parameter settings and types of image data.
Acknowledgment
The authors would like to thank Prof. R. Buckner for the 40 volume data set [48].
This work was supported in part by the National Center for Research Resources (NCRR) (P41-RR14075, R01 RR16594-01A1 and the NCRR Biomedical Informatics Research Network Morphometric Project BIRN002, U24 RR021382), in part by the National Institute for Biomedical Imaging and Bioengineering (R01 EB001550, R01EB006758), in part by the National Institute for Neurological Disorders and Stroke (R01 NS052585-01) as well as the Mental Illness and Neuroscience Discovery (MIND) Institute, and is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research under Grant U54 EB005149.
Footnotes
We note that the version of HAMMER that was available to us uses the gray/white matter segmentation from FAST [44] exclusively to produce the attribute vectors. As such, it is possible that results improve with different inputs to the attribute vector.
The selected 20 cortical labels correspond to the labels with the largest surface areas and their results are representative of the remaining of the 46 labels.
Contributor Information
Gheorghe Postelnicu, Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Boston, MA 02176 USA (postelni@nmr.mgh.harvard.edu)..
Lilla Zöllei, Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Boston, MA 02176 USA lzollei@nmr.mgh.harvard.edu..
Bruce Fischl, Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Boston, MA 02176 USA fischl@nmr.mgh.harvard.edu..
References
- 1.Maintz J, Viergever M. A survey of medical image registration. Med. Image Anal. 1998;2(1):1–36. doi: 10.1016/s1361-8415(01)80026-8. [DOI] [PubMed] [Google Scholar]
- 2.Thompson PM, Hayashi KM, de Zubicaray G, Janke AL, Rose DE, Semple J, Doddrell DM, Cannon TD, Toga AW. Proc. Int. Symp. Biomed. Imag. 2002. Detecting dynamic and genetic effects on brain structure using high-dimensional cortical pattern matching; pp. 473–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischl B, Sereno MI, Tootell R, Dale AM. High-resolution inter-subject averaging and a coordinate system for the cortical surface. Human Brain Mapp. 1999;8:272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davatzikos C, Bryan N. Using a deformable surface model to obtain a shape representation of the cortex. IEEE Trans. Med. Imag. 1996 Dec.15(6):785–795. doi: 10.1109/42.544496. [DOI] [PubMed] [Google Scholar]
- 5.Van Essen D, Drury H. Structural and functional analyses of human cerebral cortex using a surface-based atlas. J. Neurosci. 1997;17(18):7079–7102. doi: 10.1523/JNEUROSCI.17-18-07079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bajcsy R, Kovacic S. Multi-resolution elastic matching. Comput. Vis., Graphics, Image Process. 1989;46:1–21. [Google Scholar]
- 7.Davatzikos C. Spatial transformation and registration of brain images using elastically deformable models. Comput. Vis. Image Understand. 1997;66(2):207–222. doi: 10.1006/cviu.1997.0605. [DOI] [PubMed] [Google Scholar]
- 8.Christensen G, Joshi S, Miller M. Volumetric transformation of brain anatomy. IEEE Trans. Med. Imag. 1997 Dec.16(6):864–877. doi: 10.1109/42.650882. [DOI] [PubMed] [Google Scholar]
- 9.Fischl B, Salat DH, van der Kouwe AJ, Segonne F, Dale AM. Sequence-independent segmentation of magnetic resonance images. NeuroImage. 2004;23:S69–S84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 10.Beg MF, Miller MI, Trouvé A, Younes L. Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int. J. Comput. Vis. 2005 Feb.61(2):139–157. [Google Scholar]
- 11.Peckar W, Schnorr C, Rohr K, Stiehl HS. Parameter-free elastic deformation approach for 2-D and 3-D registration using prescribed displacements. J. Math. Imag. Vis. 1999;10(2):143–162. [Google Scholar]
- 12.Ferrant M, Nabavi A, Macq BM, Jolesz FA, Kikinis R, Warfield SK. Registration of 3-D intraoperative MR images of the brain using a finite element biomechanical model. IEEE Trans. Med. Imag. 2001 Dec.20(12):1384–1397. doi: 10.1109/42.974933. [DOI] [PubMed] [Google Scholar]
- 13.Thompson PM, Toga AW. A surface-based technique for warping three-dimensional images of the brain. IEEE Trans. Med. Imag. 1996 Aug.15(4):402–417. doi: 10.1109/42.511745. [DOI] [PubMed] [Google Scholar]
- 14.Broit C. Ph.D. dissertation. Univ. Pennsylvania; Philadelphia: 1981. Optimal registration of deformed images. [Google Scholar]
- 15.Collins D, LeGoualher G, Caramanos Z, Evans A, Barillot C. Proc. Int. Conf. Visualizat. Biomed. Comput. 1996. Cortical constraints for non-linear cortical registration; pp. 307–316. [Google Scholar]
- 16.Shen D, Davatzikos C. Hammer: Hierarchical attribute matching mechanism for elastic registration. IEEE Trans. Med. Imag. 2002 Nov.21(11):1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- 17.Liu T, Shen D, Davatzikos C. Deformable registration of cortical surfaces via hybrid volumetric and surface warping. NeuroImage. 2004;(22):1790–1801. doi: 10.1016/j.neuroimage.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 18.Postelnicu G, Zöllei L, Desikan R, Fischl B. Geometry driven volumetric registration. Inf. Process. Med. Imag. 2007;20:675–686. doi: 10.1007/978-3-540-73273-0_56. [DOI] [PubMed] [Google Scholar]
- 19.Joshi AA, Shattuck D, Thompson PM, Leahy RM. Brain image registration using cortically constrained harmonic mappings. IPMI. 2007:359–371. doi: 10.1007/978-3-540-73273-0_30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joshi AA, Shattuck D, Thompson PM, Leary RM. Surface-constrained volumetric brain registration using harmonic mappings. IEEE Trans. Med. Imag. 2007 Dec.26(12):1657–1669. doi: 10.1109/tmi.2007.901432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis II: Inflation, flattening, and a surface-based coordinate system. NeuroImage. 1999;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 22.Fischl B, Salat D, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale A. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 23.Segonne F, Dale AM, Busa E, Glessner M, Salvolini U, Hahn H, Fischl B. A hybrid approach to the skull-stripping problem in MRI. NeuroImage. 2004;(22):1160–1175. doi: 10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 24.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis i: Segmentation and surface reconstruction. NeuroImage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 25.Fischl B, Liu A, Dale AM. Automated manifold surgery: Constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Trans. Med. Imag. 2001 Jan.20(1):70–80. doi: 10.1109/42.906426. [DOI] [PubMed] [Google Scholar]
- 26.Segonne F, Grimson E, Fischl B. Genetic algorithm for the topology correction of cortical surfaces. IPMI. 2005:393–305. doi: 10.1007/11505730_33. [DOI] [PubMed] [Google Scholar]
- 27.Sled J, Zijdenbos A, Evans A. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. Imag. 1998 Feb.17(1):87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 28.Segonne F, Pacheco J, Fischl B. Geometrically accurate topology-correction of cortical surfaces using nonseparating loops. IEEE Trans. Med. Imag. 2007 Apr.26(4):518–529. doi: 10.1109/TMI.2006.887364. [DOI] [PubMed] [Google Scholar]
- 29.Fischl B, Dale AM, Sereno MI, Tootell R, Rosen B. A coordinate system for the cortical surface. NeuroImage. 1998;(7) [Google Scholar]
- 30.Bookstein F. Principal warps: Thin-plate splines and the decomposition of deformations. IEEE Trans. Pattern Anal. Mach. Intell. 1989 Nov.11(6):567–585. [Google Scholar]
- 31.Rueckert D, Sonoda L, Hayes C, Hill D, Leach M, Hawkes D. Non-rigid registration using free-form deformations: Applications to breast MR images. IEEE Trans. Med. Imag. 1999 Aug.18(8):712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 32.Ciarlet P. Mathematical Elasticity. Volume I: Three-Dimensional Elasticity. North-Holland; Amsterdam: 1988. [Google Scholar]
- 33.Zienkewickz O, Taylor R. The Finite Element Method. McGraw Hill; New York: 1987. [Google Scholar]
- 34.Edelsbrunner H, Shah NR. Incremental topological flipping works for regular triangulations. Algorithmica. 1996;15(3):223–241. [Google Scholar]
- 35.Edelsbrunner H, Shah N. Incremental topological flipping works for regular triangulations. Algorithmica. 1996;15:223–241. [Google Scholar]
- 36.Si H, Gaertner K. Proc. 14th Int. Meshing Roundtable. 2005. Meshing piecewise linear complexes by constrained Delaunay tetrahedralizations; pp. 147–163. [Google Scholar]
- 37.Si H. Proc. 15th Int. Meshing Roundtable. 2006. On refinement of constrained Delaunay tetrahedralizations; pp. 509–528. [Google Scholar]
- 38.Knupp P. Hexahedral and tetrahedral mesh untangling. Engineering with Computers. 2001;17(3):261–268. [Google Scholar]
- 39.Balay S, Buschelman K, Eijkhout V, Gropp WD, Kaushik D, Knepley MG, McInnes LC, Smith BF, Zhang H. PETSc Users Manual Argonne National Laboratory; Argonne, IL: 2004. Tech. Rep. ANL-95/11-Revision 2.1.5
- 40.Balay S, Gropp WD, McInnes LC, Smith BF, Arge E, Bruaset AM, Langtangen HP, editors. Modern Software Tools in Scientific Computing. Birkhäuser Press; New York: 1997. Efficient management of parallelism in object oriented numerical software libraries; pp. 163–202. [Google Scholar]
- 41.Hermosillo G, Chef'dHotel C, Faugeras O. Variational methods for multimodal image matching. IJCV. 2002;50(3) [Google Scholar]
- 42.Desikan R, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 43.Jenkinson M, Bannister P, Brady J, Smith S. Improved optimisation for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17(2):825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation maximization algorithm. IEEE Trans. Med. Imag. 2001 Jan.20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 45.Jaccard P. The distribution of flora in the alpine zone. New Phytol. 1912;11(2) [Google Scholar]
- 46.Crum W, Camara O, Hill DLG. Generalized overlap measures for evaluation and validation in medical image analysis. IEEE Trans. Med. Imag. 2006 Nov.25(11):1451–1461. doi: 10.1109/TMI.2006.880587. [DOI] [PubMed] [Google Scholar]
- 47.Van Essen D. A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature. 1997;385:313–318. doi: 10.1038/385313a0. [DOI] [PubMed] [Google Scholar]
- 48.Marcus D, Wang T, Parker J, Csernansky J, Morris J, Buckner R. Open access series of imaging studies (oasis): Cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J. Cognitive Neurosci. 2007;19:1498–1507. doi: 10.1162/jocn.2007.19.9.1498. [DOI] [PubMed] [Google Scholar]