Abstract
The hypothalamic suprachiasmatic nucleus (SCN), which in mammals serves as the master circadian pacemaker by synchronizing autonomous clocks in peripheral tissues, is composed of coupled single-cell oscillators that are driven by interlocking positive/negative transcriptional/translational feedback loops. Several studies have suggested that heme, a common prosthetic group that is synthesized and degraded in a circadian manner in the SCN, may modulate the function of several feedback loop components, including the REV-ERB nuclear receptors and PERIOD2 (PER2). We found that ferric heme (hemin, 3-100 μM) dose-dependently and reversibly damped luminescence rhythms in SCN explants from mice expressing a PER2∷LUCIFERASE (PER2∷LUC) fusion protein. Inhibitors of heme oxygenases (HOs, which degrade heme to biliverdin, carbon monoxide, and iron) mimicked heme’s effects on PER2 rhythms. In contrast, heme and HO inhibition did not damp luminescence rhythms in thymus and esophagus explants and had only a small effect on PER2∷LUC damping in spleen explants, suggesting that heme’s effects are tissue-specific. Analysis of the effects of heme’s degradation products on SCN PER2∷LUC rhythms indicated that they probably were not responsible for heme’s effects on rhythms. The heme synthesis inhibitor N-methylprotoporphyrin IX (NMP) lengthened the circadian period of SCN PER2∷LUC rhythms by about an hour. These data are consistent with an important role for heme in the circadian system.
Keywords: Circadian, Tissue Culture, Desynchronization, Luciferase, NR1D1, Clock genes
In mammals, circadian rhythms in physiology and behavior are generated by a master pacemaker in the hypothalamic suprachiasmatic nucleus (SCN), which is composed of thousands of coupled single-cell oscillators (Welsh et al., 1995, Yamaguchi et al., 2003). Rhythms in individual cells are sustained by a series of interacting positive and negative transcriptional/translational feedback loops (Ko and Takahashi, 2006). Among these, CLOCK:BMAL1 and NPAS2:BMAL1 heterodimers drive transcription of the Period (mPer1-3 in mice) and Cryptochrome (mCry1-2 in mice) genes and of the genes for the nuclear receptors REV-ERBα (NR1D1) and REV-ERBβ (NR2D2); the mCRYs and mPERs then feed back to inhibit CLOCK:BMAL1- and NPAS2:BMAL1-mediated transcription while the REV-ERBs repress Bmal1 expression.
While models of the molecular basis of circadian rhythms have focused primarily on these transcriptional/translational feedback loops, small cytosolic signaling molecules, such as Ca2+ and cAMP, have recently emerged as fundamental components of the circadian clock mechanism (Ikeda et al., 2003, Lundkvist et al., 2005, Hastings et al., 2008, O’Neill et al., 2008). Heme, an ubiquitous molecule that is synthesized in virtually every cell in the body, has long been considered essential for normal cellular function, since it acts as a prosthetic group for several proteins involved in a broad range of physiological processes (Wagener et al., 2003). The recent finding that heme functions as a reversible REV-ERB ligand to repress transcription of REV-ERB target genes (Raghuram et al., 2007, Yin et al., 2007) suggests that heme may also act as a signaling molecule, and, more specifically, as a cytosolic regulator of the circadian clock. Consistent with this idea, heme application phase-shifted the electrical activity rhythm in acute SCN slices (Artinian et al., 2001) and peripheral heme administration altered clock gene expression in mouse liver (Kaasik and Lee, 2004). Additionally, hemin treatment has been shown to synchronize clock gene expression in mouse fibroblasts (Kaasik and Lee, 2004, Rogers et al., 2008).
We investigated if heme homeostasis is necessary for normal circadian clock function in mouse tissue explants and found that pharmacologically elevating intracellular heme levels reversibly and dose-dependently damps PER2∷LUCIFERASE rhythms in the SCN. These data are consistent with an important role for heme in the mammalian circadian clock.
EXPERIMENTAL PROCEDURES
Animals
Male and female heterozygous (mPer2Luc/+) and homozygous (mPer2Luc/Luc) mPER2∷LUCIFERASE knockin mice (Yoo et al., 2004) were drawn from an in-house colony (derived from founders generously provided by Joseph Takahashi, Northwestern University) and were housed in a 12 h:12 h light:dark cycle prior to dissection. All animal protocols were approved by the Smith College Institutional Animal Care and Use Committee.
Tissue Preparation
SCN and peripheral tissue explants were prepared according to standard methods (Yoo et al., 2004). Subjects were overdosed with halothane or isoflurane anesthesia, and 300 m coronal sections through the SCN were cut on a vibratome (Campden Istruments, Lafayette, IN) in chilled Hank’s balanced salt solution (Invitrogen, Carlsbad, CA). Sections were trimmed by hand to the SCN and immediately surrounding tissue and were transferred to culture inserts (Millipore, Billerica, MA) in 35 mm dishes [BD Falcon (Franklin Lakes, NJ) or Electron Microscopy Sciences, (Hatfield, PA)]. Peripheral tissues were trimmed by hand to approximately 1-8 mm3 and were cultured on nylon mesh (Small Parts, Miami Lake, FL). Dishes contained 1.0 mL-1.2 mL Dulbecco’s modified Eagle’s medium (Sigma-Aldrich, St. Louis, MO) supplemented with 1X B-27 (Invitrogen), 4 mM L-glutamine (Invitrogen), 25 mM glucose (Sigma), 4.2 mM NaHCO3 (Sigma), 10 mM HEPES (Sigma), 25 Units/mL penicillin G sodium (Invitrogen), 34 μM streptomycin sulfate (Invitrogen), and 100 μM beetle luciferin (Promega, Madison, WI). Dishes were sealed with hot glue (for CO experiments) or with vacuum grease (Dow, Midland, MI; for all other experiments). As many as two SCN sections and four peripheral tissue sections (per tissue type) were collected from a single animal; multiple samples of the same tissue from a single animal were placed in different treatment groups.
Drug Treatments
Biliverdin hydrochloride, hemin chloride, tin protoporphyrin IX, protoporphyrin IX, and N-methylprotoporphyrin IX (all from Sigma or Frontier) were dissolved in sterile 0.085 M or 0.17 M Na3PO4 (Sigma). Ferrous ammonium sulfate [Fe(NH4)2(SO4)2] was dissolved in sterile water. Ketoconazole (Sigma) was dissolved in DMSO. Lipopolysaccharide (Sigma) was dissolved in Hank’s balanced salt solution. Tumor necrosis factor α (TNFα; Peprotech, Rocky Hill, NJ) was dissolved in water. Drugs were diluted 500-fold (ketoconazole), 10,000-fold (lipopolysaccharide), 2000-fold (TNFα), or 1000-fold (all other drugs) into culture media at the time of dissection. For all porphyrin treatments, the final Na3PO4 concentration was 170 μM, which did not change the initial pH of the media or the media’s pH after a week in culture (data not shown). Carbon monoxide was measured, mixed with air, and delivered using gas-tight syringes (Hamilton, Reno, NV). To produce equilibrium aqueous CO concentrations of 30 μM and 100 μM, 50 mL of 4%:96% and 13%:87% (v/v) CO:Air mixtures (see R. Sander, http://www.henrys-law.org), respectively, were flushed through a hole in the top of each pre-sealed culture dish before rapidly sealing the hole with hot glue; in preliminary experiments, this procedure resulted in minimal CO leakage from culture dishes over the course of a week (data not shown). For all treatment groups, tissues were washed three times with control media 5-8 days after dissection, and bioluminescence monitoring was continued.
Bioluminescence Monitoring
Integrated bioluminescence was recorded for 60 s every 10 min using a commercial luminometer (LumiCycle, Actimetrics, Wilmette, IL) housed in an air incubator at 35.8 °C.
Data Analysis
Data from the dissection day (Day 0) and from the days with media changes were excluded from analysis because of transient luminescence spikes. Data were baseline-subtracted with a 24 h running average and smoothed with a 2 h running-median filter. Rhythmicity indices (RIs; defined as the first peak after lag = 0 of the autocorrelation function) calculated for the first three cycles in vitro and for the first three cycles following the media change were used to classify arrhythmic cultures (Levine et al., 2002), which were excluded from subsequent analysis (Supplemental Table). Damped sine curves [Y(t) = A sin (2πt/P-t0) • e-td/24] were fit to pre-wash data to obtain the damping constant (d) and to 3-day pre- and post-wash segments to obtain the circadian period (P). All analyses were performed using Lumicycle Analysis (Actimetrics), SPSS, and custom-written software. The no-drug and Na3PO4 + DMSO control groups had statistically similar damping constants (t(22) = 0.332, p = .743) and post-media change RIs (t(22) = 0.972, p = .342), so they were combined into a single control group for subsequent analysis. In addition, no-drug and Na3PO4 + DMSO tissues collected on five different runs had similar damping constants (F(4,14) = 0.982, p = .449), suggesting that our run-to-run consistency was sufficient to enable us to compare samples collected on different days. Controls cultured under the conditions used for CO treatment (“Air” controls) damped more quickly (t(28) = -2.611, p = .014) and had lower post-media change RIs (t(28) = 2.393, p = .024) than controls cultured under the conditions used in the other experiments, so those groups are analyzed separately. Damping constants and rhythmicity indices were analyzed with one-way ANOVAs followed by Tukey’s HSD; the treatment groups included in a particular analysis are those presented in the corresponding figure.
RESULTS
Heme reversibly and dose-dependently damps SCN PER2∷LUCIFERASE rhythms
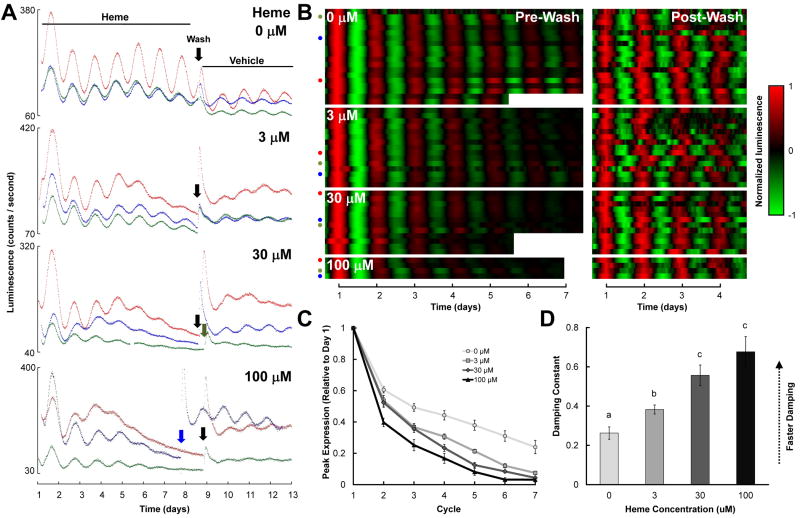
Control SCN cultures (0 μM heme) exhibited stable and robust circadian bioluminescence rhythms in vitro (Figs. 1A and 1B, top panels), while the amplitude of rhythms in slices treated with heme (administered as hemin chloride in the culture media immediately following dissection) decreased gradually over a period of several days (Figs. 1A and 1B, lower panels). The baseline-to-peak rhythm amplitude of each cycle, normalized to the peak on the day following dissection (cycle 1) decreased more rapidly in heme-treated cultures than in control cultures (Fig. 1C; treatment, F(3,51) = 15.18, p < .001; treatment × cycle, F(12,204) = 8.289, p < .001). To further quantify this effect, we fit damped sine curves to the baseline-subtracted data (Fig. 1B), and extracted from the fit curve the component (called the damping constant) that determines how rapidly rhythm amplitude decreases over time. Heme dose-dependently increased the damping constant (Fig. 1D; 30 vs. 100, p = .419; all other comparisons, p’s < .05). To verify that the effect of heme on rhythm damping was not merely due to heme toxicity, we washed tissue and replaced heme-containing media with control media six to eight days after dissection (indicated by arrows in Fig. 1A). This media change restored rhythm robustness in heme-treated cultures (quantified using rhythmicity indices; Table 1; RI, F(3,51) = 0.765, p = .519), indicating that heme’s effects were reversible.
Figure 1.
Effects of heme on PER2∷LUC rhythms in mouse SCN explants. A, B Three representative raw bioluminescence traces (A) and all baseline-subtracted, smoothed data (B) for SCNs treated with 0-100 μM heme (left panels in B); cultures were subsequently washed and transferred to control media at the time indicated by the downward arrows in A, and bioluminescence monitoring was continued (right panels in B). Tissue was dissected on day 0 (not shown). In B, each heatmap row represents a different culture, the x-axis indicates time (with t = 1 corresponding to the time of peak luminescence on day 1 in the left panels and to the time of peak luminescence on the day following the media change in the right panels), colors represent luminescence level as indicated in the key to the right, pre- and post-media change data were normalized independently, and colored dots indicate the cultures illustrated by traces in part A. C, Mean peak PER2:LUC expression during each circadian cycle prior to the media change, normalized to peak expression on the day after dissection (Cycle 1). D, Mean damping constants for recordings prior to the media change. In this and subsequent figures: groups with the same letters above error bars are statistically similar while groups with different letters are statistically different (Tukey’s HSD, p’s < .05); all error bars indicate ±SEM.
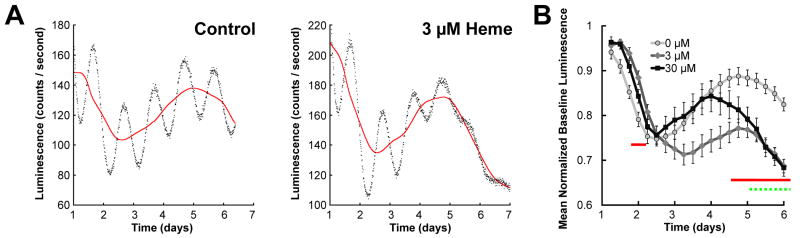
To determine whether heme treatment affected overall luminescence levels, we fit 24 h running average baselines to raw bioluminescence traces (Fig. 2A) and calculated the average normalized baseline for each treatment group (Fig. 2B; the 100 μM group was excluded from this analysis due to small sample size). Heme decreased baseline luminescence after several days, suggesting that heme may decrease mPER2 expression, but this effect did not appear to be dose-dependent (Fig. 2B). Since raw luminescence of heme-treated SCNs typically remained well above dark-count levels (~30 counts/s) even after a week of treatment (see Figure 1A), heme-mediated rhythm damping could not have been due to suppression of luminescence to levels that rendered rhythms undetectable.
Figure 2.
Effects of heme treatment on baseline luminescence in SCN explants from PER2∷LUC knockin mice. A, Example of a baseline (solid line) calculated as the 24 h. running average of the raw data (points) for a control SCN (left) and an SCN treated with 3 μM heme (right). B, Mean normalized baseline luminescence for control SCNs (0 μM, light gray line) and SCNs treated with 3 μM (dark gray line) or 30 μM (black line) heme binned every six hours. Horizontal lines indicate where 3 μM heme-treated (solid) and 30 μM heme-treated (dashed) groups differ statistically (p < .05) from the control (0 μM) group (one-way ANOVAs at each time point with Bonferroni correction, followed by Tukey’s HSD).
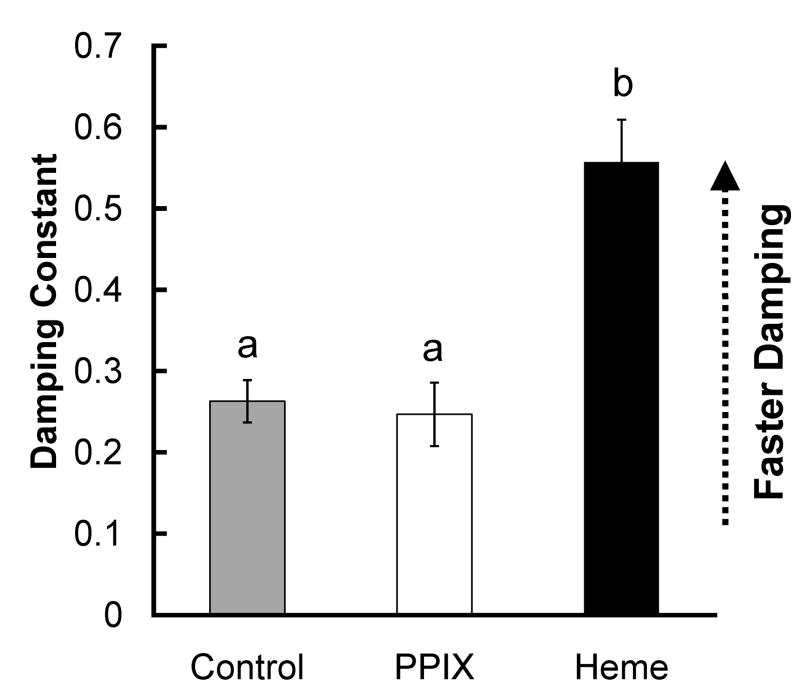
We also investigated whether heme’s effect could be due to a general effect of porphyrins. SCNs treated with 30 μM protoporphyrin IX (PPIX), a molecule identical to heme but lacking the central iron atom, did not damp more quickly than controls (Figs. 3 and S1; p = .971), suggesting that heme’s effect on damping were not due to a general effect of porphyrins.
Figure 3.
Mean damping constants for control SCN explants (gray bar) and SCN explants treated with heme (black bar) or with the control porphyrin PPIX (open bar). Data for control and 30 μM heme groups are repeated from Fig. 1.
Heme oxygenase inhibition damps SCN PER2∷LUCIFERASE rhythms
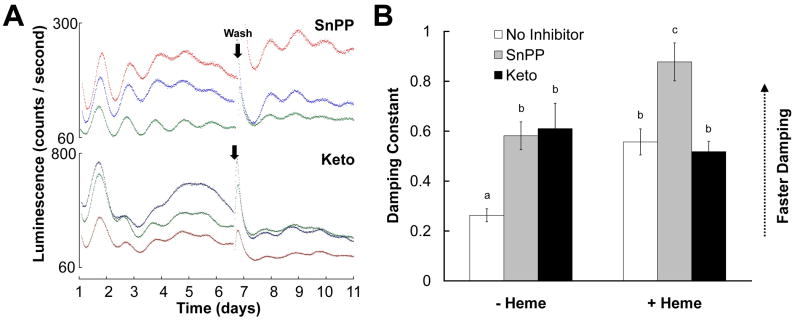
We also tested whether increasing the intracellular heme concentration indirectly via inhibition of heme breakdown (Goodman et al., 2007) could also damp PER2∷LUC rhythms in the SCN. The competitive HO inhibitor tin protoporphyrin IX (SnPP; (Drummond and Kappas, 1981) and the non-competitive HO inhibitor ketoconazole (Keto; Kinobe et al., 2006), both damped SCN PER2∷LUC rhythms relative to controls (Figs. 4 and S2; p’s < .05). Moreover, heme + SnPP damped rhythms more quickly than heme or SnPP alone (p’s < .05), but a similar combined effect was not observed for heme + keto (p’s > .940). In constrast, Keto but not SnPP shortened the circadian period of SCN PER2∷LUC rhythms by about 1 h (Supplementary Table). Following the media change, rhythms in cultures treated with keto or SnPP with or without heme were as robust as those in control cultures (Table 1; two-way ANOVA, all p’s > .05), indicating that the effects of those drugs were not due to irreversible toxicity.
Figure 4.
Effects of heme oxygenase inhibitors on PER2∷LUCIFERASE rhythms in mouse SCN explants. A, Representative raw bioluminescence data for SCNs treated with 10 μM ketoconazole (Keto) or 30 μM tin protoporphyrin IX (SnPP). B, Damping constants for SCN cultures treated with Keto (black bars), SnPP (gray bars), or no HO inhibitors (open bars) in the presence or absence of heme (x-axis). Data for control and 30 μM heme groups are repeated from Fig. 1.
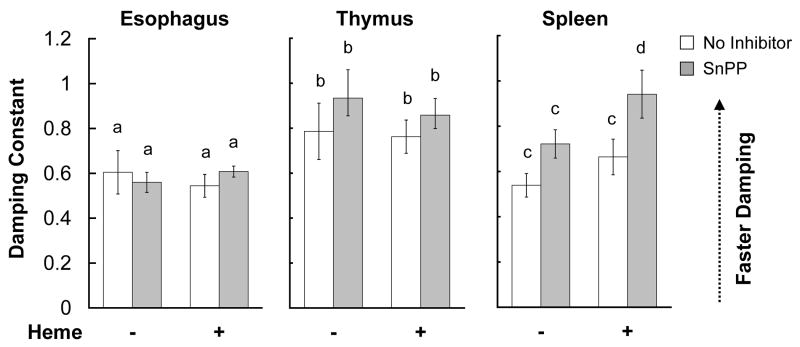
The effects of heme and of HO inhibition are tissue-specific
In addition to the SCN, peripheral tissues throughout the body maintain rhythms of clock gene expression (Yoo et al., 2004). We found no effect of 30 μM heme, 30 μM SnPP, or both together on PER2∷LUC rhythms in esophagus or thymus explants (Figs. 5, S3, S4, and S6; two-way ANOVAs, p’s > .182). We also tested the effect of a 100 μM heme on esophagus explants and found no effect (Fig. S3 and S6 and data not shown). In contrast, spleen explants treated with heme + SnPP damped more quickly than control spleen cultures (Figs. 5 and S5; control vs. SnPP + heme, p = .008; heme vs. SnPP + heme, p = .085; all other comparisons, p’s > .360). Analysis of cycle-to-cycle rhythm amplitude suggested that heme and SnPP alone might also have an effect on damping in spleen explants but not in thymus or esophagus explants (Fig. S6). For all tissues, post-media change RIs were similar in all treatment groups (two-way ANOVA, all p’s > .330), and we observed no effect of 30 μM heme on baseline luminescence in any of the tissues tested (data not shown).
Figure 5.
Damping constants of PER2∷LUC rhythms from control peripheral tissue explants and from peripheral tissue explants treated with 30 μM SnPP, 30 μM heme, or both together. Each tissue was analyzed independently, and letters above bars refer only to comparisons within a tissue.
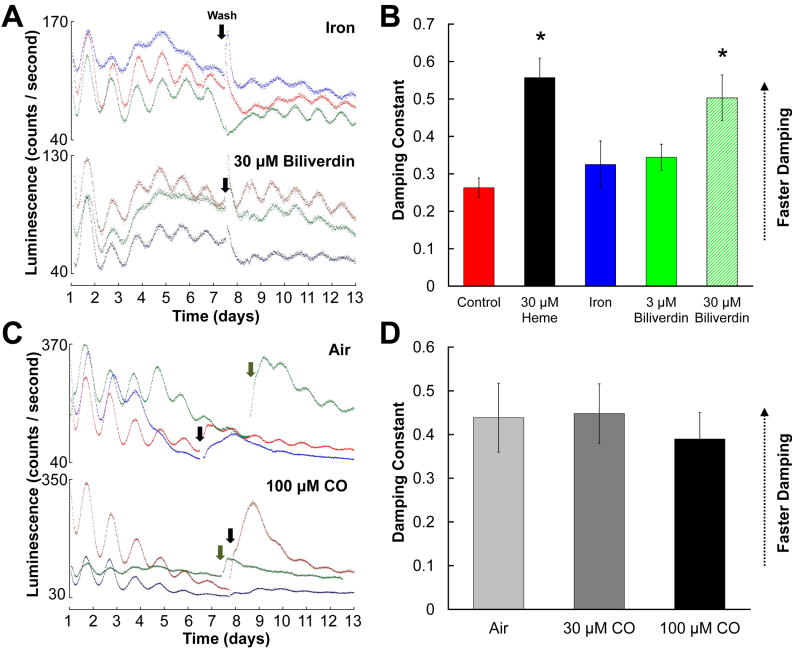
Effects of heme metabolites on PER2∷LUCIFERASE rhythms
To determine if the effects of heme and HO inhibitor treatments might be due to changes in levels of the heme breakdown products, we applied each breakdown product individually to SCN explants. Because CO treatments required different culture conditions than other treatments (see Experimental Procedures), we analyzed biliverdin and iron groups (Figs. 6A-B and S7) separately from CO groups (Figs. 6C-D and S8). PER2∷LUC rhythms in iron and 3 μM biliverdin groups damped at similar rates to the control group (p’s > .700), but a higher dose of biliverdin (30 μM) accelerated rhythm damping (p < .001). None of the treatments tested affected rhythm robustness following a medium change (Table 1; one-way ANOVA of RI, F(4,52) = 0.164, p = .956).
Figure 6.
Effects of heme metabolites on SCN PER2∷LUCIFERASE rhythms. A, C, Representative raw luminescence traces for SCN cultures treated with 30 μM iron or 30 μM biliverdin (A) or with air or 100 μM carbon monoxide (C). B, D, Mean damping constants for SCNs treated with iron or biliverdin (B) or with air or CO (D) as indicated in figure keys. In B, data for control and 30 μM heme groups are repeated from Fig. 1. (*, significantly different from controls, p < .05)
Control (“Air”) SCN slices cultured under conditions identical to those used for CO-treated cultures damped more quickly and had less robust post-media change rhythms than SCNs cultured under our original conditions, suggesting that the sealed culture conditions necessary for CO treatment were detrimental to tissue health (Supplementary Table; see Experimental Procedures). We found no effect of 30 μM or 100 μM CO on damping or post-wash rhythmicity indices (damping constant, one-way ANOVA, F(2,18) = 0.168, p = .847; post-media change RI, F(2,18) = 0.081, p = .923). At 300 μM, CO caused luminescence to drop irreversibly to dark-count levels after 2-4 days, consistent with tissue death (data not shown).
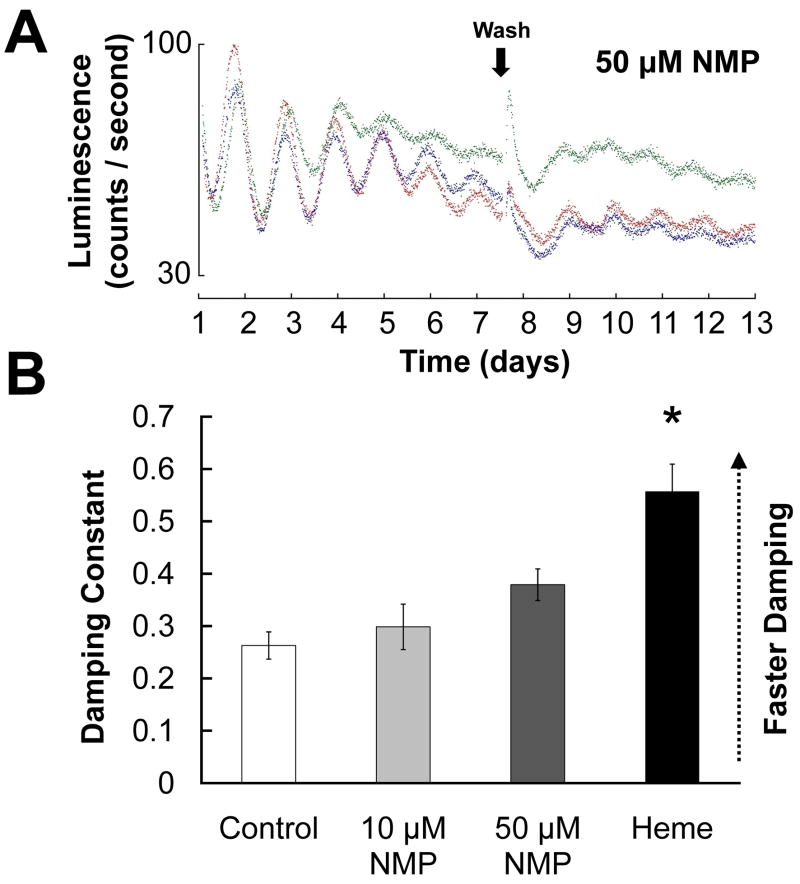
Inhibition of heme synthesis lengthens circadian period
Since treatments that increase intracellular heme levels disrupt PER2∷LUC rhythms, we examined whether heme deficiency is also associated with abnormal rhythmicity. N-methylprotoporphyrin IX (NMP) is an inhibitor of ferrochelatase, which catalyzes the last step in heme synthesis, and has been used to induce heme deficiency in primary neuronal cultures (Atamna et al., 2002). Neither 10 μM or 50 μM NMP increased the rate at which SCN PER2∷LUC rhythms damped relative to controls (Fig. 7; Tukey’s HSD, p’s > .250), but the difference in damping between 50 μM NMP and the 30 μM heme group only approached significance (p = .058). However, NMP did significantly lengthen circadian period of PER2∷LUC rhythms before (p = .012) but not after (p = .995) the media change (Supplementary Table).
Figure 7.
A, Representative raw bioluminescence traces for SCNs treated with 50 μM N-methylprotoporphyrin IX (NMP). B, Damping constants for SCNs treated with 10 μM or 50 μM NMP, as well as for control SCNs and SCNs treated with 30 μM heme (reproduced from Fig. 1). (*, significantly different from controls, p < .05) NMP also had a significant effect on circadian period (see Supplementary Table).
Heme’s effects are not due to actions at toll-like receptor 4 (TLR4)
Recently, heme was reported to activate the lipopolysaccharide (LPS) receptor TLR4 to induce tumor necrosis factor α (TNFα) secretion in cultured macrophages (Figueiredo et al., 2007). To determine whether heme’s effects could be mediated by TLR4 activation and TNFα secretion, we treated PER2∷LUC SCN explants with LPS (100 ng/mL) or TNFα (50 ng/mL). We did not observe any effects of LPS on rhythm damping [mean damping constant ± SEM for control (n = 4): 0.325 ± 0.037; for LPS (n = 4): 0.344 ± 0.052; t(6) = .307, p = .769] or circadian period (data not shown). There were similarly no effects of TNFα [mean damping constant ± SEM for control (n = 4): 0.327 ± 0.058; for TNFα (n = 4): 0.359 ± 0.055; t(6) = 0.406, p = .699]
DISCUSSION
We found that experimentally elevating heme levels, whether via direct application of exogenous heme or via heme oxygenase inhibition, damps PER2∷LUC rhythms in the mouse SCN. The effects of exogenous heme on PER2∷LUC rhythms were dose-dependent in the SCN. Heme and heme oxygenase inhibition also accelerate damping in spleen explants but not in esophagus or thymus explants; thus, while heme’s effect is not specific to the central SCN oscillator, it is tissue specific.
Interestingly, while the heme oxygenase inhibitors SnPP and Keto both caused PER2∷LUC rhythms to damp, SnPP but not Keto potentiated heme’s effect on damping. This apparent discrepancy could be due to non-specific effects of SnPP. For instance, SnPP has been reported to inhibit nitric oxide synthase (Grundemar and Ny, 1997), and nitric oxide can bind to REV-ERB and suppress its inhibitory effect on target gene transcription (Pardee et al., 2009). Thus, if heme affects rhythms by activating REVERB (Raghuram et al., 2007, Yin et al., 2007), then SnPP could further enhance heme’s effects by reducing NO levels that tonically inhibit REV-ERB activity. Alternately, SnPP, which is structurally identical to heme except for the central metal atom, could mimic heme by binding to similar sites and by activating similar biological processes.
Heme and HO inhibitor treatments are expected to respectively increase and decrease levels of the heme metabolites biliverdin, carbon monoxide, and iron, all of which could potentially alter clock function (Pinero and Connor, 2000, Barañano et al., 2001, Dioum et al., 2002). However, our results suggest that the effects of exogenous heme on rhythm damping were not mediated by these metabolites. Iron clearly had no effect on SCN luminescence rhythms. Since the volume of media in our cultures was large relative to the amount of tissue in our explants, it is unlikely that the tissue was capable of metabolizing a significant fraction of exogenous heme, and it is thus unlikely that biliverdin concentrations in any of our heme-treated cultures approached 30 μM. Therefore, the effect of heme is most likely independent of the effect we observed with 30 μM biliverdin. Consistent with this conclusion, 3 μM heme but not 3 μM biliverdin damped PER2∷LUC rhythms. The lack of an effect of iron and 3 μM biliverdin treatments is not likely due to a failure of those compounds to cross the plasma membrane. The concentration of Fe(NH4)2(SO4)2 used (30 μM) would be expected to elevate intracellular iron levels to at least 4.5-6 μM, based on analysis of rat brain slices treated briefly with a similar compound (Oubidar et al., 1996). Biliverdin has likewise been reported to readily enter the cytosol (Shu et al., 2009). We found that treating SCN explants with up to 100 μM CO did not affect PER2∷LUCIFERASE rhythms relative to controls cultured under the same conditions. However, since these conditions were detrimental to tissue health, more work is necessary to conclusively rule out a role for CO in the circadian clock.
Yamaguchi and colleagues (2003) found that prolonged blockade of sodium-dependent action potentials in the SCN both caused rhythms in individual cells to damp and caused rhythms across the SCN to become desynchronized (Yamaguchi et al., 2003), suggesting that intercellular coupling and stable intracellular rhythms may be related. At the same time, SCN neurons are capable of maintaining circadian electrical rhythms in isolation (Welsh et al., 1995), and cells in SCN slices can remain rhythmic even under conditions in which they are desynchronized (Ohta et al., 2005, Ciarleglio et al., 2009). Heme could thus function by desynchronizing stable single-cell PER2 rhythms, by damping rhythms in individual cells, or by a combination of these effects. Consistent with the latter two possibilities, we found that heme decreased baseline luminescence (Fig. 2); this effect is not likely due to an effect of heme on LUC activity, since we observed no effect of heme on baseline LUC activity in esophagus and thymus. Since desynchronization of individual oscillators would not be expected to alter baseline PER2 expression, the effect of heme on baseline luminescence suggests that heme probably does not merely cause individual cellular clocks to become uncoupled; rather, heme likely affects some process that causes overall PER2∷LUC expression levels to decrease. Single-cell imaging will be necessary to confirm this result and to determine definitively how heme affects coupling between single-cell clocks in the SCN.
One concern is that the effects observed here are due to heme’s neurotoxicity (Goldstein et al., 2003). However, heme neurotoxicity can be mimicked by inorganic iron and blocked by co-treatment with SnPP, suggesting that neurotoxicity associated with heme treatment is due to release of inorganic iron and subsequent generation of reactive oxygen species (ROSs; Goldstein et al., 2003). Since heme’s effects on PER2∷LUC damping could not be mimicked by iron and were potentiated by SnPP, it is unlikely that heme neurotoxicity underlies the effects observed here. This conclusion is supported by our findings that heme-treated and control SCNs had equally robust PER2∷LUC rhythms following a media change. Nonetheless, heme could generate sub-toxic concentrations of ROSs; consistent with this possibility, we found that under the more stressful culture conditions required for CO treatment, heme accelerated tissue death or irreversibly disrupted PER2∷LUCIFERASE rhythms (data not shown). While ROSs have traditionally been viewed as toxic biproducts of cellular function, they may also be important intracellular and intercellular signaling molecules (Thannickal and Fanburg, 2000). For instance, MAPK signaling and Ca2+ release from ryanodine-sensitive stores both appear to be involved in clock function (Ikeda et al., 2003, Akashi et al., 2008) and are both regulated by ROSs (Hidalgo et al., 2005, Muñoz et al., 2006). It is thus possible that heme could affect circadian clock function through generation of sub-toxic concentrations of ROSs and that ROSs could be important in circadian clock function even under physiological, non-pathological conditions.
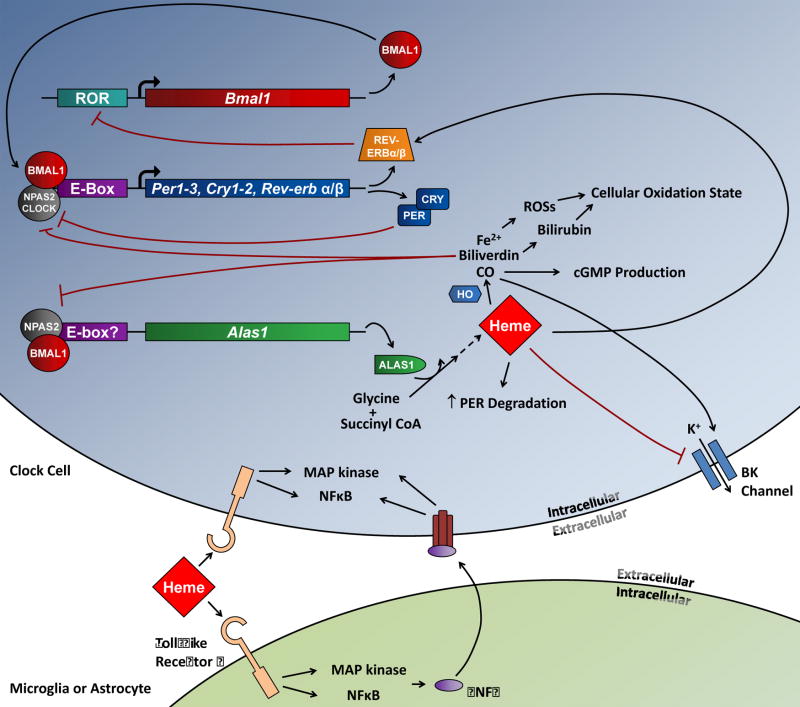
Heme could also act through several other mechanisms to affect SCN clock function (Fig. 8). In immortalized cell lines, heme has been shown to bind directly to human PER2 to induce its degradation in the proteasome (Yang et al., 2008). While the lack of an effect of heme on esophagus and thymus rhythms argues against a direct effect on PER2, heme could affect PER2 levels indirectly through other mechanisms. For instance, heme could affect the SCN circadian clock by acting on TLR4 (Figueiredo et al., 2007) either directly on clock cells or on resident microglia. This possibility initially appeared plausible in light of the findings that systemic LPS administration suppressed Per2 expression and behavioral rhythms (Okada et al., 2008) and that TNFα, a downstream mediator of TLR4 activation, altered clock gene expression in vitro and in vivo, reduced the amplitude of behavioral circadian rhythms, and altered SCN neuronal activity (Cavadini et al., 2007, Nygård et al., 2009). However, since we failed to observe effects of LPS and TNFα on SCN PER2∷LUC rhythms, those molecules probably do not mediate heme’s effect on damping. Heme can also block BK channels (Tang et al., 2003), which are known to be important for maintaining robust SCN electrophysiological rhythms in vitro and normal behavioral rhythms in vivo (Meredith et al., 2006). However, relative to wild-type mice, BK channel knockout mice do not show less robust rhythms of SCN Per2 expression in vivo (Meredith et al., 2006), while heme has clear effects on PER2 rhythms in vitro.
Figure 8.
Schematic of the circadian clock mechanism and the possible sites where heme may act to affect rhythms. CLOCK:BMAL1 or NPAS2:BMAL1 bind to E-boxes to drive expression of circadian genes; two families of those, the Per and Cry genes, dimerize and inhibit CLOCK/NPAS2:BMAL1-mediated transcription; the REV-ERBs feedback to inhibit Bmal1 expression by binding to a ROR element in the Bmal1 promoter. Expression of the rate-limiting enzyme in heme synthesis, Alas1, is also circadianly regulated by NPAS2:BMAL1, while the metabolism of heme to Fe2+. biliverdin, and CO by heme oxygenase (HO) is regulated at the post-translational level. Heme could affect rhythms by increasing PER protein degradation, inhibiting BK channels, or enhancing transcriptional repression by the REV-ERBs. Heme can also bind to toll-like receptor 4 (TLR4), perhaps on clock cells or on resident microglia, and induce downstream effects through MAPK and NFκB signaling; TNFα secretion induced by TLR4 stimulation could also feedback to affect clock cell function through those same two signaling pathways. CO could affect rhythms by decreasing NPAS2-dependent transcription, by promoting cGMP production, or by stimulating BK channels. Biliverdin is rapidly degraded to bilirubin, and both biliverdin and bilirubin could affect rhythms by altering the overall cellular oxidation state. Fe2+ could be involved in the generation of reactive oxygen species (ROSs) that could affect diverse cellular processes. Our results rule out a role for TLR4/TNFα in heme’s effect, while the lack of an effect of heme on esophagus and thymus rhythms argues against a direct effect of heme on PER2 degradation; in addition, we found no evidence that heme’s effects are mediated by its degradation products.
Finally, heme could suppress Bmal1 expression by activating the REV-ERBs (Raghuram et al., 2007, Yin et al., 2007), functionally mimicking the effect of Bmal1 knockout, which abolishes SCN mPer2 transcript rhythms in vivo and fibroblast PER2∷LUC rhythms in vitro (Bunger et al., 2000, Liu et al., 2008). Consistent with this hypothesis, the inducible overexpression of Rev-erbα in liver explants of a transgenic mouse line decreased Bmal1 expression and abolished PER2∷LUC rhythms (Kornmann et al., 2007). However, since BMAL1 in turn regulates Rev-erbα expression (Preitner et al., 2002, Triqueneaux et al., 2004), the effects of endogenous REV-ERB over-activity on molecular oscillator function are unclear. In this context, the slow time course of heme’s effects on PER2∷LUC rhythms could be due to the general robustness of clock function in the face of molecular perturbation (Liu et al., 2007). While the reason heme had no effect on rhythms in esophagus and thymus is not clear, one possibility is that heme’s downstream effector may function in a tissue-specific manner. For instance, the RORs, transcriptional activators that compete with the REV-ERBs for binding to the Bmal1 promoter, are expressed tissue-specifically (Sato et al., 2004, Guillaumond et al., 2005, Ko and Takahashi, 2006, Liu et al., 2008) and could modulate heme-bound REV-ERB’s ability to suppress clock gene expression. Further studies are clearly necessary to understand how heme affects SCN PER2∷LUC rhythms.
Both pharmacological heme depletion and a REV-ERB point mutation that prevents heme binding inhibit REV-ERB-mediated transcriptional repression in cell lines (Raghuram et al., 2007, Yin et al., 2007), suggesting that basal heme levels must be sufficient to activate REV-ERB. Given that the heme-REV-ERB dissociation constant has been estimated as 2-6 μM (Raghuram et al., 2007, Pardee et al., 2009), it is not unreasonable to assume that intracellular free heme concentrations in the SCN may be in the low micromolar range. Since as little as 3 μM heme was found to damp rhythms in the present study, it is likely that the effects observed here are physiologically relevant.
We found that inhibiting heme synthesis with NMP did not cause SCN rhythms to damp relative to controls, although the difference in damping between 30 μM heme and NMP groups only approached significance (see Results). Since heme is essential for many critical cellular processes, it is impossible to completely deplete heme without compromising tissue viability. Since 1 μM NMP decreased heme levels in primary rat hepatocytes by about 40% (Jacobs et al., 1998), we anticipate that the 10-50 μM NMP used in the present study induced substantial heme deficiency. The lack of a clear effect of NMP on rhythm damping would not be unexpected if heme’s effects on rhythms are mediated by REV-ERB, since the REV-ERBs themselves are dispensable for PER2∷LUC rhythmicity (Liu et al., 2008). Regardless, the lengthening of circadian period by NMP is consistent with an important role for heme in the circadian system. Of particular interest is that possibility that heme could be involved in determining the phases of gene expression rhythms, for which the REV-ERBs are thought to be essential (Liu et al., 2008, Ukai-Tadenuma et al., 2008). Broad transcriptional profiling of cells or tissues treated with heme will be necessary to test this possibility.
The findings that the circadian clock regulates expression of the gene for the rate-limiting enzyme (5-aminolevulinc acid synthetase 1) in heme synthesis (Alas1; (Zheng et al., 2001) and that heme oxygenase activity varies across the circadian cycle (Rubio et al., 2003) have led to speculation that heme levels may oscillate in a circadian manner. Heme could therefore participate in a novel feedback loop wherein CLOCK/NPAS2:BMAL1 drives expression of Alas1 and thus synthesis of heme, which could feed back to inhibit Bmal1 expression via REV-ERB. Since REV-ERB/heme regulate expression of several metabolic genes and since heme is synthesized from the Krebs cycle intermediate succinyl CoA, heme could be involved in coupling the circadian clock and metabolism (Raghuram et al., 2007, Yin et al., 2007). From the clinical perspective, regulation of heme catabolism by cytokines (Wagener et al., 2003) could underlie the circadian disruption associated with elevated cytokine levels in conditions such as cancer-related fatigue (Coogan and Wyse, 2008), and abnormal heme homeostasis associated with Alzheimer’s disease and aging (Atamna et al., 2002, Dwyer et al., 2009) could be involved in the circadian system disturbance observed in those conditions (Wu and Swaab, 2007). As other authors have proposed (Kaasik and Lee, 2004), the ability of heme to alter SCN function could also lead to the development of treatments for circadian system disturbance.
While heme’s role as a cofactor necessary for the function of a large number of proteins and enzymes has been well-established, more recent work has suggested that heme could be an important signaling molecule in the brain. Indeed, heme could have wide-ranging effects on neuronal physiology through its abilities to regulate gene expression (Raghuram et al., 2007, Yin et al., 2007), potassium currents (Tang et al., 2003), and protein stability (Yang et al., 2008). However, evidence of an essential role for heme signaling in any neural process has been lacking. Our data suggest that heme homeostasis is necessary for normal circadian clock function and that heme could be an important component of the circadian clock in mammals. Based on our findings, additional studies of heme’s precise role in the circadian system and of possible roles for heme in regulating other aspects of neural function are clearly warranted.
Supplementary Material
Acknowledgments
CJG previously submitted a portion of this work in partial fulfillment of an AB with honors (Amherst College, 2008). The authors wish to thank Penny Molyneux and Cynthia Franqui for technical assistance, Leandro Casiraghi for preliminary work on TNFα, the Harrington lab and Horacio de la Iglesia for comments on this manuscript, and the Smith College animal care program for animal care. This work was supported by NSF0618129 and R21CA125215 (to MEH).
List of Abbreviations
- CO
carbon monoxide
- HO
heme oxygenase
- LPS
lipopolysaccharide
- NMP
N-methylprotoporphyrin IX
- PPIX
protoporphyrin IX
- SCN
suprachiasmatic nucleus
- SnPP
tin protoporphyrin IX
- TLR4
toll-like receptor 4
- RI
rhythmicity index
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akashi M, Hayasaka N, Yamazaki S, Node K. Mitogen-Activated Protein Kinase Is a Functional Component of the Autonomous Circadian System in the Suprachiasmatic Nucleus. J Neurosci. 2008;28:4619–4623. doi: 10.1523/JNEUROSCI.3410-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artinian LR, Ding JM, Gillette MU. Carbon Monoxide and Nitric Oxide: Interacting Messengers in Muscarinic Signaling to the Brain’s Circadian Clock. Experimental Neurology. 2001;171:293–300. doi: 10.1006/exnr.2001.7781. [DOI] [PubMed] [Google Scholar]
- Atamna H, Killilea DW, Killilea AN, Ames BN. Heme deficiency may be a factor in the mitochondrial and neuronal decay of aging. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:14807–14812. doi: 10.1073/pnas.192585799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barañano DE, Ferris CD, Snyder SH. Atypical neural messengers. Trends in Neurosciences. 2001;24:99. doi: 10.1016/s0166-2236(00)01716-1. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 Is an Essential Component of the Master Circadian Pacemaker in Mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavadini G, Petrzilka S, Kohler P, Jud C, Tobler I, Birchler T, Fontana A. TNFα suppresses the expression of clock genes by interfering with E-box-mediated transcription. Proceedings of the National Academy of Sciences. 2007;104:12843–12848. doi: 10.1073/pnas.0701466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarleglio CM, Gamble KL, Axley JC, Strauss BR, Cohen JY, Colwell CS, McMahon DG. Population Encoding by Circadian Clock Neurons Organizes Circadian Behavior. J Neurosci. 2009;29:1670–1676. doi: 10.1523/JNEUROSCI.3801-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coogan AN, Wyse CA. Neuroimmunology of the circadian clock. Brain Research. 2008;1232:104. doi: 10.1016/j.brainres.2008.07.087. [DOI] [PubMed] [Google Scholar]
- Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez M-A, McKnight SL. NPAS2: A Gas-Responsive Transcription Factor. Science. 2002;298:2385–2387. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- Drummond GS, Kappas A. Prevention of neonatal hyperbilirubinemia by tin protoporphyrin IX, a potent competitive inhibitor of heme oxidation. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:6466–6470. doi: 10.1073/pnas.78.10.6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer BE, Stone ML, Gorman N, Sinclair PR, Perry G, Smith MA, Zhu X. Heme-a, the heme prosthetic group of cytochrome c oxidase, is increased in Alzheimer’s disease. Neurosci Lett. 2009;461:302. doi: 10.1016/j.neulet.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo RT, Fernandez PL, Mourao-Sa DS, Porto BN, Dutra FF, Alves LS, Oliveira MF, Oliveira PL, Graca-Souza AV, Bozza MT. Characterization of Heme as Activator of Toll-like Receptor 4. J Biol Chem. 2007;282:20221–20229. doi: 10.1074/jbc.M610737200. [DOI] [PubMed] [Google Scholar]
- Goldstein L, Teng Z-P, Zeserson E, Patel M, Regan RF. Hemin induces an iron-dependent, oxidative injury to human neuron-like cells. Journal of Neuroscience Research. 2003;73:113–121. doi: 10.1002/jnr.10633. [DOI] [PubMed] [Google Scholar]
- Goodman AI, Olszanecki R, Yang LM, Quan S, Li M, Omura S, Stec DE, Abraham NG. Heme oxygenase-1 protects against radiocontrast-induced acute kidney injury by regulating anti-apoptotic proteins. Kidney International. 2007;72:945. doi: 10.1038/sj.ki.5002447. [DOI] [PubMed] [Google Scholar]
- Grundemar L, Ny L. Pitfalls using metalloporphyrins in carbon monoxide research. Trends in Pharmacological Sciences. 1997;18:193–195. doi: 10.1016/s0165-6147(97)01065-1. [DOI] [PubMed] [Google Scholar]
- Guillaumond F, Dardente H, Giguere V, Cermakian N. Differential Control of Bmal1 Circadian Transcription by REV-ERB and ROR Nuclear Receptors. J Biol Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Maywood ES, O’Neill JS. Cellular Circadian Pacemaking and the Role of Cytosolic Rhythms. Current Biology. 2008;18:R805–R815. doi: 10.1016/j.cub.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Hidalgo C, Donoso P, Carrasco MA. The Ryanodine Receptors Ca2 + Release Channels: Cellular Redox Sensors? IUBMB Life. 2005;57:315–322. doi: 10.1080/15216540500092328. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Sugiyama T, Wallace CS, Gompf HS, Yoshioka T, Miyawaki A, Allen CN. Circadian Dynamics of Cytosolic and Nuclear Ca2+ in Single Suprachiasmatic Nucleus Neurons. Neuron. 2003;38:253–263. doi: 10.1016/s0896-6273(03)00164-8. [DOI] [PubMed] [Google Scholar]
- Jacobs JM, Sinclair PR, Sinclair JF, Gorman N, Walton HS, Wood SG, Nichols C. Formation of zinc protoporphyrin in cultured hepatocytes: effects of ferrochelatase inhibition, iron chelation or lead. Toxicology. 1998;125:95. doi: 10.1016/s0300-483x(97)00164-9. [DOI] [PubMed] [Google Scholar]
- Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- Kinobe RT, Dercho RA, Vlahakis JZ, Brien JF, Szarek WA, Nakatsu K. Inhibition of the Enzymatic Activity of Heme Oxygenases by Azole-Based Antifungal Drugs. J Pharmacol Exp Ther. 2006;319:277–284. doi: 10.1124/jpet.106.102699. [DOI] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:R271–277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-Driven and Oscillator-Dependent Circadian Transcription in Mice with a Conditionally Active Liver Clock. PLoS Biology. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine J, Funes P, Dowse H, Hall J. Signal analysis of behavioral and molecular cycles. BMC Neuroscience. 2002;3:1. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant Function of REV-ERBα and β and Non-Essential Role for Bmal1 Cycling in Transcriptional Regulation of Intracellular Circadian Rhythms. PLoS Genetics. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, Doyle Iii FJ, Takahashi JS, Kay SA. Intercellular Coupling Confers Robustness against Mutations in the SCN Circadian Clock Network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundkvist GB, Kwak Y, Davis EK, Tei H, Block GD. A Calcium Flux Is Required for Circadian Rhythm Generation in Mammalian Pacemaker Neurons. J Neurosci. 2005;25:7682–7686. doi: 10.1523/JNEUROSCI.2211-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith AL, Wiler SW, Miller BH, Takahashi JS, Fodor AA, Ruby NF, Aldrich RW. BK calcium-activated potassium channels regulate circadian behavioral rhythms and pacemaker output. Nat Neurosci. 2006;9:1041–1049. doi: 10.1038/nn1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz P, Zavala G, Castillo K, Aguirre P, Hidalgo C, Núñez MT. Effect of iron on the activation of the MAPK/ERK pathway in PC12 neuroblastoma cells. Biological Research. 2006;39:189–190. doi: 10.4067/s0716-97602006000100021. [DOI] [PubMed] [Google Scholar]
- Nygård M, Lundkvist GB, Hill RH, Kristensson K. Rapid nitric oxide-dependent effects of tumor necrosis factor-alpha on suprachiasmatic nuclei neuronal activity. NeuroReport. 2009;20:213–217. doi: 10.1097/WNR.0b013e32831f1ca2. [DOI] [PubMed] [Google Scholar]
- O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-Dependent Signaling as a Core Component of the Mammalian Circadian Pacemaker. Science. 2008;320:949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta H, Yamazaki S, McMahon DG. Constant light desynchronizes mammalian clock neurons. Nat Neurosci. 2005;8:267. doi: 10.1038/nn1395. [DOI] [PubMed] [Google Scholar]
- Okada K, Yano M, Doki Y, Azama T, Iwanaga H, Miki H, Nakayama M, Miyata H, Takiguchi S, Fujiwara Y, Yasuda T, Ishida N, Monden M. Injection of LPS Causes Transient Suppression of Biological Clock Genes in Rats. Journal of Surgical Research. 2008;145:5. doi: 10.1016/j.jss.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Oubidar M, Boquillon M, Marie C, Bouvier C, Beley A, Bralet J. Effect of intracellular iron loading on lipid peroxidation of brain slices. Free Radical Biology and Medicine. 1996;21:763. doi: 10.1016/0891-5849(96)00173-6. [DOI] [PubMed] [Google Scholar]
- Pardee KI, Xu X, Reinking J, Schuetz A, Dong A, Liu S, Zhang R, Tiefenbach J, Lajoie G, Plotnikov AN, Botchkarev A, Krause HM, Edwards A. The Structural Basis of Gas-Responsive Transcription by the Human Nuclear Hormone Receptor REV-ERBβ. PLoS Biology. 2009;7:e43. doi: 10.1371/journal.pbio.1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinero DJ, Connor JR. Iron in the Brain: An Important Contributor in Normal and Diseased States. Neuroscientist. 2000;6:435–453. [Google Scholar]
- Preitner N, Damiola F, Luis Lopez M, Zakany J, Duboule D, Albrecht U, Schibler U. The Orphan Nuclear Receptor REV-ERBα Controls Circadian Transcription within the Positive Limb of the Mammalian Circadian Oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers PM, Ying L, Burris TP. Relationship between circadian oscillations of Rev-erbα expression and intracellular levels of its ligand, heme. Biochemical and Biophysical Research Communications. 2008;368:955–958. doi: 10.1016/j.bbrc.2008.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio MF, Agostino PV, Ferreyra GA, Golombek DA. Circadian heme oxygenase actitivy in the hamster suprachiasmatic nuclei. Neuroscience Letters. 2003;353:9–12. doi: 10.1016/j.neulet.2003.08.075. [DOI] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A Functional Genomics Strategy Reveals Rora as a Component of the Mammalian Circadian Clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Shu X, Royant A, Lin MZ, Aguilera TA, Lev-Ram V, Steinbach PA, Tsien RY. Mammalian Expression of Infrared Fluorescent Proteins Engineered from a Bacterial Phytochrome. Science. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang XD, Xu R, Reynolds MF, Garcia ML, Heinemann SH, Hoshi T. Haem can bind to and inhibit mammalian calcium-dependent Slo1 BK channels. Nature. 2003;425:531–535. doi: 10.1038/nature02003. [DOI] [PubMed] [Google Scholar]
- Thannickal VJ, Fanburg BL. Reactive oxygen species in cell signaling. Am J Physiol Lung Cell Mol Physiol. 2000;279:L1005–1028. doi: 10.1152/ajplung.2000.279.6.L1005. [DOI] [PubMed] [Google Scholar]
- Triqueneaux G, Thenot S, Kakizawa T, Antoch MP, Safi R, Takahashi JS, Delaunay F, Laudet V. The orphan receptor Rev-erbα gene is a target of the circadian clock pacemaker. J Mol Endocrinol. 2004;33:585–608. doi: 10.1677/jme.1.01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukai-Tadenuma M, Kasukawa T, Ueda HR. Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat Cell Biol. 2008;10:1154. doi: 10.1038/ncb1775. [DOI] [PubMed] [Google Scholar]
- Wagener FADTG, Volk H-D, Willis D, Abraham NG, Soares MP, Adema GJ, Figdor CG. Different Faces of the Heme-Heme Oxygenase System in Inflammation. Pharmacol Rev. 2003;55:551–571. doi: 10.1124/pr.55.3.5. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M, Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Wu Y-H, Swaab DF. Disturbance and strategies for reactivation of the circadian rhythm system in aging and Alzheimer’s disease. Circadian Rhythms in Sleep Medicine. 2007;8:623. doi: 10.1016/j.sleep.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of Cellular Clocks in the Suprachiasmatic Nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- Yang J, Kim KD, Lucas A, Drahos KE, Santos CS, Mury SP, Capelluto DGS, Finkielstein CV. A Novel Heme-Regulatory Motif Mediates Heme-Dependent Degradation of the Circadian Factor Period 2. Mol Cell Biol. 2008;28:4697–4711. doi: 10.1128/MCB.00236-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Rev-erbα, a Heme Sensor That Coordinates Metabolic and Circadian Pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Yoo S-H, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong H-K, Oh WJ, Yoo OJ, Menaker M, Takahashi JS. Inaugural Article: PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. PNAS. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC. Nonredundant Roles of the mPer1 and mPer2 Genes in the Mammalian Circadian Clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.