SUMMARY
Mitotic spindle assembly and maintenance relies on Kinesin-5 motors that act as bipolar homotetramers to cross-link microtubules [1–5]. Kinesin-5 motors have been subject to extensive structure-function analysis [5], but the regulation of their activity in the context of mitotic progression remains less well understood [2]. We report that Drosophila Kinesin-5 (KLP61F) is regulated by Drosophila Wee1 (dWee1). Wee1 tyrosine kinases are known to regulate mitotic entry via inhibitory phosphorylation on Cdk1 [6–10]. Recently, we showed that dWee1 also plays a role in mitotic spindle positioning through γ-tubulin and spindle fidelity through an unknown mechanism [11]. Here we investigated whether a KLP61F-dWee1 interaction could explain the latter role for dWee1. We found that dWee1 phosphorylates KLP61F in vitro on three tyrosines within the head domain, the catalytic region that mediates movement along microtubules. In vivo, KLP61F with tyrosine-to-phenylalanine mutations fails to complement a klp61f mutant and dominantly induces spindle defects similar to ones seen in dwee1 mutants. We propose that phosphorylation of the KLP61F catalytic domain by dWee1 is important for the motor’s function. This study identifies a second substrate for a Wee1 kinase and provides evidence for phospho-regulation of a kinesin in the head domain.
RESULTS AND DISCUSSION
dWee1 and KLP61F interact at endogenous protein levels
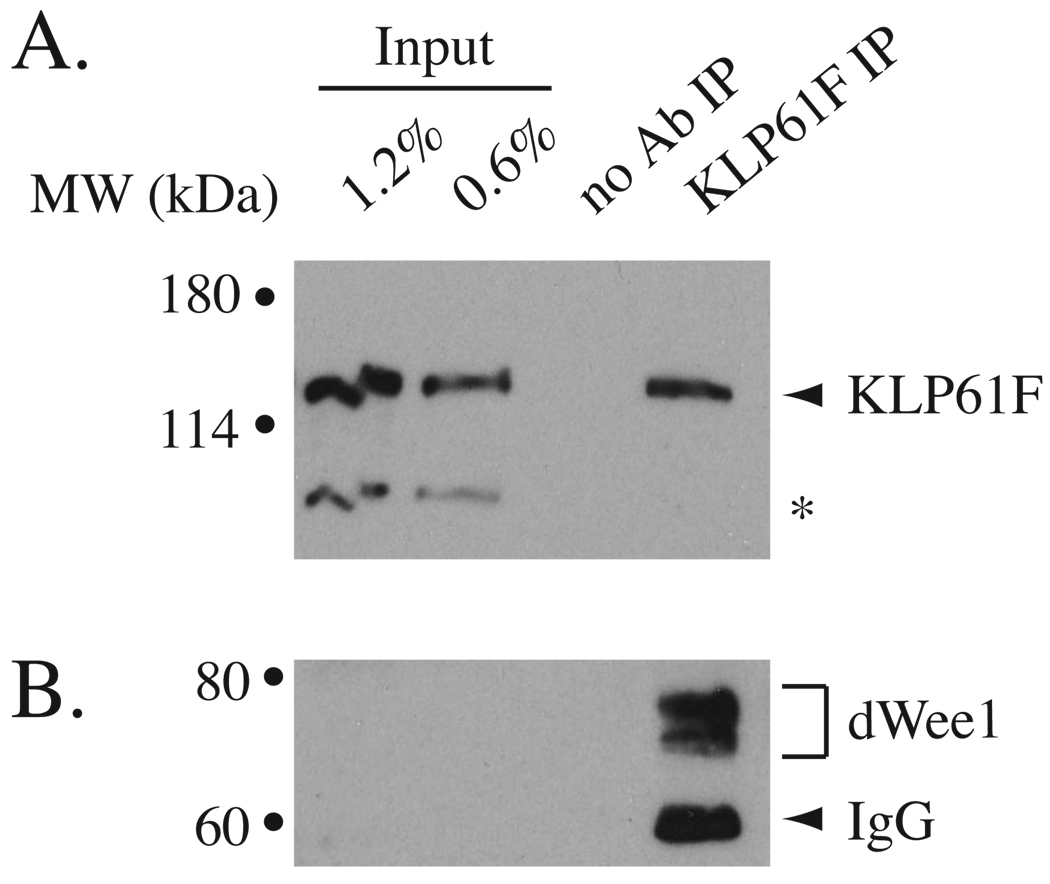
Previously, we identified KLP61F in mass spectrometric analysis of HA-dWee1-containing protein complexes [11] (data not shown). To confirm that dWee1 and KLP61F interact at physiological levels, we assayed for co-immunoprecipitation of endogenous proteins (Figure 1). The specificity of a previously described anti-KLP61F serum [12] and a newly generated anti-dWee1 antibody were first confirmed with western blots of extracts from respective mutants (Figure S1). The anti-dWee1 antibody recognized a doublet (Figure S1C), similarly seen for human Wee1 [13]. This doublet was present in immunoprecipitates obtained with the anti-KLP61F serum (Figure 1). Also, the anti-dWee1 antibody immunoprecipitated dWee1 and co-precipitated KLP61F from syncytial embryos (data not shown).
Figure 1. Endogenous KLP61F and dWee1 interact.
Extracts from syncytial wild type embryos were immunoprecipitated with anti-KLP61F serum or mock-precipitated (no Ab IP) and western blotted for KLP61F (A) or dWee1 (B). Samples equivalent to 0.6% and 1.2% of starting extracts were loaded for comparison.
GST-dWee1 phosphorylates His-KLP61F within the head domain
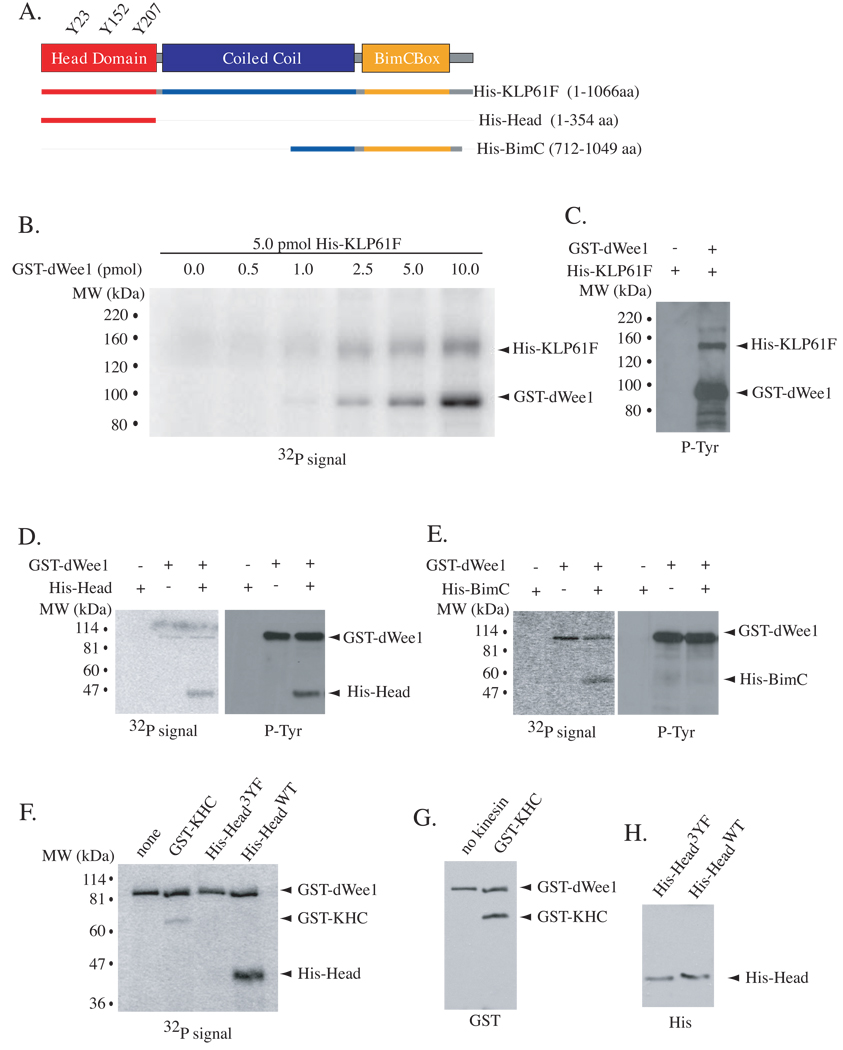
Because dWee1 is a tyrosine kinase, we next investigated whether dWee1 could phosphorylate KLP61F. Incubation of recombinant, purified His-KLP61F with GST-dWee1 resulted in phosphorylation (Figure 2B) and recognition of the former by an anti-phosphotyrosine antibody (Figure 2C). Autophosphorylation by dWee1 occurred as expected [14, 15]. Mass spectrometry was used to identify phosphorylated peptides of His-KLP61F after GST-dWee1 kinase assays. We achieved 57% coverage of KLP61F and identified 4 phosphopeptides containing a single tyrosine (see Experimental Procedures in Supplemental Data). Three of these peptides are in the head domain and contain Y23, Y152, or Y207 as their single tyrosine (Figures S1A–S1C). The fourth is in the BimC box, a conserved region of ~20 amino acids in the tail domain, and contains Y927. To confirm that these regions of KLP61F are important for phosphorylation by dWee1, we generated purified polypeptides containing either the head domain (His-HeadWT) or the BimC box (His-BimC; Figure 2A). Both were phosphorylated by GST-dWee1, but only His-HeadWT was recognized by an anti-phosphotyrosine antibody after phosphorylation by GST-dWee1 (Figures 2D–2E). Therefore, we focused our analysis on the head domain.
Figure 2. GST-dWee1 phosphorylates KLP61F on tyrosines in the head domain in vitro.
(A) Schematic representation of KLP61F with head domain (red), coiled-coil (blue) and tail domain that contains the BimC box (yellow). His-tagged polypeptides used in kinase assays are depicted. (B) His-KLP61F was incubated with varying amounts of GST-dWee1 in kinase reactions in vitro and analyzed for 32P incorporation. (C) Reaction with equimolar GST-dWee1 and His-KLP61F was western blotted with anti-phosphotyrosine antibody after in vitro kinase assays. (D–E) GST-dWee1 and His-KLP61F polypeptides were incubated in in vitro kinase reactions and analyzed for 32P incorporation (left panels) and subsequently by western blots for phosphotyrosines (right panels). (F) KLP61F head domain with Y23, Y152 and Y207 mutated to phenylalanines, His-Head3YF, was incubated with GST-dWee1 in in vitro kinase reactions and analyzed for 32P incorporation. none = no kinesin; GST-KHC = conventional Kinesin Heavy Chain head domain; His-HeadWT = wild type head domain. (G–H) Western blots for GST (G) or His tags (H) show equivalent amounts of substrates in kinase reactions in (F).
Tyrosine(s) in head domain are required for phosphorylation by dWee1
Mutation of the putative phospho-acceptor residues Y23, Y152, and Y207 in the head domain to phenylalanines resulted in greatly diminished phosphorylation by dWee1 (“3YF” mutant; Figures 2F and 2H). It is unlikely that these mutations resulted in protein mis-folding; mutant head domain binds ATP in preliminary studies (not shown) and, more importantly, full-length KLP61F with 3YF mutations shows activity in vivo (described in detail below). The head domain of Drosophila Kinesin-1 motor, Kinesin Heavy Chain (GST-KHC), lacks tyrosines at the corresponding residues and was also a poor substrate for dWee1 (Figure S1A–S1C, Figure 2F–2G). Thus, phosphorylation in the head domain by dWee1 appears specific to KLP61F. Of the three putative phospho-acceptor tyrosines in KLP61F, Y207 is conserved in metazoan Kinesin-5 motors (Figure S1C). Alignment of residues flanking Y207 of Kinesin-5 motors and Y15 of Cdk1 homologs from Drosophila, human and Xenopus reveal several conserved residues (Figure S1D). These may comprise a Wee1 consensus if Y207 serves as a phospho-acceptor for Wee1 in other Kinesin-5 motors.
Potential phospho-acceptor tyrosine(s) are important in vivo
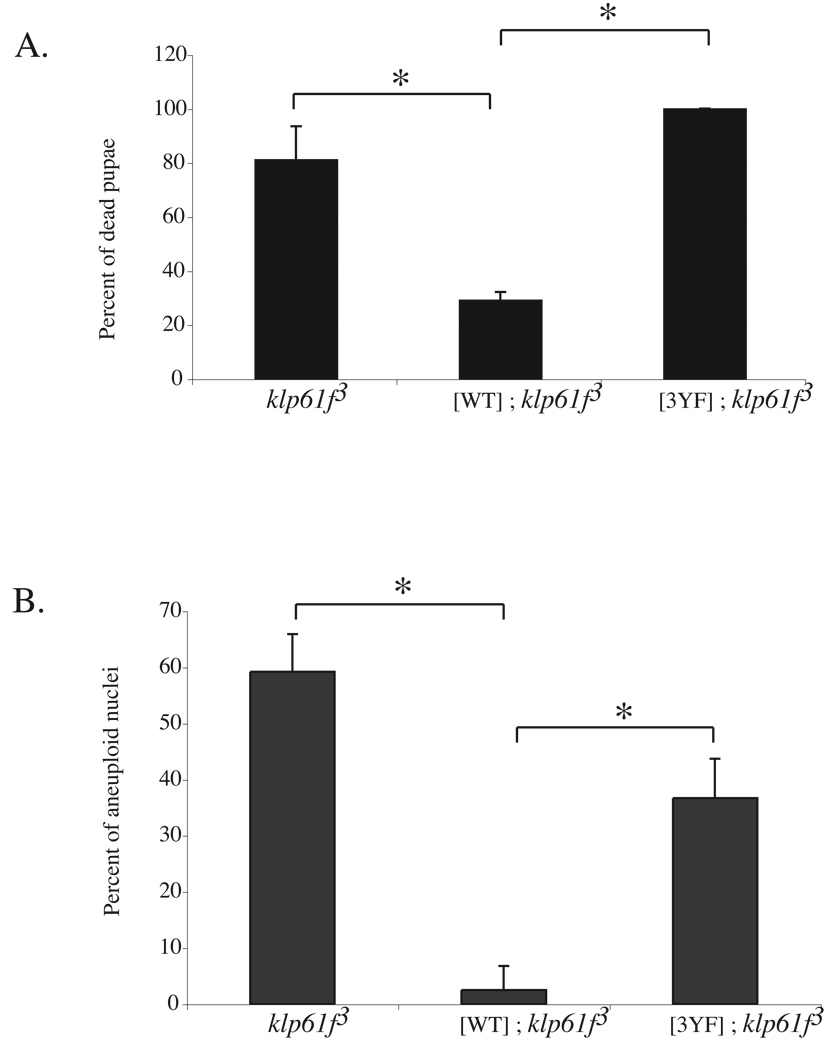
klp61f3 is a well characterized loss-of-function allele with a transposon insertion in the 5’UTR that greatly reduces protein expression [16, 17] (Figure S1A). Neuroblasts from klp61f3 homozygous larvae have mitotic spindle defects, chromosome segregation failure, and polyploid nuclei that result in death at the pupal stage [16, 17]. A full-length Myc-KLP61FWT transgene expressed constitutively from the ubiquitin promoter has been shown to rescue the lethality and cytological defects of klp61f3 homozygous larvae [16]. To determine the importance of Y23, Y152, and Y207 in vivo, we used an identical expression system to express a full-length Myc-KLP61F3YF. Of the 5 independent transgenic lines generated, we analyzed one that expresses Myc-KLP61F3YF at similar levels to Myc-KLP61FWT (Figure S1A). Unlike Myc-KLP61FWT, Myc-KLP61F3YF did not rescue the lethality of klp61f3 homozygotes or the incidence of polyploidy in larval neuroblasts (Figures 3A–3B), suggesting that Myc-KLP61F3YF has reduced function. These data indicate that Y23, Y152, and/or Y207 are important for KLP61F function in larval cells.
Figure 3. Phospho-accepting tyrosines in the head domain are important in vivo.
(A–B) Pupal lethality of klp61f3 mutants (A) and level of aneuploidy in klp61f3 mutant neuroblasts (B) are rescued by a Myc-KLP61FWT transgene ([WT]; klp61f3) but not a Myc-KLP61F3YF transgene ([3YF]; klp61f3). Data is represented as percentage of dead pupae (mean +/− SD from 3 embryo collections) in (A) and as percentage of aneuploid nuclei (mean +/− SD of nuclei from 3 larval brains) in (B). * indicates p < 0.01.
Embryos with reduced klp61f have spindle defects
Next, we asked whether Y23, Y152, and Y207 are also important for KLP61F function in embryonic syncytial cycles where dwee1 has been implicated in maintaining spindle fidelity [11]. Drosophila embryogenesis begins with 13 nuclear divisions driven by maternally contributed products. These divisions occur synchronously in a common cytoplasm called a syncytium and alternate between S and M phases without gap phases. The first nine syncytial divisions occur in the interior of the embryo (interior divisions), after which nuclei migrate outward such that cycles 11–13 occur in a monolayer directly beneath the cortex (cortical divisions). Drosophila embryos that lack maternally provided dwee1 die as syncytial embryos, consistent with other Wee1 homologs being essential for metazoan embryogenesis [18–20].
Embryos from Drosophila females that are hemizygous for the strongest extant allele of dwee1, dwee1ES1, (to be called “dwee1 mutant embryos”) show multiple spindle defects [10, 11]. As expected, these defects included ones known to result from elevated Cdk1 activity and premature mitotic entry. Surprisingly, these defects also included two “dwee1-specific defects” that could not be explained by premature mitotic entry or bulk elevation in Cdk1 activity: multiple microtubule organizing centers (MTOCs) within a single spindle and microtubule spurs that extend to neighboring spindles leading to collisions [11].
To investigate a possible link between dWee1 and KLP61F during syncytial cycles, we wanted to compare mitotic spindles in dwee1 mutant embryos to mitotic spindles in embryos with reduced KLP61F. Homozygotes of strong klp61f mutants do not survive to adulthood [17], but a recent study showed that syncytial embryos from klp61f3 heterozygous mothers show a phenotype, namely, abnormal spindle length [1]. Therefore, we examined these embryos (to be called “klp61f embryos”) for possible additional spindle defects. We found that klp61f embryos have reduced KLP61F (Figure S3) and ~20% of spindles were abnormal (Table 1). While the incidence of spindle defects was unexpectedly high for embryos that typically survive to adulthood, the majority of these problems occurred during the interior divisions. Defective syncytial nuclei are typically culled into the interior yolk mass and do not contribute to subsequent embryogenesis [21], which may explain the viability of klp61f embryos. The most frequent defects were anastral and monopolar spindles (Figures 4B, 4C; quantified in 4D; Table 1 includes a complete listing of all spindle defects). Such defects have been reported in larval neuroblasts from klp61f mutants or embryos injected with anti-KLP61F antibodies [17, 22].
Table 1.
Percent of Spindle Defects in Syncytial Embryos
| Wild-Type | klp61f | [WT]g | [3YF]h | |
|---|---|---|---|---|
| Interior Divisions (Cycles 1–9) | n=201 | n=532 | n=287 | n=410 |
| Monastrala | 0.5 | 1.88 | 0 | 0 |
| Anastralb | 0.5 | 4.89 * | 0.7 * | 0 |
| Promiscious MTsc | 0 | 0.75 | 0.35 | 0.49 |
| Multi MTOCd | 1.99 | 3.76 | 5.57 | 11.75 * |
| Monopolare | 0 | 6.77 ** | 0.35 ** | 8.29 ** |
| TOTAL | 2.99 | 18.05 | 6.97 | 20.53 |
| Cortical Divisions (Cycles 11–13) | n=118 | n=104 | n=191 | n=175 |
| Monopolar | 0.85 | 0 | 0.52 | 1.14 |
| Promiscuous MTs (Meta) | 0 | 1.83 | 4.19 | 1.71 |
| Promiscuous MTs (Ana) | 0 | 0 | 2.09 | 6.86 ^ |
| TOTAL | 0.85 | 1.83 | 7.78 | 9.71 |
| TOTAL DEFECTS | 3.84 | 19.88 | 14.75 | 30.24 |
n represents the number of spindles assayed from 30 (interior) or 3 (cortical) embryos
bipolar spindles with only one centrosome
bipolar spindles with no centrosomes
microtubule spurs or interactions between spindles
bipolar spindles with multiple MTOCs
monopolar spindles
statistically compared to wild type embryos
statistically compared to klp61f embryos
statistically compared to [WT] embryos
MT = microtubules
0.001>p
0.01>p
0.05>p
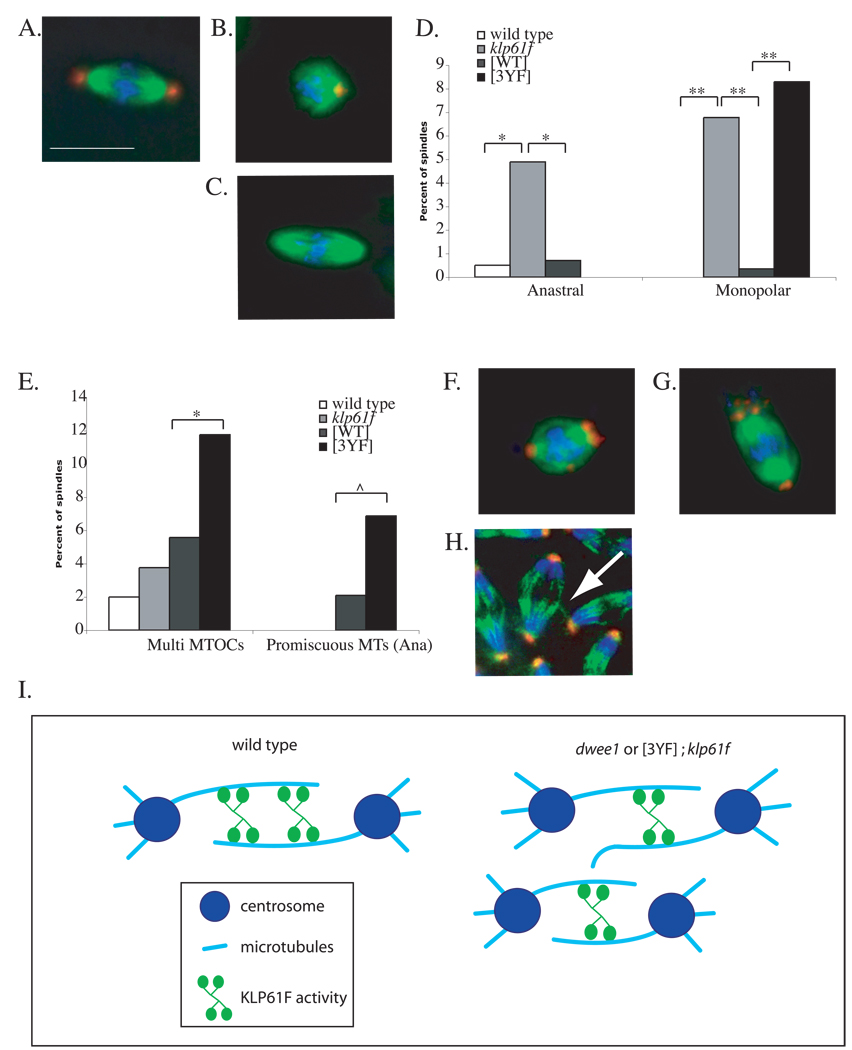
Figure 4. Mitotic spindle defects in embryos containing KLP61FWT or KLP61F3YF.
Embryos from mothers that are wild type, klp61f3 heterozygotes (klp61f), klp61f3 heterozygotes with Myc-KLP61FWT ([WT]) and klp61f3 heterozygotes with Myc-KLP61F3YF ([3YF]) were fixed and stained for DNA (blue), α-tubulin (green), and centrosomin (red). Representative images show a normal spindle (A) and defects significantly different among the genotypes: a monopolar spindle (B), an anastral spindle (C), and spindles with multiple MTOCs (F–G) from interior divisions, and a promiscuous microtubule interaction during anaphase in cortical divisions (H). Scale bar = 5 µm. Defects in (A–C) and (F–H) are quantified in (D) and (E) respectively. See Table 1 for numbers. * p < 0.01, ** p < 0.001, and ^ p < 0.05. (I) A model for regulation of KLP61F to maintain mitotic spindle integrity and prevent promiscuous MT interactions. dWee1 may regulate KLP61F activity to bundle parallel (not shown) and/or anti-parallel microtubules to create a more robust spindle (left). Without dWee1 regulation, KLP61F activity is reduced on the spindle, leading to an unstable spindle with microtubule spurs. These microtubule spurs can then interact with neighboring spindles in a syncytium (spur-spindle interaction is not depicted). Reduced KLP61F activity on the spindle is depicted as reduced protein levels on the spindle, but it remains possible that similar level of protein associate with the spindle but display reduced activity.
Potential phospho-acceptor tyrosines are important for the prevention of monopolar spindles
Restoring KLP61F with a maternal transgene encoding Myc-KLP61FWT rescued both anastral and monopolar spindles in klp61f3 embryos (“[WT]” in Figure 4D). To date, it remains unclear what aspect of KLP61F function normally prevents anastral spindles [17]. Nonetheless, anastral spindles in klp61f3 embryos were similarly reduced when mothers carried a transgene encoding Myc-KLP61F3YF (“[3YF]” in Figure 4D). This suggests that Myc-KLP61F3YF is at least partially active. Consistent with this idea, Myc-KLP61F3YF also localized to mitotic spindles similar to endogenous KLP61F and Myc-KLP61FWT(Figure S3B) [12]. These data, together with the preliminary finding that His-Head3YF can bind ATP in vitro (data not shown), suggest strongly that the 3YF mutations do not create a general protein-folding problem for KLP61F.
The Myc-KLP61F3YF transgene, however, did not rescue the monopolar spindle defect in klp61f3 embryos (“[3YF]” in Figure 4D). The role of KLP61F in the prevention of monopolar spindles has been explained by its ability to antagonize minus-end directed motors [4, 22]. In the absence of KLP61F, the latter brings separated centrosomes together; this collapses the spindle from a bipolar to a monopolar structure [22]. Consistent with the idea that monopolar spindles in klp61f3 embryos result from reduced KLP61F, such defects were rescued in [WT]; klp61f3 embryos (Figure 4D). Monopolar spindles, however, were not rescued in [3YF]; klp61f3 embryos (Figure 4D). Thus, potential phospho-acceptor tyrosines in the KLP61F head domain appear to be important for this aspect of KLP61F function.
KLP61F3YF induces spindle defects in a dominant manner
The presence of maternal Myc-KLP61F3YF produced two additional spindle defects in klp61f embryos. These defects were also seen in dwee1 mutant embryos and are ‘dwee1-specific’, that is, independent of Cdk1 mis-regulation [11]. One defect was multiple MTOCs per spindle, as defined by staining for centrosomin (Figures 4F and 4G). The incidence of spindles with multiple MTOCs was not significantly different among embryos from wild type, klp61f3/+ and klp61f3/+ mothers carrying the Myc-KLP61FWT transgene (“wild type”, “klp61f” and “[WT]” respectively in Figure 4E). In contrast, embryos from klp61f3/+ mothers carrying the Myc-KLP61F3YF transgene displayed a significantly higher incidence of spindles with multiple MTOC (p <0.01) (“[3YF]” in Figures 4E; Table 1). Spindles with multiple MTOCs were also seen in dwee1 mutant embryos both during interior and cortical divisions ([11] and data not shown).
The second dwee1-specific defect induced by Myc-KLP61F3YF in klp61f3 embryos was promiscuous microtubules, which included microtubule spurs and microtubules interacting between neighboring spindles, during cortical divisions (Figure 4H). Since embryo size remains constant, increases in nuclear density leads to crowding of spindles. During cortical divisions, cortical actin descends to form ‘furrows’ separating neighboring spindles. These actin furrows retract during metaphase and are absent in subsequent stages of mitosis, and yet spindles do not collide [23]. We had proposed that dWee1 plays a role, via phospho-regulation of γ-tubulin, in positioning nuclei/spindles within the protection of these furrows to prevent spindle collisions [11]. However, in dwee1 mutant embryos, spindles collide throughout mitosis [11], suggesting that dWee1 acts in another, furrow-independent mechanism that normally prevents spindle interactions after metaphase.
During cortical divisions, we saw promiscuous microtubules in metaphase in klp61f3 embryos whether or not the mother carried a transgene; the incidences were not significantly different among klp61f3, [WT]; klp61f3 and [3YF]; klp61f3 embryos (Table 1). However, in anaphase, [3YF]; klp61f3 embryos, but not [WT]; klp61f3 embryos, showed a significant increase in spindles with promiscuous microtubules (p < 0.05) (Figure 4E, Table 1). This could be because in [WT]; klp61f3 embryos, regulation by dWee1 ensures optimal activation of transgenic KLP61F and proper spindle integrity (Model in Figure 4I). In [3YF]; klp61f3 embryos, the level of transgenic KLP61F is similar to that in [WT]; klp61f3 embryos, but the lacks regulation by dWee1. Further more, it appears that having elevated KLP61F that cannot be regulated (in [3YF]; klp61f3) is worse than having reduced KLP61F that can be regulated because klp61f3 embryos do not show anaphase spindle interactions.
The incidence of promiscuous microtubules in [3YF]; klp61f3 embryos at ~7% was lower than the ~28% in dwee1 mutant embryos (Table 1) [11]. This could be because dWee1 acts through both KLP61F and nuclear/spindle positioning [11] to prevent collisions. Also, the presence of endogenous KLP61F may prevent a stronger effect by Myc-KLP61F3YF. Since KLP61F forms tetramers [2, 4], Myc-KLP61F3YF may complex with endogenous KLP61F. The resulting complex could be compromised in function and could also explain why Myc-KLP61F3YF acts in a dominant negative manner.
These results lead us to conclude that mutation of tyrosines in KLP61F, which are important for in vitro phosphorylation by dWee1, phenocopies spindle defects seen in dwee1 mutant embryos in vivo. We suggest that in the absence of phosphorylation by dWee1, Myc-KLP61F3YF is less functional. This would explain the failure of Myc-KLP61F3YF to rescue monopolar spindles in klp61f3 embryos or to rescue polyploidy in klp61f3 mutant larvae. Reduced ability of KLP61F to cross-link and translocate along microtubules would result in loss of spindle bipolarity and chromosome segregation failure. In addition, a decreased ability of Myc-KLP61F3YF to cross-link microtubules may lead to reduced organization within the spindle. Resulting microtubule spurs could then interact with neighboring spindles in a syncytium, especially during anaphase when spindles are no longer protected by actin furrows.
Multiple roles for dWee1
Based on these and previous data, dWee1 appears to regulate mitosis in three ways during syncytial divisions. It (negatively) regulates Cdk1 to time the entry into mitosis [10]; it regulates γ-TuRC, perhaps indirectly, to position the mitotic spindle directly beneath the cortex [11]; finally, it (positively) regulates KLP61F to preserve spindle integrity and prevent microtubule interaction between neighbors (Figure 4I). Regulation of KLP61F by dWee1, we propose, is important for bundling parallel and/or anti-parallel microtubules and, consequently, generating a robust bipolar spindle. Without dWee1 regulation, KLP61F activity would be reduced, leading to unstable spindles, microtubule spurs, and interaction between neighboring spindles in a syncytium.
Outside the syncytium, mitotic regulation by dWee1 also appears to be important, particularly, during somatic cell cycles when the phospho-acceptor tyrosines of KLP61F are required for larval viability and chromosome segregation in neuroblasts. Consistent with this idea, dWee1 is also necessary to prevent mitotic defects in neuroblasts [10]. dwee1, however, is dispensable for larval growth as homozygous or hemizygous dwee1 mutants survive to adulthood, whereas Myc-KLP61F3YF cannot support larval growth. We speculate that, another kinase may partially substitute for dWee1 in the larva. Precedence for overlapping function between Wee1 and another kinase (Myt1) is seen in Cdk1 regulation [24].
Implications of phosphorylation within the KLP61F head domain
Our studies show that the head domain of KLP61F is phosphorylated by dWee1. Regulation of a specific kinesin via phosphorylation in this domain may appear unlikely since it is highly conserved among all kinesins. However, we found that modification by dWee1 may be specific to the KLP61F head domain since the KHC head domain was a poor substrate. Previous studies show that domains outside of the Kinesin-5 head domain are also phosphorylated [25–28]. Cdk1 phosphorylates Kinsin-5 on a threonine in the “BimC box”, and is important for spindle localization of the motor [3, 26, 29]. Nek6, a NIMA kinase, also phosphorylates Kinesin-5 in the tail domain, possibly to regulate its function at the spindle poles [28]. The coiled-coil domain of Xenopus Kinesin-5 is also phosphorylated on a non-conserved serine by Aurora kinase, but does not appear to be important for spindle assembly [27, 29]. If and how these phosphorylation events affect activity of the head domain remains unknown. Direct regulation via phosphorylation within catalytic regions is not unheard of. Indeed, Wee1 kinases inhibit Cdk1 activity by phosphorylation in the ATP-binding domain [30]. The structure of KLP61F remains to be solved, but in the structure for human Kinesin 5, the amino acid that correspond to Y207 of KLP61F, is located near regions involved in nucleotide-sensing [31] (Supplemental Figure 4). The amino acid that corresponds to another potential phospho-acceptor, Y152, is near regions of KLP61F important for microtubule interaction [32] (Supplemental Figure 4). Exactly how phosphorylation in the head domain by dWee1 alters KLP61F activity remains to be investigated, but it is intriguing that two of the potential phospho-acceptor tyrosines may reside near regions important for kinesin activity.
Supplementary Material
ACKNOWLEDGEMENTS
We thank David Sharp, Jon Scholey, Mark Winey and Pat O’Farrell for critical comments. We thank Jon Scholey for His-KLP61F, Andrea Pereira for Myc-KLP61F fly stock and Pwum2-KLP61F plasmid, Jane Atienza for anti-KLP61F serum and Tim Megraw for anti-CNN antibody. We thank Andreas Hoenger for help with the Eg5 structure display. This work was supported by a NIH RO1 grant to T. T. S. (GM66441) and a NIH pre-doctoral fellowship to K. G. (F31 CA117042-02).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL DATA
Supplemental Data include Supplemental Experimental Procedures and four figures, and can be found with this article online at http://www.
REFERENCES
- 1.Brust-Mascher I, Sommi P, Cheerambathur DK, Scholey JM. Kinesin-5-dependent Poleward Flux and Spindle Length Control in Drosophila Embryo Mitosis. Mol Biol Cell. 2009 doi: 10.1091/mbc.E08-10-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kashina AS, Rogers GC, Scholey JM. The bimC family of kinesins: essential bipolar mitotic motors driving centrosome separation. Biochim Biophys Acta. 1997;1357:257–271. doi: 10.1016/s0167-4889(97)00037-2. [DOI] [PubMed] [Google Scholar]
- 3.Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, Mitchison TJ, Scholey JM. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J Cell Biol. 1999;144:125–138. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tao L, Mogilner A, Civelekoglu-Scholey G, Wollman R, Evans J, Stahlberg H, Scholey JM. A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr Biol. 2006;16:2293–2302. doi: 10.1016/j.cub.2006.09.064. [DOI] [PubMed] [Google Scholar]
- 5.Valentine MT, Fordyce PM, Block SM. Eg5 steps it up! Cell Div. 2006;1:31. doi: 10.1186/1747-1028-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Booher RN, Deshaies RJ, Kirschner MW. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gould KL, Nurse P. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- 8.McGowan CH, Russell P. Human Wee1 kinase inhibits cell division by phosphorylating p34cdc2 exclusively on Tyr15. EMBO J. 1993;12:75–85. doi: 10.1002/j.1460-2075.1993.tb05633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mueller PR, Coleman TR, Dunphy WG. Cell cycle regulation of a Xenopus Wee1-like kinase. Mol Biol Cell. 1995;6:119–134. doi: 10.1091/mbc.6.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stumpff J, Duncan T, Homola E, Campbell SD, Su TT. Drosophila Wee1 kinase regulates Cdk1 and mitotic entry during embryogenesis. Curr Biol. 2004;14:2143–2148. doi: 10.1016/j.cub.2004.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stumpff J, Kellogg DR, Krohne KA, Su TT. Drosophila Wee1 interacts with members of the gammaTURC and is required for proper mitotic-spindle morphogenesis and positioning. Curr Biol. 2005;15:1525–1534. doi: 10.1016/j.cub.2005.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barton NR, Pereira AJ, Goldstein LS. Motor activity and mitotic spindle localization of the Drosophila kinesin-like protein KLP61F. Mol Biol Cell. 1995;6:1563–1574. doi: 10.1091/mbc.6.11.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe N, Arai H, Nishihara Y, Taniguchi M, Hunter T, Osada H. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc Natl Acad Sci U S A. 2004;101:4419–4424. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murakami MS, Copeland TD, Vande Woude GF. Mos positively regulates Xe-Wee1 to lengthen the first mitotic cell cycle of Xenopus. Genes Dev. 1999;13:620–631. doi: 10.1101/gad.13.5.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katayama K, Fujita N, Tsuruo T. Akt/protein kinase B-dependent phosphorylation and inactivation of WEE1Hu promote cell cycle progression at G2/M transition. Mol Cell Biol. 2005;25:5725–5737. doi: 10.1128/MCB.25.13.5725-5737.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heck MM, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LS. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J Cell Biol. 1993;123:665–679. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson PG, Fuller MT, Borisy GG. Monastral bipolar spindles: implications for dynamic centrosome organization. J Cell Sci. 1997;110(Pt 4):451–464. doi: 10.1242/jcs.110.4.451. [DOI] [PubMed] [Google Scholar]
- 18.Price D, Rabinovitch S, O'Farrell PH, Campbell SD. Drosophila wee1 has an essential role in the nuclear divisions of early embryogenesis. Genetics. 2000;155:159–166. doi: 10.1093/genetics/155.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murakami MS, Moody SA, Daar IO, Morrison DK. Morphogenesis during Xenopus gastrulation requires Wee1-mediated inhibition of cell proliferation. Development. 2004;131:571–580. doi: 10.1242/dev.00971. [DOI] [PubMed] [Google Scholar]
- 20.Tominaga Y, Li C, Wang RH, Deng CX. Murine Wee1 plays a critical role in cell cycle regulation and pre-implantation stages of embryonic development. Int J Biol Sci. 2006;2:161–170. doi: 10.7150/ijbs.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sibon OC, Kelkar A, Lemstra W, Theurkauf WE. DNA-replication/DNA-damage-dependent centrosome inactivation in Drosophila embryos. Nat Cell Biol. 2000;2:90–95. doi: 10.1038/35000041. [DOI] [PubMed] [Google Scholar]
- 22.Sharp DJ, Yu KR, Sisson JC, Sullivan W, Scholey JM. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat Cell Biol. 1999;1:51–54. doi: 10.1038/9025. [DOI] [PubMed] [Google Scholar]
- 23.Sullivan W, Theurkauf WE. The cytoskeleton and morphogenesis of the early Drosophila embryo. Curr Opin Cell Biol. 1995;7:18–22. doi: 10.1016/0955-0674(95)80040-9. [DOI] [PubMed] [Google Scholar]
- 24.Jin Z, Homola E, Tiong S, Campbell SD. Drosophila myt1 is the major cdk1 inhibitory kinase for wing imaginal disc development. Genetics. 2008;180:2123–2133. doi: 10.1534/genetics.108.093195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blangy A, Lane HA, d'Herin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- 26.Sawin KE, Mitchison TJ. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc Natl Acad Sci U S A. 1995;92:4289–4293. doi: 10.1073/pnas.92.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giet R, Uzbekov R, Cubizolles F, Le Guellec K, Prigent C. The Xenopus laevis aurora-related protein kinase pEg2 associates with and phosphorylates the kinesin-related protein XlEg5. J Biol Chem. 1999;274:15005–15013. doi: 10.1074/jbc.274.21.15005. [DOI] [PubMed] [Google Scholar]
- 28.Rapley J, Nicolas M, Groen A, Regue L, Bertran MT, Caelles C, Avruch J, Roig J. The NIMA-family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. J Cell Sci. 2008;121:3912–3921. doi: 10.1242/jcs.035360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cahu J, Olichon A, Hentrich C, Schek H, Drinjakovic J, Zhang C, Doherty-Kirby A, Lajoie G, Surrey T. Phosphorylation by Cdk1 increases the binding of Eg5 to microtubules in vitro and in Xenopus egg extract spindles. PLoS ONE. 2008;3:e3936. doi: 10.1371/journal.pone.0003936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atherton-Fessler S, Parker LL, Geahlen RL, Piwnica-Worms H. Mechanisms of p34cdc2 regulation. Mol Cell Biol. 1993;13:1675–1685. doi: 10.1128/mcb.13.3.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner J, Anderson R, Guo J, Beraud C, Fletterick R, Sakowicz R. Crystal structure of the mitotic spindle kinesin Eg5 reveals a novel conformation of the neck-linker. J Biol Chem. 2001;276:25496–25502. doi: 10.1074/jbc.M100395200. [DOI] [PubMed] [Google Scholar]
- 32.Woehlke G, Ruby AK, Hart CL, Ly B, Hom-Booher N, Vale RD. Microtubule interaction site of the kinesin motor. Cell. 1997;90:207–216. doi: 10.1016/s0092-8674(00)80329-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.