Abstract
A number of phosphatidylcholine (PC) cations spanning a mass range of 400 to 1000 Da are investigated using electrospray ionization mass spectrometry coupled with traveling wave ion mobility spectrometry (TWIMS). A high correlation between mass and mobility is demonstrated with saturated phosphatidylcholine cations in N2. A significant deviation from this mass-mobility correlation line is observed for the unsaturated PC cation. We found that the double bond in the acyl chain causes a 5% reduction in drift time. The drift time is reduced at a rate of ~1% for each additional double bond. Theoretical collision cross sections of PC cations exhibit good agreement with experimentally evaluated values. Collision cross sections are determined using the recently derived relationship between mobility and drift time in TWIMS stacked ring ion guide (SRIG) and compared to estimate collision cross-sections using empiric calibration method. Computational analysis was performed using the modified trajectory (TJ) method with nonspherical N2 molecules as the drift gas. The difference between estimated collision cross-sections and theoretical collision cross-sections of PC cations is related to the sensitivity of the PC cation collision cross-sections to the details of the ion-neutral interactions. The origin of the observed correlation and deviation between mass and mobility of PC cations is discussed in terms of the structural rigidity of these molecules using molecular dynamic simulations.
INTRODUCTION
Lipids are essential biological components and have critical roles for cell structure, energy storage, and metabolic control.1 Characterizing their structures is an essential part of lipid analysis. In addition, searching for lipid molecules is a valuable strategy for finding traces of extinct or extant life elsewhere in outer space. Lipids and biomembranes can be preserved for a long period; thus, detailed characterization of these biomarker compositions allows for the assessment of major contributing species.2 Lipids offer records of modern and ancient life, environmental conditions, and changes in history. However, the variety and in situ alteration of lipids also increases complexity, making them difficult to characterize fully.3
The separation and characterization of phospholipids using tandem ion mobility mass spectrometry (IM-MS) has been investigated by several research groups.4–7 Utilizing matrix-assisted laser desorption ionization (MALDI) with IM-MS, phospholipid ions have been separated from other biomolecule ions.4,8 Separation can be achieved based on their distinct correlation between mass and ion mobility. Phospholipids in tissue samples have been directly analyzed using MALDI-IM-MS.4,5 These studies have reported that phospholipid ions have slower mobility than peptide, carbohydrate, and nucleotide ions with similar masses.4,7–9 In general, peptides, nucleotides, and carbohydrates form globular conformations in the gas phase due to intramolecular Coulombic interactions.10–12 However, such interactions are difficult to achieve for phospholipid molecules because their major components are aliphatic acyl chains. Recently, Jackson et al. reported the effects of various head and tail groups of phospholipids on mass-mobility correlations using MALDI-IM-MS.6 They report a slight increase in the mobility of phospholipids as the degree of unsaturation on the acyl chain increases.
The correlation between the mass and mobility of molecular ions has been used to separate and characterize ions related to the mobility of gas phase ion molecules. In the early 1970s, Griffin et al.13 showed that mass and mobility are strongly correlated for structurally related compounds. In the late 1980s, Karpas and Berant demonstrated distinct mass-mobility correlations of acetyls, aromatic amines, and aliphatic amines drifting in various drift gases including He, N2, CO2, and air.14,15 Clemmer and co-workers have demonstrated distinct mass-mobility correlations for peptides with molecular weights of 500 to 2500 Da.16 Separating and characterizing phosphorylated peptides from their non-phosphorylated counterparts based on different mass-mobility correlations have been also demonstrated using IMS.17–19
Our laboratory has investigated the distinct mass-mobility correlations of amino acids and carboxylic acids drifting in N2 and CO2.20,21 Recently, we experimentally observed a high correlation between mass and mobility of tertiary and quaternary ammonium cations in N2.22 This observed correlation was investigated using classical ion-neutral collision dynamic theories and computational calculation using a modified trajectory method (TJ method). From these theoretical investigations, the ammonium cations in the mass range from 60 Da to 150 Da are on the borderline between being dominated by long range versus short range interactions with N2. In addition, all potential terms, ion quadrupole, ion induced dipole, and Van der Waals potential are important considerations for determining the collision cross-sections of the ions in N2.
In this paper, we measure drift times for a number of phosphatidylcholines (PC) spanning a mass range of 400 to 1000 Da in N2 using a commercial traveling wave ion mobility spectrometry (TWIMS) coupled with orthogonal acceleration time-of-flight (oa-TOF) mass spectrometry (Waters Synapt HDMS). Of particular interest is the possible dependence of mass-mobility correlations on the symmetry, length, and degree of saturation of the acyl chains. Despite a wide range of TWIMS applications in various chemistry fields,17,23–27 studies have only begun to understand the principal physics behind the TWIMS drift time and ion mobility. A number of studies have employed the empiric calibration method to estimate mobilities and collision cross-sections of analyte ions from the drift times in TWIMS.17,23,24,27,28 Recently, Shvartsburg and Smith quantitatively revealed the relationship between drift time and ion mobility in TWIMS.29
Jarrold and co-workers have proposed a TJ method based on a soft-core ion-neutral interaction potential to interpret collision cross-sections of ion molecules.30 A modified TJ method for the ion-neutral interaction to account for the potential associated with the non-spherical drift gas N2 has been applied to predict collision cross-sections of PC cations and to test the sensitivity of these cross-sections in order to detail the structural rigidity of these molecules.22 Results from the estimated collision-cross sections using empiric calibration are compared with the evaluated relationship between TWIMS drift time and mobility by Shvartsburg and Smith.29 The origin of the observed correlation and deviation between PC mass and mobility is discussed.
EXPERIMENTAL SECTION
Chemicals and Reagents
All phosphatidylcholines studied in this work were purchased from Avanti Polar Lipids (Alabaster, AL) and were used without further purification. All solvents (water, methanol, and formic acid) were HPLC grade and were purchased from EMD Chemicals Inc. (Gibbstown, NJ). Calibrant peptides (GGGGGG and AAAAAA), cytochrome C, and trypsin from porcine pancreas were purchased from Sigma-Aldrich (St. Louis, MO). Samples were prepared by dissolving known quantities of molecules in a solvent consisting of 1:1 water and methanol with 0.1% formic acid by volume to yield sample concentrations in the range of 50 μM. Trypsin digest of cytochrome C was prepared by incubating 200 μM of cytochrome C with 6 μg of trypsin from porcine pancreas in 1 mL of water containing 25 mM ammonium bicarbonate (NH4HCO3) at 37 °C for 4 hours. The trypsin was then removed using a Millipore Microcon centrifugal filter fitted with an Ultracel YM-10 membrane. The sample solution was diluted to an appropriate concentration for ESI with 1:1 water/methanol and 0.1% formic acid by volume. Phospholipid and peptide ions examined in this study are listed in Table S1 along with their respective molecular weights.
Phosphatidylcholines examined in this study are named by their acyl chain length and number of double bonds. For example, 1-steroyl-2-oleoyl-sn-phosphatidylcholine (SOPC), which comprises two 18 carbon acyl chains and one double bond, is referred to as 18:0-18:1 PC.
Electrospray Ionization Traveling Wave Ion Mobility Mass Spectrometer
Experiments were performed on a Synapt HDMS traveling wave ion mobility orthogonal acceleration time-of-flight (TW-IM-oa-TOF, Waters, Manchester, U.K.) in positive ion mode. The details of the instrument have been described elsewhere.31,32 Source temperature of 100 °C, capillary voltage of 3 kV, desolvation temperature of 250 °C, and cone voltage of 30 V were set as parameters for ESI. Other parameters of the instrument were optimized to achieve the best separation of phospholipids without the roll-over effect.31 Nitrogen drift gas was introduced to the TWIMS stacked ring ion guide (SRIG) at a 25 mL/min flow rate, which corresponds to 0.39 Torr. The traveling wave (T-wave) height and velocity were optimized as 8 V and 300 m/s, respectively. For each sample, 150 spectra were obtained and averaged for analysis. The drift times of the singly charged phospholipid cations and peptides were determined from the location of the ion mobility peak maxima extracted manually using MassLynx (v 4.1) software (Waters corp. Milford, MA). Resolution of the instrument was found to be ~0.8 ms in full width at half maximum (FWHM) with drift times for the ions studied and the parameters employed in this study.
Collision Cross-Section Calibration
Previously published collision cross-sections of singly charged peptide hexaglycine, hexaalanine, and trypic digest of cytochrome C in helium drift gas were used to create a calibration curve.16 Recently published PC collision cross-sections determined in helium drift gas were also used for the calibration.9 The calibration procedure was adopted from Thalassinos et al.17 The effective drift time (td″) of the calibrant was corrected for mass independent and mass dependent time. The published collision cross-section of the calibrant was scaled by reduced mass in N2. The effected drift time was plotted against the corrected published collision cross-section (ΩD′). The plot was used to fit a linear and power trend. The equation from the fitting result was used to estimate collision cross-sections of phospholipids with reduced mass.
Computational Modeling
Collision cross-sections of ions were calculated using the modified TJ method,22 which consists of two potential terms representing Van der Waals and ion-induced dipole interactions characterized by Lennard-Jones parameters and neutral polarizability, respectively.30 The modified TJ method describes the interaction between ions and an N2 drift gas that expands applicability beyond cases of ions drifting in He (details of this modification can be found elsewhere).22 In brief, we set the polarizability of neural gas for N2 (1.710×10−24 cm2). Due to the linear geometry of N2, two more consequences were taken into account: ion-quadrupole interaction and molecule orientation. The ion-quadrupole interaction is expressed in simple summations of partial charges of negative q (0.4825e) to each nitrogen atom and one positive 2q at the center of the nitrogen molecule. The orientations of N2 are sampled along the x-, y-, and z-axis; the averaged interaction potential is evaluated using Boltzmann weighting.
In order to consider the effect of structural fluctuation on the collision cross-section at room temperature, we performed NVT molecular dynamics (MD) simulations using a Nose-Hoover thermostat at 300 K. The inter-atom interactions are described with the all-atom CHARMM PARAM27 force field33 using the Large-scale Atomic/Molecular Massively Parallel Simulator (LAMMPS) code.34 We adopted the “sp2 C-sp3 C-sp2 C” angle parameter and the “sp2 C-sp2 C-sp3 C-sp2 C” dihedral parameters from reference 35, which were optimized using 1,4-pentadiene. The partial charge distribution of protonated phosphate (O3P-O-H) was optimized using Mulliken charge distributions from density functional theory (DFT) calculations (Table S2), since the common CHARMM force field only has a partial charge distribution of negatively charged phophate (O3P-O−). The systems are pre-equilibrated for 100 ps, and the conformations are sampled every one ps from the 200 ps simulations. We note that such a procedure allows for canonical sampling of the conformations at 300 K and 400 K. We analyzed the collision cross-sections and potential energies of all sampled conformations of PC.
RESULTS
Saturated Phosphatidylcholine Cations
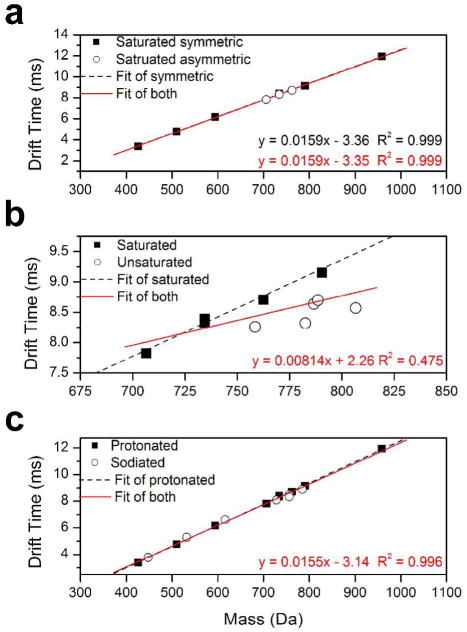
The drift times, td, of the PC cations were determined as described above. The drift times were then corrected with the mass dependent flight time, defined as the time that an ion spent in the TOF.17,28 Measured and corrected drift times for the PC cations chosen for this study are found in Table S1. The corrected drift time from TWIMS was plotted against the mass to charge (m/z) of the ion, and the plot was used to fit a linear trend. The plot of drift time versus mass for singly protonated PC cations is shown in Figure 1 along with the linear fit to the data. As seen in Figure 1a, all saturated PC cations investigated in this study (400 –1000 Da) exhibit a good correlation (R2 > 0.999) between mass and drift time (i.e., ion mobility). In particular, symmetry of the two acyl chains in the phospholipid does not affect the common mass mobility correlation of a saturated PC cation.
Figure 1.
(a) Plot of drift time of saturated phosphatidylcholine (PC) cations in traveling wave ion mobility spectrometer versus ion mass. Experimentally determined data for symmetric PC and asymmetric PC cations are shown as solid squares and empty circles, respectively. The black dash and red solid lines are the linear fit to the symmetric PC cation data set and to both symmetric and asymmetric PC cation data set, respectively. (b) Plot of drift time of PC cations spanning mass range 700 – 800 Da in traveling wave ion mobility spectrometer versus ion mass. Experimentally determined data for saturated PC and unsaturated PC cations are shown as solid squares and empty circles, respectively. The black dash and red solid lines are the linear fit to the saturated PC cation data set and to both saturated and unsaturated PC cation data set, respectively. (c) Plot of drift time of protonated and sodiated PC cations in traveling wave ion mobility spectrometer versus ion mass. Experimentally determined data for protonated PC and sodiated PC cations are shown as solid squares and empty circles, respectively. The black dash and red solid lines are the linear fit to the protonated PC cation data set and to both protonated and sodiated PC cation data set, respectively.
Unsaturated Phosphatidylcholine Cations
The usual acyl chain length of membrane phospholipids vary from 18 to 20 carbon atoms.36 Most unsaturated phospholipids contain one acyl chain with one or more cis-double bonds and a saturated one as a second acyl chain.36 We have selected unsaturated PC cations with these characteristics to investigate the dependence of mass-mobility correlations on the presence of double bonds in the acyl chains of membrane phospholipids (Table S1).
Figure 1b shows a plot of corrected drift time versus mass for PC cations from 700 Da to 810 Da along with the linear fit to the data. A good correlation (R2 = 0.984) is still observed for the saturated PC cations within the mass range. However, a poor correlation between mass and mobility from unsaturated and saturated PC cations is also observed (R2 = 0.475). Unsaturated PC cations show higher mobilities (i.e., faster drift time) compared to saturated PC cations. Corrected drift times of 16:0–18:2 PC (MW 759) and 16:0–20:4 PC (MW 783) are measured as 8.26 ms and 8.32 ms, respectively. They traveled in the SRIG faster than smaller saturated PC cations such as 16:0-16:0 and 18:0-14:0 (MW 735), which have 8.39 ms and 8.33 ms drift times, respectively. The corrected drift time of 16:0–22:6 PC (MW 807) is measured as 8.57 ms. Compared to the 8.71 ms and 9.15 ms, which are corrected drift times of two smaller saturated PC cations, 18:0-16:0 PC (MW 763) and 18:0-18:0 PC (MW 791), respectively, 16:0-22:6 PC travels across the SRIG faster.
The presence of a cis-double bond causes the acyl chain to bend. In addition, a double bond causes a relatively rigid acyl chain structure compared to that of the saturated acyl chain. It is inferred that these two factors cause smaller collision cross-sections and thus faster mobility than unsaturated PC cations.
Sodiated Phosphatidylcholine Cations
Figure 1c shows the plot of drift time versus mass for protonated and sodiated PC cations. The sodiated PC cations investigated in this study exhibit a good mass mobility correlation (R2 = 0.996) with protonated PC ions. A recent investigation by Kim et al. reported that short range interactions are most important for the collision cross-sections of molecular ions larger than 150 Da.22 The PC cations (400 – 1000 Da) investigated in this study are larger than ions that Kim et al.22 investigated (60 – 250 Da). Thus, the importance of short range interactions is emphasized for collision cross-sections of PC cations. In numerous cases, metal cations have been shown to cause specific peptide structures in the gas phase through Columbic interactions with backbone amide, carboxyl, amine, and functional groups.37,38 In contrast, PC is composed of two esterified acyl chains and one phosphorylcholine attached to glycerol.1 In the sodiated PC cations, sodium cation interacts solely with the phosphate group without inducing a noticeable conformation change of PC. Thus, a good correlation between mass and mobility is observed from PC cations regardless of whether they are protonated or sodiated.
Estimated Collision Cross-Sections of Ions Using T-Wave Calibration
A number of studies have employed empiric calibration methods to estimate collision cross-sections of ions using a set of calibrant ions.17,23,24,27,28 To understand the structural characteristics related to collision cross-sections of PC cations, the calibration method was applied to estimate collision cross-sections. Figure 2a shows the calibration plots of ΩD′ versus td″ for 14 singly charged peptides and 6 PC cations (Table S1). Due to the different natures of peptide and PC ions in the gas phase, we fit only the peptide calibrants first. Then we compared the fit of peptide calibrants to the fit result from the combined peptide and PC calibrants. Both linear fit and power fit to the calibrants were performed, and both fittings exhibit a high correlation coefficient (R2 = 0.98 and 0.99, respectively). Thalassinos et al. reported that linear fit is appropriate for calibration with small peptides. However, a slightly higher correlation was observed for power fitting in the present study. Nearly identical calibration curves were obtained from both fits for peptide and combined peptide and PC calibrants. The nature of ions in the gas phase influenced the different mass mobility correlations. However, the empiric calibration considered only the relationship between ΩD′ and td″. Thus, utilizing appropriate ΩD for calibration is more important than the chemical category of the calibrant. Figure 2b summarizes the estimated collision cross-sections of protonated PC cations. The estimated collision cross-section values of PC cations are found in Table 1.
Figure 2.

(a) Plot of corrected empiric cross-sections versus effective drift times for 14 peptides and 4 phosphatidylcholines (PC). For each peptide and PC the singly charged cation is used. Linear trend and power trend lines are shown as solid and dash lines, respectively. (b) A plot of the estimated cross sections versus the ion mass for PC cations investigated in this study. The estimated collision cross sections from linear trend and power trend are shown as solid squares and empty circles, respectively.
Table 1.
Collision cross-sections of phosphatidylcholine cations in N2 drift gas estimated and evaluated using empiric calibration method and equations from Shavartsburg and Smith29, respectively. Theoretically determined collision cross-sections in N2 and He are also listed.
| ΩD (Å2) |
||||||
|---|---|---|---|---|---|---|
| PC | mass | Estimated (linear fit) | Estimated (power fit) | Evaluated | a Theoretical (in N2) | a Theoretical (in He) |
| 5:0-5:0 | 426 | 143.4 | 136.1 | 244.8 | 255.3 | 162.5 |
| 8:0-8:0 | 510 | 171.0 | 159.5 | 289.2 | 301.3 | 197.3 |
| 11:0-11:0 | 594 | 193.1 | 183.0 | 327.3 | 335.0 | 223.8 |
| 14:0-16:0 | 706 | 214.9 | 210.7 | 367.0 | 361.1 | 248.2 |
| 16:0-16:0 | 734 | 221.9 | 220.4 | 379.9 | 400.7 | 277.1 |
| 18:0-14:0 | 734 | 221.1 | 219.3 | 378.4 | 363.7 | 254.4 |
| 16:0-18:2 | 758 | 220.1 | 218.0 | 376.7 | 388.2 | 265.8 |
| 18:0-16:0 | 762 | 225.5 | 225.7 | 386.7 | 364.1 | 254.2 |
| 16:0-20:4 | 782 | 220.7 | 218.9 | 377.8 | 357.0 | 250.6 |
| 18:0-18:2 | 786 | 224.5 | 224.4 | 385.0 | 387.1 | 270.3 |
| 18:0-18:1 | 788 | 225.3 | 225.5 | 386.4 | 396.2 | 274.4 |
| 18:0-18:0 | 790 | 230.5 | 233.2 | 396.2 | 395.3 | 275.3 |
| 16:0-22:6 | 806 | 223.6 | 223.2 | 383.3 | 366.8 | 258.0 |
| 24:0-24:0 | 958 | 258.7 | 280.4 | 451.2 | 423.4 | 305.1 |
averaged over 200 conformations at 300K.
Determination of Collision Cross-Sections of Ions
Shavartsburg and Smith derived equations to describe the quantitative relationship between drift time and ion mobility in TWIMS.29 Under the condition that KEmax < s, where K is ion mobility, Emax is maximum electric field (E), and s is wave velocity, the mobility of an ion is related to the average ion velocity in TWIMS as29
| (1) |
where E(x) is a half-sinusoidal traveling wave function and b is the waveform baseline width. Note that the equation ignores the focusing field and restricts the dynamics to axial coordinates of the SRIG. The rearrangement of Equation (1) with drift length L and the corrected drift time td′ yields
| (2) |
Once K is determined, the reduced mobility K0 can be determined according to
| (3) |
where P and T are the experimental pressure and temperature, respectively. Finally, the collision cross-section of an ion is evaluated by the relation39,40
| (4) |
where N0 is the number density at standard state (273 K and 760 Torr), q is the charge on the ion, μ is the reduced mass of ion and N2, kB is the Boltzmann constant, T is the temperature in the drift region, and ΩD is the collision cross-section. The evaluated collision cross-sections of the examined PC cations are listed in Table 1. Note that a significant difference is found between the estimated ΩD and the evaluated ΩD. The evaluated ΩD values are on average ~ 42% larger than the estimated ΩD values from both power and linear fit. It is of note that the collision cross-sections of calibrants are determined in He16,41 while the drift gas used in TWIMS is N2. A strong contribution of short range interaction between ion and neutral is expected for ΩD of an ion at the mass range of PC.22 Yet, a considerable contribution is still considered from long range interactions of ion-neutral, linear shape, and larger mass in N2 for the determination of ΩD of an ion. It is inferred that the observed difference of ΩD values are caused by lack of these terms in the calibration procedure.
Calculated Collision Cross-Sections of Ions Using the Trajectory Method
The ΩD of the PC cations investigated in this study were calculated using the TJ method in N222 and He (Table 1).30 The MD simulation trajectories of the PC cation for 200 ps reveal that the two acyl chains undergo large structural fluctuation due to the thermal energy at 300 K. Figure 3a shows the time profile of the C-C distance between the carbon atoms at the end of each chain of the 18:0-18:0 PC during 200 ps of dynamics. The distance between two carbon atoms fluctuates in the range of 5 to 25 Å within a 5 to 20 ps time period. In order to account for the sufficient amount of conformational change required for ΩD calculation, we need to sample the conformations at every 1ps. Scatter plot of collision cross sections versus relative energy for each PC exhibits a single domain (Figure S1.) The proteins and polypeptides commonly form multiple iso-energetic conformations in gas phase due to possible multiple interactions among functional groups.12 However, PC cations, which comprise ionic head group and aliphatic two acyl chains, lack such complicated interactions. No clear sub-set of geometry through the whole set of MD simulations is observed. Therefore, the ΩD values are determined using the TJ method from 200 structures on MD simulation trajectories, and all of them are employed to determine the average ΩD. 22,30
Figure 3.

(a) Time profile of the distance between the carbon atoms at the end of each acyl chain of 18:0-18:0 phophatidylcholine during 200 ps of the molecular dynamics simulation. The fluctuation is ranging from ~5Å to ~25Å with the time period of 5–20 ps. Approximately 17 times of fluctuation is observed from this trajectory. (b) Plot of experimentally determined collision cross sections (ΩD) of phosphatidylcholine (PC) cations in N2 against theoretically determined ΩD using the modified TJ method for N2 drift gas. The theoretical ΩD is obtained by averaging ΩD for 200 structures from MD simulations. The solid line is y = x. (c) Plot of theoretical ΩD in He over theoretical ΩD in N2 versus mass of PC cations.
Figure 3b shows the plot of ΩD for PC cations evaluated using the Shavartsburg and Smith29 equations versus the theoretical ΩD in N2 calculated using the modified TJ method. The theoretical ΩD values of PC cations exhibit good agreement with the experimentally evaluated values. The agreement is within 6.2% in the worst-case deviation with 3.2% deviation on average. This shows that the experimental collision cross-sections of analyte ions can be determined using Synapt HDMS and the relationship between SRIG drift time and mobility derived by Shavartsburg and Smith.29 In contrast, poor agreement was observed from the estimated ΩD of PC cations from the linear fit and power fit calibration curves with deviations of 71.2% and 73% on average, respectively, using a calibration curve, which was created by the DTIMS data in helium drift gas.
DISCUSSION
Effect of Drift Gas on Ion Mobility
The difference between the estimated ΩD of the PC cation using empiric calibration and the evaluated ΩD using Equations (2) to (4) can be explained by the different polarizabilities, sizes, and shapes of He and N2 molecules. In drift tube ion mobility spectrometry (DTIMS), the drift time, which corresponds to the effective drift time in TWIMS, td″, is inversely proportional to the ion mobility, K:
| (5) |
where V is voltage across the drift tube. The relationship between K and ΩD is described as42
| (6) |
where Teff is an effective temperature of ion. The corrected collision cross-section, ΩD′, for the empiric calibration is defined by taking into account reduced mass and charge state of ions.17,28
| (7) |
From Equations (5) through (7), we obtain the proportional relationship between td″ and ΩD′. As discussed earlier, the default drift gas of TWIMS of Synapt HDMS is N2.17,32,43 Calibration methods commonly employ the empirical ΩD determined in He.16 Thus, the corrected collision cross-section, Ω′D,N2, using the reduced mass in N2, μN2, is related to the ΩD in He as
| (8) |
This relationship works if ΩD,He ≅ ΩD,N2. Hill and co-workers have demonstrated the high dependence of ΩD of ions on drift gas.44,45 Beegle et al. demonstrated the different polarizability effects of the drift gas molecule on the ΩD of the ion molecule using a series of homologous Gly peptides.44 As the size of the Gly peptide increases, the difference between ΩD in N2 and in He decreases. The short range interaction for ΩD becomes more important as the ion size increases.22 Thus, the pre-assumption for the empiric calibration is valid when the size of the ion is very large; geometric factors of neutral and long range ion-neutral interactions are completely negligible for the determination of ΩD.28 Theoretically calculated ΩD in N2 and He further support this argument. Figure 3c shows the plot of the theoretical ΩD in He divided by ΩD in N2 versus the mass of PC cations. As the size of the cation increases from 426 Da to 959 Da, the agreement between the two theoretical ΩD values increases from 64% to 72%. As a result, for the mass range of the PC cations (400 – 1000 Da), estimating ΩD from the drift time measured in N2 using the empiric calibration method, which uses DTIMS data in helium drift gas, is not valid.
Geometrical Effect on the Collision Cross-Sections of Phosphatidylcholine Cations
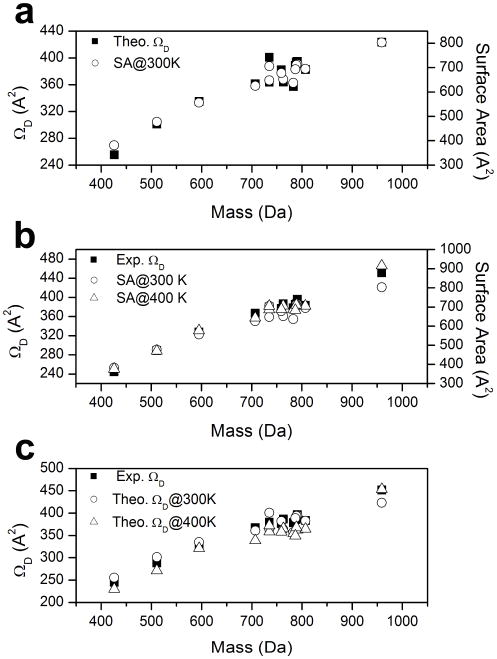
Figure 4a shows the plot of theoretical characteristic ΩD of PC cations versus ion mass compared with the corresponding surface area of PC cations in N2 at 300 K using the Maximal Speed Molecular Surface (MSMS) program.46,47 Note that high similarity is observed from the characteristics of relative ΩD from theoretical calculation and the relative surface areas of PC cations. This implies that the ΩD for each PC cation is largely influenced by the short range Van der Waals interaction between the ion and the neutral N2 molecule. The molecular weight and specific geometry of the ions dominate the short range Van der Waals interaction, which affects the collision cross-section of the ion.22
Figure 4.
(a) Plots of theoretically determined collision cross sections (ΩD) and surface areas of phosphatidylcholine (PC) cations in N2 versus ion mass. The calculated average ΩD of the 200 ion comformations are shown as solid squares (left y-axis). The calculated surface areas of PC cations in N2 at 300 K are shown as empty circles (right y-axis). (b) Plots of experimentally evaluated ΩD and surface areas of phosphatidylcholine (PC) cations in N2 versus ion mass. The ΩD of PC cations are shown as solid squares (left y-axis). The calculated surface areas of PC cations in N2 at 300 K and 400 K are shown as empty circles and empty triangle, respectively (right y-axis). (c) Plots of experimentally evaluated ΩD and theoretically determined ΩD calculated at 300 K and 400 K. The experimental ΩD of PC cations are shown as solid squares. The theoretically determined ΩD of PC cations in N2 at 300 K and 400 K are shown as empty circles and empty triangle, respectively.
The mobility of ion, K, becomes field-dependent at a high electric field.48 The field dependence of K depends on the nature properties of ion-neutral interactions. In general, high field behavior of an ion is observed when the ion acquires enough energy from E to change the nature of the ion-neutral collisions.48 The total average energy of the ions can be determined from the Wannier energy formula as follows:
| (9) |
where M is the mass of a drift gas molecule, and vd is the drift velocity of an ion.49 The thermal kinetic energy is 3/2kT, and the field energy is 1/2Mvd2. The low field behavior of an ion is achieved when
| (10) |
Ion mobility spectrometers typically operate at low electric fields. The typical E/N range for the low field is a few Townsend (Td = 10−17 Vcm2).20,48 Although the applied voltage in the TWIMS is as low as 8 V in this study, due to the low pressure of the SRIG (0.39 Torr in this study), the average E/N is ~80 Td. This is an order of magnitude larger than common IMS operating field. In addition, E/N increases to as much as ~230 Td at Emax of traveling wave.
The primary effect of the strong field of TWIMS is to heat the ions,48 which increases their internal energy through ion-neutral collisions. This collisional activation can result in conformation changes of the ions. Figure 4b shows the plots of the experimental ΩD of PC cations evaluated using the Shavartsburg and Smith29 equations versus ion mass compared with the corresponding surface area of PC cations in N2 at 300 K and 400 K. In general, the characteristic ΩD value of the PC cation is found to be similar to the relative surface area of PC cation at both 300 K and 400 K. However, slightly greater similarity is observed between the characteristic relative ΩD and the relative surface areas of PC cations at 400 K as the size of PC cation increases (24:0-24:0 PC). Although the experiment was performed at ~300K, field heating induced a shift in ion conformation distribution to slightly higher energy state for large PC. Figure 4c shows the plots of the experimental ΩD of saturated PC cations versus ion mass compared with the theoretical ΩD of PC cations in N2 at 300 K and 400 K. Overall, the experimental ΩD value of the PC cation is found between the ΩD values calculated at two different temperatures. This implies that field heating of the ion in the TWIMS is less than 100 K. It is noteworthy that the theoretical ΩD calculated at 300 K exhibits a better agreement to the experimental ΩD. However, better agreement to the experimental ΩD is found from the theoretical ΩD at 400 K with the largest PC cation (24:0-24:0 PC). For small PC cations, a shift in ion conformation distribution by field heating is not expected due to the steric restrict of the short acyl chains. However, larger PC cations with longer acyl chains are easily affected by field heating to shift its ion conformation distribution to a higher energy state.
As stated earlier, 8 V of voltage was employed during the current measurements of PC cation mobilities. To ensure that electric field effect is limited to ion conformation distribution and negligible for the determination of the mobility of ions at this field strength, a series of test measurements were conducted. The reduced mobility measurements were repeated for three PC cations (5:0-5:0 PC, 14:0-16:0 PC, and 18:0-18:0 PC) with the applied voltage of 7, 7.5, and 8.5 V. Agreement was seen between all corresponding mobilities with an average observed deviation of ~1% (Table S3). These results verified that although the electric field in TWIMS in this study can induce conformational change of the PC cation, it is sufficiently low to avoid any dependence of the measured mobilities on the electric field to gas density ratio.
Mass Mobility Correlations of Phophatidylcholine Cations
Saturated PC cations investigated in this study exhibit a good correlation between mass and mobility (Figure 1a). However, deviations from the correlation are observed in unsaturated PC cations (Figure 1b). In the previous study, we discussed the importance of Van der Waals potential for determining the collision cross-section of an ion as the ion size increases.22 This implies that strong mass-mobility correlation is highly affected by the geometry of the ion. In order to understand the mass mobility correlation of saturated PC cations and deviations of unsaturated PC cations from the correlation plot, we investigate structures of PC cations with corresponding ΩD.
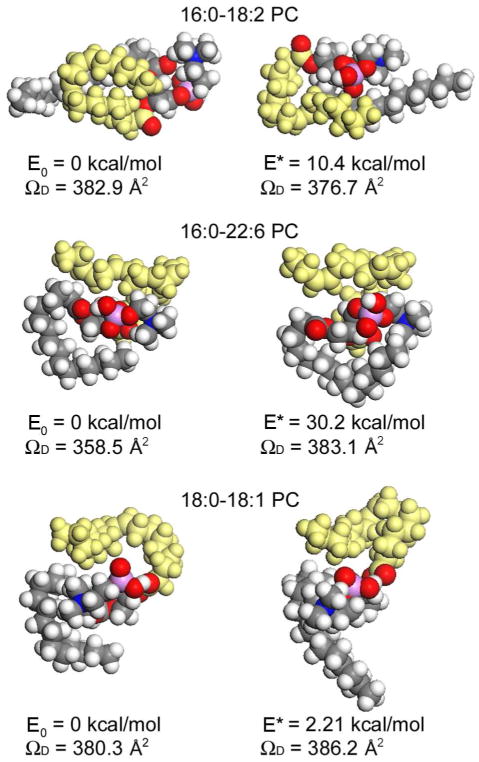
The minimum energy (E0) structures of some of the saturated PC cations examined in this study are shown in Figure 5a. The structures with the closest ΩD to the experimental values are also shown along with their corresponding relative energy values (E*). The plot of ΩD for the PC cations experimentally evaluated using the Shavartsburg and Smith29 equations versus the theoretical ΩD values of E0 and E* is shown in Figure 5b. Slightly larger ΩD values are observed from PC cations with longer acyl chains (≥18 carbon) compared to the ΩD values calculated from minimum energy structures. In contrast, smaller ΩD values are observed from PC cations with short acyl chains (≤16 carbon). Note that PC cations with an acyl chain longer than 16 carbons form globular structures that are energetically favored. It is inferred that intramolecular Van der Walls interactions of acyl chains drives the globular conformation to be preferred for large PC cations. However, extended structures are energetically favored for those with a shorter acyl chain (fewer than 16 carbons), whose steric effects prevent them from forming globular conformations in the gas phase. In contrast to peptide or protein ions, PC cations do not have strong intramolecular interactions to stabilize certain conformations. The energy difference between the E0 structure and the E* structure is only 19 kcal/mol on average. Thus, the conformations of these ions may fluctuate while traveling in the SRIG. As discussed earlier, the internal energy of an ion increases with collisional activations from the traveling wave electric field, which results in continual excitation of the ion.29 The shift in ion conformations occurs toward slightly excited (E*) state. Especially for larger molecules (18 or more carbon acyl chains), a significant increase in ΩD occurs, since the increase in internal energy of these large molecules increases the importance of the entropy. As a result, the structural similarity of the saturated PC cations is maintained with moderately extended structures regardless of the length and symmetry of the PC acyl chains (Figure 5a); this promotes a good correlation between mass and mobility.
Figure 5.
(a) MD simulated structures of saturated phosphatidylcholine cations at minimum energy state (E0) are shown. The structures of the closest ΩD to the experimental values are also shown along with corresponding relative energy values (E*). (b) Plot of experimentally determined collision cross sections (ΩD) of phosphatidylcholine (PC) cations in N2 against theoretically determined ΩD at E0 and E* using the modified TJ method for N2 drift gas. The solid line is y = x.
Characterizing Unsaturated Phosphatidylcholines from Mass Mobility Correlation
Unsaturated PC cations exhibit significantly deviated mobility values from the mass mobility correlation plot of saturated PC cations (Figure 1b). The drift time is reduced by ~5% for the unsaturated PC cation with one double bond; the drift time reduces further at a rate of ~1% for the additional double bond. Jackson et al. recently reported a ~0.5% reduction in drift time for each additional double bond of phospholipids in DTIMS. In the present study, the larger difference in the mobility of unsaturated PC cations compared to saturated PC cations results from the different rate of conformation changes in TWIMS. Figure 6 shows structures of selected unsaturated PC cations at E0 along with the conformations at E*. For unsaturated PC cations, a smaller shift in ΩD is observed from the ΩD of the most stable conformation compared to saturated PCs of similar mass. For those PC cations with more than 16 carbon acyl chains, saturated PC cations exhibit a ~10 % difference in ΩD on average, while unsaturated PC cations show only ~5% difference on average (Table 2). As observed in Figure 6, the major change in conformation occurs at the saturated acyl chain, while the conformation of the unsaturated acyl chain (yellow) maintains a bent structure. The presence of cis-double bonds in an acyl chain prevents the unsaturated acyl chain from extending by activation. As a result, less fluctuation in the ion structure occurred among unsaturated PC cations in the TWIMS. Unsaturated PC cations show smaller ΩD values than saturated PC cations, which can form more extended conformations. This is logical given that globular structures of unsaturated PC cations are more compact and therefore have smaller collision cross–sections. This allows us to characterize unsaturated PC cations based on their mobility, and thus collision cross-sections, using TWIMS.
Figure 6.
MD simulated structures of unsaturated phosphatidylcholine cations at minimum energy state (E0) are shown. The structures of the closest ΩD to the experimental values are also shown along with corresponding relative energy values (E*). Unsaturated acyl chain is colored in yellow.
Table 2.
Theoretically determined collision cross-sections (ΩD) of phosphatidylcholine cations at minimum energy state (E0). The differences of ΩD (ΔΩD) and potential energy (ΔE) from the PC structure at E0 to experimentally determined ΩD.
| PC | ΩD, E0 (Å2) | ΔΩD (%) | ΔE (kcal/mol) |
|---|---|---|---|
| 5:0-5:0 | 267.08 | 9.1 | 15.2 |
| 8:0-8:0 | 295.44 | 2.1 | 10.6 |
| 11:0-11:0 | 311.24 | −4.9 | 34.7 |
| 14:0-16:0 | 369.21 | 0.6 | 10.3 |
| 16:0-16:0 | 390.55 | 2.8 | 15.9 |
| 18:0-14:0 | 343.25 | −9.3 | 11.4 |
| 16:0-18:2 | 382.85 | 1.6 | 10.4 |
| 18:0-16:0 | 343.38 | −11.2 | 28.7 |
| 16:0-20:4 | 333.7 | −11.7 | 26.8 |
| 18:0-18:2 | 362.37 | −5.9 | 10.6 |
| 18:0-18:1 | 380.32 | −1.6 | 2.21 |
| 18:0-18:0 | 368.93 | −6.9 | 31.5 |
| 16:0-22:6 | 358.53 | −6.5 | 30.2 |
| 24:0-24:0 | 388.94 | −13.8 | 30.4 |
CONCLUSIONS
A high correlation between mass and mobility in N2 is observed from a number of saturated PC cations in TWIMS. A significant deviation from this mass mobility correlation is observed with unsaturated PC cations. Theoretical investigation using a modified TJ method indicates that the empiric calibration method is not suitable to estimate collision cross-sections for PC cations. Instead, we evaluate collision cross-sections using a quantitative relationship between drift time and mobility derived by Shavartsburg and Smith.29 In addition to the lack of intramolecular interactions among PC cations, collisional excitation of the ions in the SRIG induces a shift in ion conformational distribution. The unsaturated acyl chain remains bent, while the saturated acyl chain extends under the electric field, which causes larger collision cross-sections for saturated PCs and smaller ones for unsaturated PCs. The initial double bond in the acyl chain yields an approximately 5% reduction in drift time, with further drift time reduction at the rate of ~1 % for each additional double bond. As a result, greater separation and characterization of unsaturated PC cations can be achieved using TWIMS.
Supplementary Material
Acknowledgments
This research was carried out at the Jet Propulsion Laboratory, California Institute of Technology, under a contract with the National Aeronautics and Space Administration (NASA), The University of California Los Angeles Mass Spectrometry and Proteomics Technology Center, and the Material and Process Simulation Center, Beckman Institute, California Institute of Technology. Financial support through NASA’s Astrobiology Science and Technology Instrument Development, Planetary Instrument Definition and Development, and Mars Instrument Development programs is gratefully acknowledged. JAL acknowledges support from the NIH (RR20004).
References
- 1.Gurr MI, Hardwood JL. Lipid Biochemistry. 4. Chapman and Hall; New York: 1991. [Google Scholar]
- 2.Simoneit BRT, Summons RE, Jahnke LL. Orig Life Evol Biosph. 1998;28:475–483. doi: 10.1023/a:1006508012904. [DOI] [PubMed] [Google Scholar]
- 3.Hemming FW, Hawthorne JN. Lipid Analysis. BIOS Scientific Oxford; 1996. [Google Scholar]
- 4.Woods AS, Ugarov M, Egan T, Koomen J, Gillig KJ, Fuhrer K, Gonin M, Schultz JA. Anal Chem. 2004;76:2187–2195. doi: 10.1021/ac035376k. [DOI] [PubMed] [Google Scholar]
- 5.Jackson SN, Ugarov M, Egan T, Post JD, Langlais D, Schultz JA, Woods AS. J Mass Spectrom. 2007;42:1093–1098. doi: 10.1002/jms.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson SN, Ugarov M, Post JD, Egan T, Langlais D, Schultz JA, Woods AS. J Am Soc Mass Spectrom. 2008;19:1655–1662. doi: 10.1016/j.jasms.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLean JA, Ridenour WB, Caprioli RM. J Mass Spectrom. 2007;42:1099–1105. doi: 10.1002/jms.1254. [DOI] [PubMed] [Google Scholar]
- 8.Fenn LS, McLean JA. Anal Bioanal Chem. 2008;391:905–909. doi: 10.1007/s00216-008-1951-x. [DOI] [PubMed] [Google Scholar]
- 9.Fenn LS, Kliman M, Mahsut A, Zhao SR, McLean JA. Anal Bioanal Chem. 2009;394:235–244. doi: 10.1007/s00216-009-2666-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gidden J, Bowers MT. J Am Soc Mass Spectrom. 2003;14:161–170. doi: 10.1016/S1044-0305(02)00866-8. [DOI] [PubMed] [Google Scholar]
- 11.Fenn LS, McLean JA. Chem Commun. 2008:5505–5507. doi: 10.1039/b810421b. [DOI] [PubMed] [Google Scholar]
- 12.Jarrold MF. Annu Rev Phys Chem. 2000;51:179–207. doi: 10.1146/annurev.physchem.51.1.179. [DOI] [PubMed] [Google Scholar]
- 13.Griffin GW, Dzidic I, Carroll DI, Stillwel Rn, Horning EC. Anal Chem. 1973;45:1204–1209. [Google Scholar]
- 14.Berant Z, Karpas Z. J Am Chem Soc. 1989;111:3819–3824. [Google Scholar]
- 15.Karpas Z, Berant Z. J Phys Chem. 1989;93:3021–3025. [Google Scholar]
- 16.Valentine SJ, Counterman AE, Clemmer DE. J Am Soc Mass Spectrom. 1999;10:1188–1211. doi: 10.1016/S1044-0305(99)00079-3. [DOI] [PubMed] [Google Scholar]
- 17.Thalassinos K, Grabenauer M, Slade SE, Hilton GR, Bowers MT, Scrivens JH. Anal Chem. 2009;81:248–254. doi: 10.1021/ac801916h. [DOI] [PubMed] [Google Scholar]
- 18.Ruotolo BT, Gillig KJ, Woods AS, Egan TF, Ugarov MV, Schultz JA, Russell DH. Anal Chem. 2004;76:6727–6733. doi: 10.1021/ac0498009. [DOI] [PubMed] [Google Scholar]
- 19.Ruotolo BT, Verbeck GF, Thomson LM, Woods AS, Gillig KJ, Russell DH. J Proteome Res. 2002;1:303–306. doi: 10.1021/pr025516r. [DOI] [PubMed] [Google Scholar]
- 20.Johnson PV, Kim HI, Beegle LW, Kanik I. J Phys Chem A. 2004;108:5785–5792. doi: 10.1021/jp051274h. [DOI] [PubMed] [Google Scholar]
- 21.Kim HI, Johnson PV, Beegle LW, Beauchamp JL, Kanik I. J Phys Chem A. 2005;109:7888–7895. doi: 10.1021/jp051274h. [DOI] [PubMed] [Google Scholar]
- 22.Kim H, Kim HI, Johnson PV, Beegle LW, Beauchamp JL, Goddard WA, Kanik I. Anal Chem. 2008;80:1928–1936. doi: 10.1021/ac701888e. [DOI] [PubMed] [Google Scholar]
- 23.Ruotolo BT, Giles K, Campuzano I, Sandercock AM, Bateman RH, Robinson CV. Science. 2005;310:1658–1661. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- 24.Ruotolo BT, Hyung SJ, Robinson PM, Giles K, Bateman RH, Robinson CV. Angew Chem-Int Edit. 2007;46:8001–8004. doi: 10.1002/anie.200702161. [DOI] [PubMed] [Google Scholar]
- 25.Smith DP, Giles K, Bateman RH, Radford SE, Ashcroft AE. J Am Soc Mass Spectrom. 2007;18:2180–2190. doi: 10.1016/j.jasms.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bagal D, Zhang H, Schnier PD. Anal Chem. 2008;80:2408–2418. doi: 10.1021/ac7020163. [DOI] [PubMed] [Google Scholar]
- 27.Williams JP, Scrivens JH. Rapid Commun Mass Spectrom. 2008;22:187–196. doi: 10.1002/rcm.3346. [DOI] [PubMed] [Google Scholar]
- 28.Ruotolo BT, Benesch JLP, Sandercock AM, Hyung SJ, Robinson CV. Nat Protoc. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 29.Shvartsburg AA, Smith RD. Anal Chem. 2008;80:9689–9699. doi: 10.1021/ac8016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. J Phys Chem. 1996;100:16082–16086. [Google Scholar]
- 31.Giles K, Pringle SD, Worthington KR, Little D, Wildgoose JL, Bateman RH. Rapid Commun Mass Spectrom. 2004;18:2401–2414. doi: 10.1002/rcm.1641. [DOI] [PubMed] [Google Scholar]
- 32.Pringle SD, Giles K, Wildgoose JL, Williams JP, Slade SE, Thalassinos K, Bateman RH, Bowers MT, Scrivens JH. Int J Mass Spectrom. 2007;261:1–12. [Google Scholar]
- 33.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 34.Plimpton S. J Comput Phys. 1995;117:1–19. [Google Scholar]
- 35.Feller SE, Gawrisch K, MacKerell AD. J Am Chem Soc. 2002;124:318–326. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 36.Alberts B, Bray D, Hopkin K, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Essential Cell Biology. 2. Garland Science; New York: 2004. [Google Scholar]
- 37.Kohtani M, Kinnear BS, Jarrold MF. J Am Chem Soc. 2000;122:12377–12378. [Google Scholar]
- 38.Wyttenbach T, Liu DF, Bowers MT. J Am Chem Soc. 2008;130:5993–6000. doi: 10.1021/ja8002342. [DOI] [PubMed] [Google Scholar]
- 39.Revercomb HE, Mason EA. Anal Chem. 1975;47:970–983. [Google Scholar]
- 40.Eiceman GA, Karpas Z. Ion Mobility Spectrometry. CRC Press; Boca Raton, FL: 1994. [Google Scholar]
- 41.Fenn LS, Kliman M, Mahsut A, Zhao SR, McLean JA. 2009 doi: 10.1007/s00216-009-2666-3. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mason EA, O'hara H, Smith FJ. J Phys B. 1972;5:169–176. [Google Scholar]
- 43.Scarff CA, Thalassinos K, Hilton GR, Scrivens JH. Rapid Commun Mass Spectrom. 2008;22:3297–3304. doi: 10.1002/rcm.3737. [DOI] [PubMed] [Google Scholar]
- 44.Beegle LW, Kanik I, Matz L, Hill HH. Int J Mass Spectrom. 2002;216:257–268. doi: 10.1016/S1044-0305(01)00366-X. [DOI] [PubMed] [Google Scholar]
- 45.Matz LM, Hill HH, Beegle LW, Kanik I. J Am Soc Mass Spectrom. 2002;13:300–307. doi: 10.1016/S1044-0305(01)00366-X. [DOI] [PubMed] [Google Scholar]
- 46.Connolly ML. J Am Chem Soc. 1985;107:1118–1124. [Google Scholar]
- 47.Sanner MF, Olson AJ, Spehner JC. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 48.Mason EA. In: Plasma Chromatography. Carr TW, editor. Plenum Press; New York: 1984. pp. 43–93. [Google Scholar]
- 49.Wannier GH. Bell System Technical Journal. 1953;32:170–254. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.