Abstract
Alzheimer's disease is the most common cause of dementia in the elderly. Although several genetic defects have been identified in patients with a family history of this disease, the majority of cases involve individuals with no known genetic predisposition. A mutant form of ubiquitin, termed Ub+1, has been selectively observed in the brains of Alzheimer's patients, including those with nonfamilial Alzheimer's disease, but it has been unclear why Ub+1 expression should be deleterious. Here we show that Ub+1 is an efficient substrate for polyubiquitination in vitro and in transfected human cells. The resulting polyubiquitin chains are refractory to disassembly by deubiquitinating enzymes and potently inhibit the degradation of a polyubiquitinated substrate by purified 26S proteasomes. Thus, expression of Ub+1 in aging brain could result in dominant inhibition of the Ub-proteasome system, leading to neuropathologic consequences.
The conserved protein ubiquitin (Ub) functions as a covalent proteolytic signal. Conjugation of intracellular proteins to a polyUb chain is a signal for targeting to 26S proteasomes, which degrade these substrates while sparing Ub (1, 2). The first Ub is linked to the substrate through an isopeptide bond that joins the C terminus of Ub (G76) to an internal lysine residue of the substrate. To generate an efficient proteasomal targeting signal, additional Ubs are linked to the first one by means of K48-G76 isopeptide bonds between successive Ub molecules (3–5). The Ub-proteasome system regulates the levels of a host of vital regulatory proteins in eukaryotic cells, including cyclins, cyclin-dependent kinase inhibitors, transcription factors, and oncoproteins (1, 2). In accordance with a requirement for the proper homeostasis of such substrates, a functional Ub-proteasome system is essential for the viability of eukaryotic cells (1, 2, 6).
The molecular pathological hallmarks of Alzheimer's disease (AD) are intracellular neurofibrillary tangles and extracellular amyloid (Aβ) plaques (7). The Aβ in plaques arises from specific processing of amyloid precursor protein that is positively regulated by the presenilins PS1 and PS2 (8, 9). PS1 and PS2 recently have been shown to be substrates of the Ub-proteasome system (10–13). Although certain mutations in the genes encoding amyloid precursor protein and PS1/PS2 are known to confer a genetic predisposition to AD, such mutations are absent in the majority of patients with AD (7). The Ub-proteasome system was first implicated in AD through the immunohistochemical observation of Ub-conjugated tau protein in neurofibrillary tangles (14, 15). However, although the presence of Ub in abnormal aggregates is characteristic of AD and several other neurodegenerative diseases (14–16), the origin of these structures and their mechanistic relationship to disease onset and progression remain unclear.
Recently, van Leeuwen and coworkers (17, 18) discovered that a mutant form of Ub, termed Ub+1, is selectively expressed in the brains of AD patients. Expression of Ub+1 occurs through a phenomenon known as molecular misreading, in which non-DNA-encoded dinucleotide deletion(s) are found in mRNA within or adjacent to GAGAG motifs (17–19). Van Leeuwen et al. (17) localized a dinucleotide deletion to the first coding unit of a polyUb transcript, causing replacement of the C-terminal residue of Ub (G76) with a 20-residue (+1) extension. Those investigators used several approaches to show that Ub+1 was present in the brains of AD patients, including those without known predisposing factors (17). In contrast, Ub+1 was present at a significantly reduced level in the brains of nondemented control patients (17, 18). Although these results demonstrated a striking correlation between Ub+1 expression and AD pathogenesis, it was unclear why Ub+1 expression should be deleterious. Here we demonstrate a mechanism by which Ub+1 expression could lead to dominant inhibition of the Ub-proteasome system.
Materials and Methods
Proteins and Enzymes.
Ub+1 was chemically synthesized by using ≈0.5 g fluorenylmethoxycarbonyl-Gln-Wang resin functionalized at 0.2 mmol/g (20). The overall yield after purification was 5–10%. The Ub-ribosomal fusion protein Ub-CEP52 was prepared similarly (20). Ub5+1 was synthesized by using purified recombinant ubiquitin conjugating enzyme-25K to conjugate preassembled, K48-linked Ub4 (18 μM) to K48 of Ub+1 (26 μM). The conditions of this reaction and the synthesis of Ub4 have been described (5). Ub5+1 was purified by cation exchange chromatography (5). Purified recombinant glutathione S-transferase (GST)-Ub carboxyl-terminal hydrolase-D (UCH-D) was a gift of M. Bownes (University of Edinburgh). Isopeptidase T (21) and ubiquitin activating enzyme (E1) (5) were purified to homogeneity from bovine erythrocytes.
Extract Preparation.
Human embryonic kidney 293T cells were lysed in a buffer containing 20 mM Tris (pH 7.6), 0.1% NP-40, 0.5 mM EDTA, 1 mM PMSF, 5 μg/ml leupeptin, 10 μg/ml soybean trypsin inhibitor, 50 μM tosyl lysyl chloromethyl ketone, and either 0.5 mM DTT (for assays of deubiquitination) or 1 mM N-ethyl maleimide (to detect polyUb chains). Lysates were centrifuged at 13,000 × g for 10 min; the supernatant (soluble extract) was assayed.
Deubiquitination Assays.
Incubations with GST-UCH-D contained 20 nM enzyme and 4 μM of either Ub+1 or Ub-CEP52. In some cases the enzyme was preinactivated with N-ethyl maleimide. Incubations (pH 7.6, 37°C) were quenched after 2 h; reactants/products were detected by blotting with Ub antibodies. Assays of purified isopeptidase T (pH 7.6 and 37°C) contained 20 μM of either Ub5+1 or Ub4 and 0.1 to 1 μM enzyme. Incubations were quenched after 30 min and reactants/products were detected by Western blotting with +1 or Ub antibodies. For assays of deubiquitination in 293T cell extracts, soluble lysate was prepared by using DTT (above). The lysate was diluted to a protein concentration of 1 mg/ml and incubated at 37o with 1 μM of Ub4 or Ub5+1. In the experiment shown in Fig. 3D, Ub4 and Ub5+1 were pretreated with a purified deubiquitinating enzyme, YUH-1. In the case of Ub4, this treatment removes an extra residue (D77) in the proximal Ub that resulted from the synthetic procedure (5). In the case of Ub5+1, YUH-1 (equivalent to UCH-D) has no effect (see Results). Aliquots were removed at increasing times up to 3 h and analyzed by blotting with Ub antibodies.
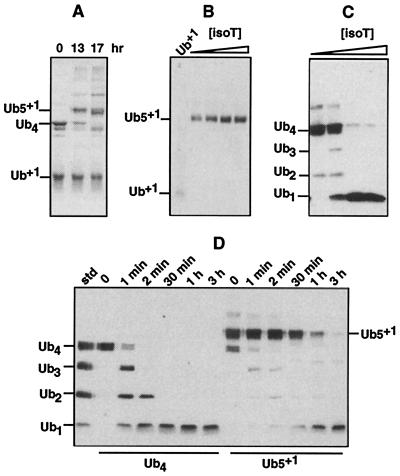
Figure 3.
Ub+1-capped chains resist disassembly. (A) Synthesis of Ub5+1 (Coomassie-stained gel). The conjugation reaction (see Materials and Methods) was sampled at the indicated times. (B) Disassembly of Ub5+1 by isopeptidase T (+1 blot). First lane: Ub+1 standard. Next four lanes: Ub5+1 was incubated with increasing concentrations of purified isopeptidase T (left to right: none, 0.1 μM, 0.5 μM, 1 μM) as described in Materials and Methods. (C) Disassembly of Ub4 by isopeptidase T (Ub blot). Conditions were identical to last four lanes of B, except the chain was Ub4. (D) Disassembly of Ub4 versus Ub5+1 in mammalian cell lysate (Ub blots). Soluble lysate from 293T cells, prepared using DTT, was incubated with Ub4 or Ub5+1 for the indicated times (see Materials and Methods). First lane, polyUb chain standard. A trace of Ub4, present as a contaminant in Ub5+1, was rapidly disassembled in the Ub5+1 incubation.
Proteasome Assays.
26S proteasomes (2.5 nM) purified from bovine erythrocytes (22) were incubated with 20 nM of the synthetic substrate 35S-Ub5–dihydrofolate reductase in the presence of MgATP, with or without increasing concentrations of Ub4 or Ub5+1 (5). Degradation was monitored as the appearance of acid-soluble radioactivity at a time (20 min) determined to be in the linear range of the assay. Ki values were calculated by assuming competitive inhibition, using Km = 17 nM (Km value determined for the proteasome preparation used in these experiments).
E1 Assay.
E1 was assayed based on its ability to form a thiol ester adduct with 125I-Ub, which was detected by autoradiography after resolution by SDS/PAGE under nonreducing conditions. Purified E1 was incubated with 2 μM 125I-Ub in the presence of MgATP. To monitor the binding of Ub+1, increasing concentrations of the unlabeled synthetic protein were mixed with 125I-Ub before initiating adduct formation by adding 0.1 μM E1 (23). For the control, increasing concentrations of unlabeled wild-type Ub were used. The level of the labeled adduct was quantitated by PhosphorImage analysis.
Ub+1 and Ub Plasmids.
Synthetic genes encoding Ub+1, Ub+1 with a C-terminal H6 tag, and Ub, were generated by PCR and cloned into the ClaI/EcoRV sites of the tetracycline-off pTet-Splice vector from Life Technologies, Grand Island, NY (24). In Ub+1-H6 the final amino acid was replaced with the sequence AAALEHHHHHH. As part of the cloning procedure, residue 2 was mutated from Gln to Ala in all three coding sequences.
Transfection.
Subconfluent 293T cells were cotransfected with the indicated pTet-Splice vector together with pTet-tTAk encoding a tet-repressible hybrid activator (4 μg each plasmid/well on 6-well plates, Lipofectamine method). Transfection efficiency was 10–20%. Cells were lysed in buffer containing N-ethyl maleimide (above) after 26 h of transfection using 40 μl of buffer per well of a 6-well plate.
Nickel Binding.
Soluble lysate from transfected 293T cells (above) containing between 200 and 675 μg of protein (see the figure legends) was applied to 10 μl of charged His-bind resin (Novagen). The resin was washed with lysis buffer, and then eluted with SDS/PAGE sample buffer. Aliquots of the starting material, unbound fraction, and bound fraction were analyzed by blotting with anti-Ub antibodies.
Antibodies and Western Blotting.
Polyclonal Ub antibodies (25) were affinity-purified. Polyclonal antibodies specific for Ub+1 were raised in rabbits against synthetic +1 peptide; the IgG fraction was used in blotting experiments. Immunocomplexes were detected by using enhanced chemiluminescence.
Results
Ub+1 Does Not Inhibit Ub Conjugation.
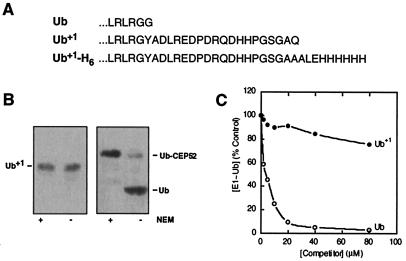
Because Ub is synthesized as a fusion either with itself or with a ribosomal protein, Ub activation and conjugation strictly depend on the exposure of G76 through processing by deubiquitinating enzymes (4, 26). In the case of Ub+1, G76 is replaced with the 20-residue +1 peptide (Fig. 1A; ref. 17). To test the potential of Ub+1 to enter the conjugation phase of the Ub-proteasome system, we first addressed whether Ub+1 was processed by a purified deubiquitinating enzyme, UCH-D (20). We found that the +1 peptide of Ub+1 was not removed by a GST-fused form of UCH-D (Fig. 1B Left), whereas the same enzyme efficiently processed a Ub-ribosomal fusion protein (Fig. 1B Right). Ub+1 also failed to undergo processing in a crude extract prepared from human embryonic kidney 293T cells (data not shown) despite the presence of a high level of deubiquitinating activity in this extract (see below).
Figure 1.
Ub+1 is refractory to processing and activation. (A) Unique sequence of Ub+1. The sequences of the extreme C termini of Ub, Ub+1 (17), and Ub+1-H6 (see Materials and Methods) are shown. (B) Processing (Ub blots). (Left) Ub+1 was incubated (see Materials and Methods) with GST-UCH-D that had been preinactivated (or not) with N-ethyl maleimide (NEM) (as indicated). (Right) Control: the same enzyme processed Ub-CEP52, a chemically synthesized Ub-ribosomal fusion protein. (C) Activation. Increasing concentrations of Ub (○) or Ub+1 (●) were mixed with 2 μM 125I-Ub before initiating assays of E1-Ub thiol ester formation with purified E1 (see Materials and Methods).
To test whether Ub+1 could interfere with Ub conjugation, we determined whether Ub+1 could bind to E1. E1 catalyzes the obligatory first step in all ubiquitination reactions and forms a catalytic thiol ester intermediate with Ub (1, 2). Any Ub+1 binding to E1 can be monitored through inhibition of E1-Ub thiol ester formation (23). As shown in Fig. 1C (●), Ub+1 exhibited negligible binding in comparison to wild-type Ub (○). The lack of Ub+1 processing by cellular deubiquitinating enzymes (Fig. 1B) and the failure of Ub+1 to bind E1 (Fig. 1C) are in agreement with earlier demonstrations that G76 is a key specificity element in Ub processing and activation (4, 26–30). It is therefore most unlikely that Ub+1 will inhibit the conjugation of wild-type Ub to cellular substrates.
Assembly of Ub+1-Capped PolyUb Chains in Cultured Mammalian Cells.
Although Ub+1 did not inhibit the conjugation phase of the proteolytic system, we reasoned that inhibition of the Ub-proteasome system could still occur through the accumulation of aberrant polyUb chains containing Ub+1. In general, a substrate must be conjugated to a K48-linked polyUb chain to be efficiently targeted to proteasomes (1–5). In the pathway for recycling of Ub, the chain is first released from the substrate; the resulting unanchored chain ultimately is disassembled by isopeptidase T (Ubp14 in yeast), a deubiquitinating enzyme whose activity requires an unblocked G76 residue at the proximal chain terminus (21, 30). (The proximal terminus refers to that end of the chain that could normally be conjugated to a substrate protein.) Inhibition of the disassembly of unanchored polyUb chains is deleterious, principally because the chains compete with polyubiquitinated substrates for binding to proteasomes (5, 30).
Besides arising as intermediates in the degradation of polyubiquitinated substrates, unanchored chains can be assembled de novo (4, 30, 31). Because Ub+1 retains K48 and other lysine residues implicated in polyUb chain assembly (4), it could act as a substrate for conjugation of wild-type Ub; the resulting Ub+1-capped polyUb chains may escape disassembly by isopeptidase T because of the absence of G76 in Ub+1. Similarly, when yeast cells are engineered to overexpress a C-terminally truncated version of Ub that lacks the C-terminal GG dipeptide, stable polyubiquitinated forms of this truncated protein accumulate and inhibit proteasomes, leading to a growth defect (30). Expression of Ub+1 thus could lead to the accumulation of stable polyUb chains, causing inhibition of proteasomes.
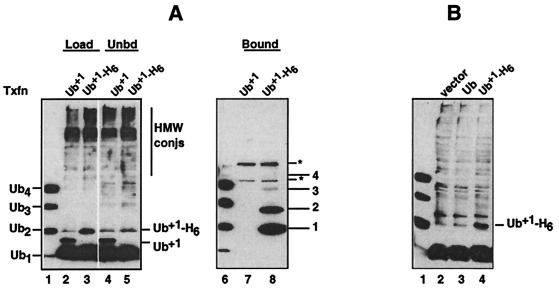
To demonstrate intracellular assembly of Ub+1-capped chains, we transfected 293T cells with a gene encoding Ub+1, with or without a C-terminal hexahistidine tag (Fig. 1A). Western blot analysis of transfected cell lysates (with Ub antibodies) showed that each protein was expressed (Fig. 2A, lanes 2 and 3); the bands designated Ub+1 and Ub+1-H6 in Fig. 2 were absent in lysates of cells transfected with empty vector or a control plasmid specifying wild-type Ub (Fig. 2B). In cells transfected with the Ub+1-H6 gene, analysis of proteins retained on nickel beads showed the presence of higher forms of Ub+1 with the mobilities expected for Ub2+1 and Ub3+1, with a faint band at the position of Ub4+1 (Fig. 2A, lane 8). Specificity was demonstrated by the absence of these species in the bound fraction from cells that received the untagged Ub+1 gene (Fig. 2A, lane 7). We conclude that Ub+1 is indeed assembled into polyUb chains within cells, perhaps by a chain assembly pathway that efficiently uses free, monomeric Ub as a substrate (31). The abundance of Ub+1-capped chains, although low, is striking in view of the weak expression of Ub+1 (Fig. 2A, lanes 2 and 3) and the short duration of the experiment (26 h). Our results therefore suggest that Ub+1-capped chains could accumulate to significant levels in aging brain even though molecular misreading is an inefficient process (17–19).
Figure 2.
Assembly of Ub+1-capped chains in mammalian cells (Ub blots). (A) Experiment. Human embryonic kidney 293T cells were transfected with plasmids specifying unmodified Ub+1 (lanes 2, 4, and 7) or Ub+1-H6 (lanes 3, 5, and 8). (Left) 200 μg of soluble extract protein was incubated with 10 μl of nickel beads; 10% of the starting lysate (lanes 2 and 3) and unbound fraction (lanes 4 and 5) were analyzed. The entire bound fraction deriving from 675 μg of lysate protein was analyzed in lanes 7 and 8. * denote cross-reacting bands that bound to the beads. Lanes 1 and 6, mixture of authentic polyUb chains. Ub+1-H6 (band 1) and presumptive Ub+1-H6-capped chains (bands 2–4) are indicated on the right. Conjs-conjugates. (B) Controls. Human 293T cells were transfected with empty vector (lane 2), vector specifying wild-type Ub (lane 3), and vector specifying Ub+1-H6 (lane 4). Soluble extracts were analyzed as in lanes 2 and 3 of A; lane 1, chain standard. The band in lanes 2 and 3, which comigrates with Ub+1-H6 and which does not bind to nickel beads (lanes 3 versus 5, A), is probably wild-type Ub2.
Ub+1-Capped Chain Strongly Resists Disassembly.
As discussed above, the facile accumulation of Ub+1-capped chains in 293T cells (Fig. 2) may reflect resistance to disassembly by cellular deubiquitinating enzymes because of replacement of G76 with the +1 peptide. To test this hypothesis directly, we used a purified Ub conjugating enzyme to link preassembled Ub4 to K48 in Ub+1 (see Materials and Methods). The purified product, termed Ub5+1, carried one Ub+1 moiety at its proximal terminus (Fig. 3A). We first determined whether Ub5+1 could be disassembled by isopeptidase T, which is the principal enzyme responsible for the disassembly of unanchored polyUb chains (21, 30). As shown in Fig. 3B, Ub5+1 was not disassembled by purified isopeptidase T at any concentration of this enzyme tested. In contrast, isopeptidase T readily processed the same concentration of Ub4 (Fig. 3C). Ub5+1 also was disassembled ≈100-fold more slowly than Ub4 in 293T cell extract (Fig. 3D), indicating that other deubiquitinating enzymes (26) are also deficient in dismantling Ub+1-capped chains. The extreme stability of Ub5+1 (Fig. 3) suggests a high likelihood that Ub+1-capped chains will accumulate in cells, even though they are likely to be generated at a slow rate (above).
Ub+1-Capped PolyUb Chain Potently Inhibits 26S Proteasomes.
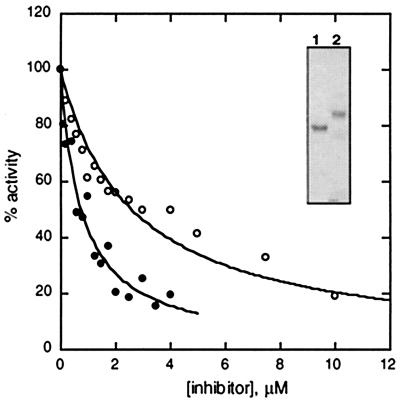
We next tested the ability of Ub5+1 to inhibit the degradation of a polyubiquitinated model substrate (5) by purified 26S proteasomes. As shown in Fig. 4 (●) Ub5+1 was a potent inhibitor, with a Ki of 350 nM. The potency of Ub5+1 was approximately 3-fold greater than that of Ub4 (Fig. 4, ● versus ○). The proteasomal signaling potential of K48-linked polyUb chains is length-dependent; Ub5 is expected to bind about 2-fold more tightly than Ub4 (5). The slightly augmented affinity of Ub5+1, in comparison to a normal chain of the same length, may reflect a small effect caused by the absence of a carboxylate at residue 76 (5).
Figure 4.
Ub5+1 potently inhibits 26S proteasomes. Purified 26S proteasomes were incubated with 35S-Ub5 dihydrofolate reductase, with or without the indicated chain inhibitor (○, Ub4; ●, Ub5+1) as described in Materials and Methods. Initial rates of degradation are expressed relative to a control without inhibitor. The lines are fits assuming competitive inhibition (see Materials and Methods), with Ki = 350 ± 30 nM (Ub5+1) or 1.2 ± 0.08 μM (Ub4). (Inset) Chain inhibitors (Coomassie-stained gel): 1 μg Ub4 (lane 1); 0.75 μg Ub5+1 (lane 2).
Discussion
Our finding that Ub+1 is polyubiquitinated to form stable polyUb chains provides a mechanistic paradigm for interpreting prior findings (14–17). Our results indicate that unmodified Ub+1 is unlikely to be detrimental (Fig. 1); in contrast, polyubiquitinated forms of Ub+1 potently inhibit proteasomes (Fig. 4), providing a likely mechanism of toxicity. Age-dependent accumulation of Ub+1-capped chains is predicted based on facile polyubiquitination of Ub+1 (Fig. 2) and strong resistance of the resulting chains to disassembly (Fig. 3). Taken together, our results demonstrate a mechanism whereby an AD/age-related change in the properties of the Ub-proteasome system could lead to neuropathological consequences. Because inhibition by the mechanism described here relies on the modification of the mutant protein by wild-type Ub, Ub+1 should act in a dominant negative fashion. This feature is critical in view of the low efficiency of the misreading process (17–19). Van Leeuwen and coworkers (17) estimated that frame-shifted UbB transcripts comprise between 1% and 10% of the total UbB transcripts in the brains of AD patients. Our finding that Ub+1-capped polyUb chains are 100-fold more resistant to disassembly than normal chains suggests that the aberrant chains could accumulate to significant levels in the brain despite the low efficiency of molecular misreading.
We have not yet detected Ub+1-capped chains in brain extracts from AD patients, a result that we attribute to slow postmortem disassembly of the aberrant chains (compare Fig. 3D). Consistent with this explanation, we found that normal polyUb chains and Ub-conjugated proteins are virtually absent from such extracts (data not shown). The detection of polyubiquitinated Ub+1 in neurons is thus likely to require the development of a specific cell culture or animal model. The availability of such a model also should allow demonstration of the intracellular proteasome inhibition that should occur upon accumulation of capped chains that are four or more Ubs long (5, 30).
The presenilins, which positively regulate the γ-secretase (Aβ generating) cleavage of amyloid precursor protein (8, 9), are substrates of the Ub-proteasome system (10–13). It is therefore possible that a progressive, age-dependent inhibition of 26S proteasomes by Ub+1-capped chains could contribute to AD pathogenesis through effects on presenilin levels. Additionally, proteasome inhibitors cause the formation of Ub-containing “aggresomes” in cultured cells (13)—inclusions that may be functionally related to the filamentous Ub-containing inclusions seen in the brains of patients with neurodegenerative diseases (14–17). Finally, inhibition of proteasomes would inhibit the degradation of misfolded proteins of the endoplasmic reticulum, leading to an increased demand on resident chaperones of this compartment (reviewed in ref. 32) and the possibility of increased amyloid precursor protein processing through competitive effects at the level of BiP/Grp78 (33). Inhibition of 26S proteasomes thus might contribute to both Aβ and tau pathology. The presence of Ub+1 in tau-related neurofibrillary tangles in progressive supranuclear palsy (34) suggests that a similar mechanism could contribute to pathogenesis in other neurodegenerative diseases. We note that gracile axonal dystrophy is caused by an intragenic deletion in the gene encoding a deubiquitinating enzyme (35). Inactivation of this enzyme could lead to elevated levels of linear polyUb, which also inhibits proteasomal proteolysis (5). Inhibition of proteasomes thus could contribute to neuropathogenesis through pathways that are related to, but distinct from, the mechanism documented here.
Acknowledgments
We thank M. Vassileva for assistance in preliminary studies and M. Bownes for the GST-UCH-D clone. We are grateful to D. Finley, R. Cohen, J. Bender, and M. Matunis for comments on the manuscript. This research was supported by a grant from the National Institutes of Health (DK46984). R.L. is a Research in Aging Queen Elizabeth the Queen Mother Research Fellow; Y.A.L. is a Fellow of The Leukemia and Lymphoma Society; and C.J. was the recipient of an award from the Biotechnology and Biological Sciences Research Council.
Abbreviations
- AD
Alzheimer's disease
- E1
ubiquitin activating enzyme
- GST
glutathione S-transferase
- Ub
ubiquitin
- UCH-D
Ub carboxyl-terminal hydrolase-D
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.170173897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.170173897
References
- 1.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Varshavsky A. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 3.Chau V, Tobias J W, Bachmair A, Marriott D, Ecker D J, Gonda D K, Varshavsky A. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 4.Pickart C M. In: Ubiquitin and the Biology of the Cell. Peters J-M, Harris J R, Finley D, editors. New York: Plenum; 1998. pp. 19–64. [Google Scholar]
- 5.Thrower J S, Hoffman L, Rechsteiner M, Pickart C M. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciechanover A, Finley D, Varshavsky A. Cell. 1984;37:57–66. doi: 10.1016/0092-8674(84)90300-3. [DOI] [PubMed] [Google Scholar]
- 7.Price D L, Sisodia S S. Annu Rev Neurosci. 1998;21:479–505. doi: 10.1146/annurev.neuro.21.1.479. [DOI] [PubMed] [Google Scholar]
- 8.De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. Nature (London) 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 9.Wolfe M S, Xia W, Ostaszewsi B L, Diehl T S, Kimberly W T, Selkoe D J. Nature (London) 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 10.Kim T-W, Pettingell W H, Hallmark O G, Moir R D, Wasco W, Tanzi R E. J Biol Chem. 1997;272:11006–11010. doi: 10.1074/jbc.272.17.11006. [DOI] [PubMed] [Google Scholar]
- 11.Steiner H, Capell A, Pesold B, Citron M, Kloetzel P M, Selkoe D J, Romig H, Mendla K, Haass C. J Biol Chem. 1998;273:32322–32331. doi: 10.1074/jbc.273.48.32322. [DOI] [PubMed] [Google Scholar]
- 12.Marambaud P, Ancolio K, Lopez-Perez E, Checler F. Mol Med. 1998;4:147–157. [PMC free article] [PubMed] [Google Scholar]
- 13.Johnston J A, Ward C L, Kopito R R. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morishima-Kawashima M, Hasegawa M, Takio K, Suzuki M, Titani K, Ihara Y. Neuron. 1993;10:1151–1160. doi: 10.1016/0896-6273(93)90063-w. [DOI] [PubMed] [Google Scholar]
- 15.Lowe J, Mayer R J, Landon M. Brain Pathol. 1993;3:55–65. doi: 10.1111/j.1750-3639.1993.tb00726.x. [DOI] [PubMed] [Google Scholar]
- 16.Sisodia S S. Cell. 1998;95:1–4. doi: 10.1016/s0092-8674(00)81743-2. [DOI] [PubMed] [Google Scholar]
- 17.van Leeuwen F W, de Kleijn D P V, Van den Hurk H H, Neubauer A, Sonnemans M A F, Sluijs J A, Koycu S, Ramdjielal R D J, Salehi A, Martens G J M, et al. Science. 1998;279:242–247. doi: 10.1126/science.279.5348.242. [DOI] [PubMed] [Google Scholar]
- 18.van Leeuwen F W, Burbach J P, Hol E M. Trends Neurosci. 1998;21:331–335. doi: 10.1016/s0166-2236(98)01280-6. [DOI] [PubMed] [Google Scholar]
- 19.van Leeuwen F, van der Beek E, Seger M, Burbach P, Ivell R. Proc Natl Acad Sci USA. 1989;86:6417–6420. doi: 10.1073/pnas.86.16.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Layfield R, Franklin K, Landon M, Walker G, Wang P, Ramage R, Brown A, Love S, Urquart K, Muir T, et al. Anal Biochem. 1999;274:40–49. doi: 10.1006/abio.1999.4234. [DOI] [PubMed] [Google Scholar]
- 21.Wilkinson K D, Tashayev V L, O'Connor L B, Larsen C N, Kasperek E M, Pickart C M. Biochemistry. 1995;34:14535–14546. doi: 10.1021/bi00044a032. [DOI] [PubMed] [Google Scholar]
- 22.Hoffman L, Pratt G, Rechsteiner M. J Biol Chem. 1992;267:22362–22368. [PubMed] [Google Scholar]
- 23.Whitby F G, Xia G, Pickart C M, Hill C P. J Biol Chem. 1998;273:34983–34991. doi: 10.1074/jbc.273.52.34983. [DOI] [PubMed] [Google Scholar]
- 24.Shockett P, Difilippantonio M, Hellman N, Schatz D G. Proc Natl Acad Sci USA. 1995;92:6522–6526. doi: 10.1073/pnas.92.14.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haas A L, Bright P M. J Biol Chem. 1985;260:12464–12473. [PubMed] [Google Scholar]
- 26.Wilkinson K D, Hochstrasser M. In: Ubiquitin and the Biology of the Cell. Peters J-M, Harris J R, Finley D, editors. New York: Plenum; 1998. pp. 99–126. [Google Scholar]
- 27.Johnson E S, Bartel B, Seufert W, Varshavsky A. EMBO J. 1992;11:497–505. doi: 10.1002/j.1460-2075.1992.tb05080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodgins R R W, Ellison K S, Ellison M J. J Biol Chem. 1992;267:8807–8812. [PubMed] [Google Scholar]
- 29.Pickart C M, Kasperek E M, Beal R, Kim A. J Biol Chem. 1994;269:7115–7123. [PubMed] [Google Scholar]
- 30.Amerik A Y, Swaminathan S, Krantz B A, Wilkinson K D, Hochstrasser M. EMBO J. 1997;16:4826–4838. doi: 10.1093/emboj/16.16.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mastrandrea L D, You J, Niles E G, Pickart C M. J Biol Chem. 1999;274:27299–27306. doi: 10.1074/jbc.274.38.27299. [DOI] [PubMed] [Google Scholar]
- 32.Mori K. Cell. 2000;101:451–454. doi: 10.1016/s0092-8674(00)80855-7. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y, Turner R S, Gaut J R. J Biol Chem. 1998;273:25552–25555. doi: 10.1074/jbc.273.40.25552. [DOI] [PubMed] [Google Scholar]
- 34.Fergusson J, Landon M, Lowe J, Ward L, van Leeuwen F W, Mayer R J. Neurosci Lett. 2000;279:69–72. doi: 10.1016/s0304-3940(99)00917-9. [DOI] [PubMed] [Google Scholar]
- 35.Saigoh K, Wang Y-L, Suh J-G, Yamanishi T, Sakai Y, Kiyosawa H, Harada T, Ichihara N, Wakana S, Kikuchi T, Wada K. Nat Genet. 1999;23:47–51. doi: 10.1038/12647. [DOI] [PubMed] [Google Scholar]