Abstract
Background
The African trypanosome Trypanosoma brucei is covered with a dense layer of Variant Surface Glycoprotein (VSG), which protects it from lysis by host complement via the alternative pathway in the mammalian bloodstream. Blocking VSG synthesis by the induction of VSG RNAi triggers an unusually precise precytokinesis cell-cycle arrest.
Methodology/Principal Findings
Here, we characterise the cells arrested after the induction of VSG RNAi. We were able to rescue the VSG221 RNAi induced cell-cycle arrest through expression of a second different VSG (VSG117 which is not recognised by the VSG221 RNAi) from the VSG221 expression site. Metabolic labeling of the arrested cells showed that blocking VSG synthesis triggered a global translation arrest, with total protein synthesis reduced to less than 1–4% normal levels within 24 hours of induction of VSG RNAi. Analysis by electron microscopy showed that the translation arrest was coupled with rapid disassociation of ribosomes from the endoplasmic reticulum. Polysome analysis showed a drastic decrease in polysomes in the arrested cells. No major changes were found in levels of transcription, total RNA transcript levels or global amino acid concentrations in the arrested cells.
Conclusions
The cell-cycle arrest phenotype triggered by the induction of VSG221 RNAi is not caused by siRNA toxicity, as this arrest can be alleviated if a second different VSG is inserted downstream of the active VSG221 expression site promoter. Analysis of polysomes in the stalled cells showed that the translation arrest is mediated at the level of translation initiation rather than elongation. The cell-cycle arrest induced in the presence of a VSG synthesis block is reversible, suggesting that VSG synthesis and/or trafficking to the cell surface could be monitored during the cell-cycle as part of a specific cell-cycle checkpoint.
Introduction
African trypanosomes are masters of extracellular survival in the mammalian bloodstream, where they multiply in the face of continuous host immune attack both from antibodies and the complement system. Of critical importance for the bloodstream form trypanosome is a dense protective layer of Variant Surface Glycoprotein (VSG), which shields invariant surface receptors from recognition [1]. Eventually the host mounts an effective antibody response against a given VSG variant, whereby B-cell responses against the predominant VSG play a critical role [2]. However, as new VSG switch variants continuously arise within the population, these escape recognition and form the next wave of infection. This highly sophisticated strategy of antigenic variation (reviewed in: [3], [4], [5]) allows the trypanosome to maintain a chronic infection.
An individual trypanosome encodes a vast repertoire of more than 1500 VSGs which are highly divergent in sequence [6]. In fact, it has been estimated that in T. brucei 927 about 60% of the VSGs are unique, with the rest occurring in very small subfamilies [7]. Despite this great dissimilarity at the sequence level, VSGs with different amino acid sequences have a highly conserved tertiary structure [8], [9]. This conservation in VSG shape presumably allows a trypanosome switching from one VSG type to another to form a protective coat composed of different VSG types. There are about 5×106 VSG dimers per cell, which are attached to the cell surface through a glycosylphosphatidylinositol (GPI) anchor [10]. This makes the VSG layer on the trypanosome cell surface a very dense but highly fluid barrier. Extremely high rates of VSG endocytosis allow the trypanosome to continuously exchange the VSG on its surface [11]. Trypanosome motility coupled with these high rates of endocytosis allow the trypanosome to rapidly remove VSG-antibody complexes, providing protection from low titres of anti-VSG antibodies [12].
We have shown previously that VSG is essential in bloodstream form T. brucei, even in vitro in the absence of antibodies or complement. Blocking VSG synthesis results in a very striking and precise precytokinesis cell-cycle arrest with no re-initiation of S phase [13]. The precision of this cell-cycle arrest argues that VSG synthesis is monitored as part of a cell-cycle checkpoint, whereby progression is halted in the absence of sufficient VSG [13]. The unusually tight nature of this precytokinesis cell cycle arrest phenotype is unique in bloodstream form T. brucei, as other bloodstream form RNAi cytokinesis mutants described thus far have phenotypes whereby cells continue to attempt cytokinesis while subsequently re-entering S-phase in a new cell-cycle [14], [15], [16], [17], [18], [19]. For example, depletion of the GPI8 catalytic subunit of the GPI: protein transamidase complex in bloodstream form T. brucei (resulting in the accumulation of unanchored VSG) results in a precytokinesis arrest [19]. Alternatively, inhibition of synthesis of a variety of flagellar proteins in bloodstream form T. brucei including the basal body and flagellar protein KMP-11 [16], [18] or aurora kinase-1 (TbAUK1) and related proteins results in an inhibition of cytokinesis [14], [15]. However these different precytokinesis cell-cycle arrest phenotypes are all imprecise, as the arrested cells repeatedly re-enter S-phase, and show characteristic multinuclear, multikinetoplast and multiflagellar phenotypes.
In contrast, cells in which VSG221 RNAi has been induced are precisely stalled precytokinesis with two nuclei and two kinetoplasts, and show no indication of cleavage furrow initiation or re-entry into S-phase. The fact that the arrested cells induced by VSG RNAi do not re-enter S-phase, and the precision of the precytokinesis block suggest that VSG synthesis or transport could be sensed through a mechanism that interacts with the trypanosome cell-cycle. It is likely that in the absence of VSG synthesis or transport to the cell surface, a checkpoint is activated which accurately stops cell-cycle progression, preventing further cell growth and an increase in cell volume, which would cause a dilution of the cell surface VSG.
Here we demonstrate that the precise precytokinesis arrest triggered by the induction of VSG RNAi, is due to a block in VSG synthesis rather than toxic effects caused by large amounts of siRNAs derived from the ablated VSG transcript. We show that the VSG RNAi induced cell-cycle arrest could be rescued if a second different VSG, which is not recognised by the VSG RNAi, was introduced into the same VSG expression site. Strikingly, we show that blocking VSG synthesis triggered a global down-regulation of protein synthesis down to less than 1–4% normal levels. This translation arrest was correlated with disassociation of ribosomes from the endoplasmic reticulum (ER) and a drastic reduction in polysomes, arguing that the translation arrest was operating at the level of translation initiation. Additionally, we show that the precise precytokinesis cell-cycle arrest observed was reversible, suggesting that VSG synthesis or transport to the cell surface could be monitored as part of a cell-cycle checkpoint.
Results
The cell-cycle arrest triggered by VSG221 RNAi could be rescued by expression of VSG117
Blocking VSG synthesis by inducing VSG RNAi results in a very striking and specific cell-cycle arrest whereby cells stall prior to cytokinesis without reinitiating S-phase [13]. However, a concern with this experimental approach, is that the induction of VSG RNAi could result in high levels of siRNA generated from the RNAi mediated degradation of the highly abundant VSG transcript, which is efficiently ablated down to 1–2% normal levels [13]. In order to determine if toxic effects of the VSG221 siRNA were causing the growth arrest, we generated trypanosomes expressing two different VSGs from the same VSG expression site (Fig. 1A). The parental T. brucei 221VB1.1 line expresses a telomeric VSG221 gene, and contains a construct allowing the induction of VSG221 RNAi. These cells cease growth very rapidly in the presence of tetracycline induced VSG221 RNAi (Fig. 1B) [13]. In this cell line, a construct containing a VSG117 gene was inserted immediately downstream of the promoter of the VSG221 expression site to produce the T. brucei 221VP117 cell line (Fig. 1A).
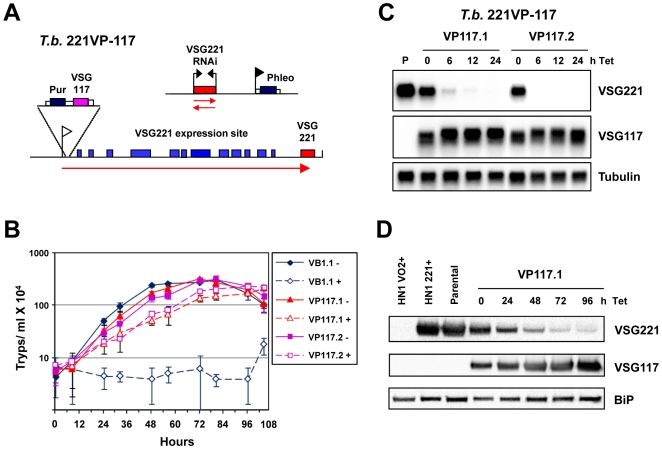
Figure 1. T. brucei expressing both VSG117 and VSG221 from the active VSG221 expression site escapes VSG221 RNAi induced cell-cycle arrest.
A) Schematic of the T. brucei 221VP-117 cell line. A construct containing a VSG117 gene linked to a puromycin resistance gene (Pur) and flanked by tubulin and VSG221 RNA processing signals (white boxes) is inserted immediately downstream of the promoter (white flag) of the active VSG221 expression site. Various expression site associated genes within the VSG221 expression site are indicated with blue boxes and the telomeric VSG221 gene with a red box. This cell line also contains a VSG221 RNAi construct linked to a phleomycin resistance gene (dark blue box) driven by an rDNA promoter (black flag). In the presence of tetracycline, transcription of a VSG221 fragment (red box) from opposing tetracycline inducible T7 promoters (black arrows) results in VSG221 RNAi. Relevant transcription is indicated with red arrows. B) T. brucei expressing both VSG117 and VSG221 from the active VSG221 expression site escapes growth arrest in the presence of VSG221 RNAi. The parental T. brucei VB1.1 cell line (VB1) expresses only VSG221 from the active VSG221 expression site and was incubated in the presence (+) or absence (−) of tetracycline to induce VSG221 RNAi. The T. brucei 221VP-117 clones VP117.1 and VP117.2 were also grown in the presence or absence of tetracycline in order to induce VSG221 RNAi. The standard deviation of triplicate counts is indicated with error bars. C) Northern blot analysis of T. brucei 221VP-117 cell lines in the presence of VSG221 RNAi. RNA from the parental (P) T. brucei 221VB1.1 cell line was compared with RNA from the T. brucei 221VP-117 clones VP117.1 and VP117.2 which had VSG221 RNAi induced with tetracycline (Tet) for the time in hours (h) indicated above. The blots were probed for VSG221, VSG117 or tubulin as a loading control. D) Western blot analysis of the T. brucei 221VP-117 cell line VP117.1 after the induction of VSG221 RNAi for the time in hours indicated above. Protein lysates from T. brucei HN1(VO2+) expressing VSGVO2, T. brucei HN1(221+) expressing VSG221 or the parental T. brucei 221VB1.1 cell line expressing VSG221 were compared with lysates from T. brucei 221VP-117 clone VP117.1 in the presence of VSG221 RNAi induced for the time in hours indicated above. The Western blot was probed with antibody against VSG221, VSG117 or BiP as a loading control.
Two independent T. brucei 221VP117 clones were generated which express two VSGs from the same VSG expression site. Approximately equal amounts of both VSG117 and VSG221 transcript and protein (Fig. 1C and 1D) were produced in these cells, and both VSGs were present on the cell surface (result not shown). Although these two VSG genes were expressed from the same VSG expression site, the amount of transcript and protein produced from each VSG appeared to be about half normal levels (Fig. 1C and 1D). When VSG221 transcript was knocked down by the induction of VSG221 RNAi, both VSG117 transcript and protein increased. This indicates that the maximum amount of VSG transcript that can be expressed is restricted, possibly due to either a limiting VSG transcript stabilising protein or a restricting aspect of the RNA processing machinery. Double-expressing trypanosomes had been thought to express more VSG than normal, and it had been speculated that these trypanosomes expressing two different VSGs on their surface could have a larger volume than normal [20]. However, in agreement with our finding that “double-expressor” trypanosomes expressing both VSG117 and VSG221 on their surface had levels of total VSG comparable to parental trypanosomes expressing just VSG221, we did not find that the “double-expressors” had a significantly larger cell volume (Fig. S1).
As VSG117 and VSG221 are dissimilar in sequence, VSG117 transcript is not targeted by VSG221 RNAi. As expected, after the induction of VSG221 RNAi with tetracycline for 6 hours, the VSG221 transcript had almost completely disappeared, while levels of the VSG117 transcript did not decrease (Fig. 1C). Disappearance of the VSG221 transcript was followed by decrease of VSG221 protein from the cells where VSG221 RNAi had been induced (Fig. 1D), presumably as a result of protein turnover, and dilution through cell growth. The induction of VSG221 RNAi in the T. brucei 221VP117 cells did not lead to significant growth arrest, showing that the VSG117 protein can compensate for lack of VSG221 (Fig. 1B). These results demonstrate that the drastic growth arrest induced in the parental T. brucei 221VB1.1 cells in the presence of VSG221 RNAi was not a consequence of siRNA toxicity, as VSG221 siRNA was also being produced in the rescued T. brucei 221VP117 lines in the presence of tetracycline. In contrast, these results argue that the precise precytokinesis arrest observed after the induction of VSG RNAi is a direct consequence of blocking VSG synthesis. Attempts to establish if high level expression of a different surface protein on the surface of bloodstream form T. brucei could complement for lack of VSG synthesis were unsuccessful. We inserted a copy of a procyclin gene in the active VSG221 expression site, however high level expression of procyclin appeared to be toxic in bloodstream form T. brucei (S.T. and G.R. unpublished results).
Blocking VSG synthesis triggers a global translation arrest
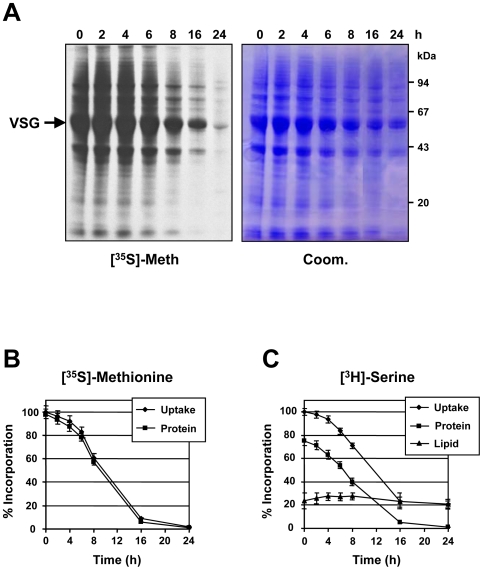
As blocking VSG synthesis triggers a cell-cycle arrest and a block in cell growth, we investigated if further metabolic activities were compromised. Total protein synthesis was followed by metabolic labeling of T. brucei 221VG1.1 cells with [35S]-methionine where VSG RNAi was induced for different times over a 24 hour period. Surprisingly, total protein synthesis decreased dramatically in a time dependent manner, and was reduced to less than 1% after 24 hours (Fig. 2A and 2B). In contrast, the total amount of protein as detected using Coomassie stained gels remained largely unchanged (Fig. 2A). As incorporation of [35S]-methionine into protein decreased, uptake also decreased, consistent with the cell maintaining a normal intracellular level of the amino acid even under conditions where protein synthesis was not draining the intracellular pool (Fig. 2B). In parallel, we investigated [3H]-serine uptake and incorporation into protein and lipid (Fig. 2C, Fig. S2). After the induction of a VSG synthesis block, the decrease in incorporation of [3H]-serine into protein resembled that seen monitoring incorporation of [35S]-methionine. However, incorporation of [3H]-serine into the lipid fraction remained unchanged over 24 hours. Using a variety of approaches, we have found that lipid synthesis was unimpaired in T. brucei 221VG1.1 stalled by the induction of VSG221 RNAi for up to 24 hours (T. K. S. unpublished results). As a consequence, the uptake of [3H]-serine, while decreasing as a consequence of the translation arrest, reached equilibrium at about 20% of its original level. This remaining serine uptake supplies the unchanged serine utilisation in the formation of serine containing phospholipids i.e. phosphatidylserine (T. K. S. unpublished results).
Figure 2. Blocking VSG synthesis triggers a global translation arrest in T. brucei.
A) T. brucei 221VG1.1 cells had VSG synthesis blocked by the induction of VSG221 RNAi for the time indicated in hours (h) prior to labeling with [35S]-methionine for one hour. Proteins were separated on an SDS-PAGE gel. The left panel shows [35S]-labeled proteins detected by fluorography ([35S]-Meth.). On the right is the corresponding Coomassie stained gel (Coom.). Protein sizes are indicated in kiloDaltons (kDa). B) Triplicate samples of the [35S]-methionine labeled cells were processed to determine the mean rate) of [35S]-methionine uptake and incorporation into total protein after the induction of VSG221 RNAi for the time indicated in hours (h) with the standard deviation indicated with error bars. C) VSG221 RNAi was induced in T. brucei 221VG1.1 cells for the time indicated in hours (h) prior to labeling for one hour with [3H]-serine. Replicate aliquots of the labeled cells were processed, and incorporation of [3H]-serine radiolabel into either the whole cell (uptake), total protein or lipid fractions was determined. The values show the means and standard deviations (indicated with error bars) of three separate labeling experiments, whereby the values at time 0 are normalised to 100%.
One mechanism for mediating a translation block is by a perturbation of amino acid transport resulting in a reduction in intracellular amino acid concentrations. We therefore measured amino acid pools in uninduced T. brucei 221VG1.1 cells compared with cells where VSG221 RNAi had been induced for 24 hours (Table 1). No drastic reductions in amino acid levels were observed with the exception of levels of serine and glutamine, which were reduced to approximately 33% and 22% of normal levels respectively. Although this reduction is significant, it is unlikely to result in the observed extreme translation block, which is down to less than 1–4% normal levels. The intracellular concentrations of both glycine and arginine surprisingly increase upon induction of VSG RNAi, which may be associated with the general slow down in other specific metabolic pathways. It should also be noted that these amino acids are also used for cellular functions other than protein synthesis. Serine is used for phospholipid biosynthesis and glutamine as a donor for amino-transferases while glycine may be used in the glycine cleavage system and arginine is utilised in polyamine de novo synthesis.
Table 1. Intracellular amino acid concentrations in T. brucei in the presence (+) or absence (−) of tetracycline induced VSG RNAi for 24 hours.
| Intracellular amino acid concentration (µM)a | |||
| Amino Acid | VSG RNAi (−) | VSG RNAi (+) | % of wild type |
| Asp | 113.5±5.8 | 86.3±4.9 | 76 |
| Glu | 243.1±8.5 | 169.0±6.4 | 70 |
| Gly | 306.2±9.3 | 545.6±13.8 | 178 |
| Ala | 68.9±3.6 | 61.8±4.6 | 90 |
| Val | 158.8±4.5 | 135.1±4.0 | 85 |
| Ileu | 207.8±6.9 | 174.1±4.6 | 84 |
| Leu | 145.6±4.5 | 126.7±3.8 | 87 |
| Gln | 841.3±7.6 | 184.1±4.5 | 22b |
| Ser | 94.4±3.0 | 31.1±1.3 | 33b |
| Thr | 45.9±2.2 | 33.3±1.8 | 73 |
| Arg | 69.9±3.5 | 112.3±9.4 | 161 |
| Lys | 55.6±7.8 | 53.7±6.2 | 97 |
| Tyr | 82.6±5.6 | 72.4±5.6 | 88 |
| Phe | 27.0±1.9 | 21.4±2.2 | 79 |
| Try | 4.7±0.4 | 5.9±0.7 | 126 |
| Meth | 379.8±8.7 | 354.8±6.7 | 93 |
| Pro | 79.7±3.4 | 79.3±4.1 | 100 |
Amino acid concentrations within the cells were determined as described in Materials and Methods. The intracellular concentration was calculated using the cell volume 5.89 µl/108 cells [59]. Values are means±SD of three separate determinations normalised using the internal standard norleucine.
Significant difference (P<0.05).
We wanted to investigate if the observed translation arrest was a downstream nonspecific effect of a lethal RNAi phenotype. We therefore additionally performed [35S]-methionine metabolic labeling experiments in bloodstream form T. brucei stalled after the induction of RNAi against clathrin [21], the flagellar protein PFR2 [18], tubulin [22] and actin [23] (Fig. S3). RNAi mediated inhibition of these different proteins in bloodstream form T. brucei results in inhibition of growth within 24 hours, with eventual cell death. Inhibition of tubulin and PFR2 synthesis results in an imprecise precytokinesis arrest and cells with multiple nuclei and multiple flagella [18], [22]. Knock-down of clathrin and actin interferes with vesicular trafficking, producing an enlarged flagellar pocket or “big-eye” phenotype and rapid cell death [21], [23] [24].
Although inhibition of tubulin synthesis for 24 hours quickly arrests cells which later die, there was no evidence for significantly reduced translation in these stalled cells (Fig. S3). Inhibition of PFR2 synthesis resulted in translation reduced to about 40% normal levels, which is still at least ten to forty fold higher than that observed after blocking VSG synthesis. Inhibition of clathrin synthesis resulted in a reduction in translation to about 19% normal levels, and blocking actin synthesis resulted in reduction in levels of translation to about 9% normal. However, knock-down of these last two proteins would also be expected to interfere with endocytosis, and therefore with VSG recycling [12], [21], [23], [24]. It is therefore unclear if the observed translation arrest in these cells was a consequence of a disruption of deposition of VSG on the cell surface. The fact that RNAi mediated knock-down of tubulin effectively stalls cell growth without a significant decrease in translation, indicates that reduced protein synthesis is not always a consequence of a lethal RNAi phenotype. In addition, there are a number of examples of conditional knock-outs of metabolic enzymes generating a lethal phenotype prior to disruptions to translation associated with a dying cell. These include conditional knock-outs of phosphatidylinositol synthase [25], myo-inositol-3-phosphate synthase [26], and cytidyl ethanolamine-phosphate transferase [27].
RNA metabolism in cells stalled by the induction of VSG221 RNAi
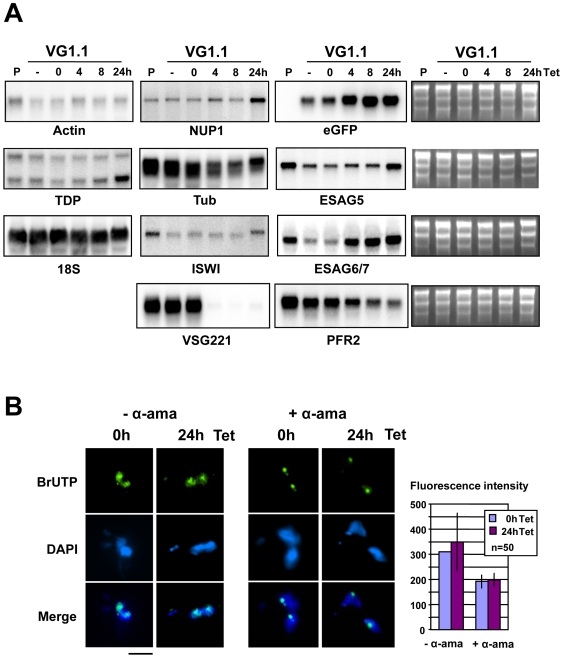
We next investigated if this total block in protein synthesis was caused by changes in RNA transcript stabilities or a global transcription arrest in the stalled T. brucei VG1.1 cells. Northern blot analysis of T. brucei 221VG1.1 [13] showed rapid ablation of VSG221 transcript within 4 hours of induction of VSG221 RNAi (Fig. 3A). The blots were subsequently hybridised with probes for a range of housekeeping genes including the chromatin proteins TDP1 [28] and TbISWI [29], NUP1 [30], structural proteins like the flagellar protein PFR2 [31], actin, tubulin and the 18S ribosomal RNA as well as eGFP (which is present in the active VSG221 expression site [13]), ESAG5 and ESAG6/7. We did not observe any drastic decreases in transcript levels, certainly not down to levels that would explain the observed global translation arrest (Fig. 3A, Fig. S4). The minor decrease in PFR2 and tubulin transcript after the induction of VSG RNAi could be a consequence of feedback mechanisms down-regulating transcripts in cells that are stalled in the cell-cycle and therefore not in need of these structural proteins. In addition, we found no evidence that induction of VSG221 RNAi resulted in a decrease in the level of transcripts derived from the active VSG221 expression site.
Figure 3. Transcription analysis of cells where VSG synthesis is blocked.
A) Northern blot analysis of T. brucei 221VG1.1 cells where VSG221 RNAi had been induced for the time indicated above in hours (h). The parental (P) T. brucei 90-13 cell line does not contain the VSG221 RNAi construct. The T. brucei 221VG1.1 cell line was either not incubated with tetracycline (− Tet) or had VSG221 RNAi induced with tetracycline for the times indicated above. Blots were hybridised with probes for Actin, NUP1, eGFP (present in the active VSG221 ES), TDP1, tubulin (Tub), ESAG5, 18S rRNA, ISWI, ESAG6/7, VSG221 and PFR2. Ethidium stains of the gels are indicated on the right to indicate total RNA loaded. B) Transcription analysis of T. brucei 221VG1.1 where VSG synthesis was blocked by the induction of VSG221 RNAi with tetracycline (Tet) for 0 or 24 hours (h). Cells were incubated with BrUTP to label nascent transcripts and subsequently incubated with an anti-BrdUTP antibody, and a secondary antibody coupled to Alexa 488. DNA was stained with DAPI. A normal precytokinesis cell (0 h) is compared with a precytokinesis cell arising after the induction of VSG RNAi for 24 hours (24 h). The experiment was performed in the absence of α-amanitin (− α-ama) to visualise total transcription, or in the presence of 200 µg ml−1 α-amanitin to inhibit transcription by RNA polymerases II and III and visualise transcription by RNA polymerase I (+ α-ama). The scale bar indicates 4 µm. Quantitation of transcription as fluorescence in the FITC channel is in arbitrary units using 50 cells per time point, with standard deviation indicated with error bars.
Recently, a novel stress response has been described in T. brucei where RNAi mediated silencing of the signal-recognition particle (SRP) receptor results in silencing of spliced leader RNA transcription and a significant reduction in total mRNA [32]. However, no reduction in spliced leader RNA was found in T. brucei 221VG1.1 cells after the induction of VSG221 RNAi for up to 24 hours (Fig. S5). Therefore the translation arrest observed after the induction of VSG221 RNAi must be operating through a different stress-response pathway than that involved in the spliced leader RNA silencing (SLS) response.
To investigate if transcription was arrested in the stalled cells, we monitored levels of total transcription in T. brucei 221VB1.1 by incorporating BrUTP into nascent transcripts in permeabilised cells (Fig. 3B). The incorporated BrUTP nucleotide analogue can be detected using an antibody against BrdUTP [33]. We compared levels of BrUTP incorporation in precytokinesis cells in the absence or presence of VSG221 RNAi. After 24 hours tetracycline induction of VSG221 RNAi, arrested cells had levels of total transcription that appeared comparable to that in untreated cells immediately prior to cytokinesis (Fig. 3B, -α-ama). The toxin α-amanitin inhibits RNA polymerase II and III transcription at concentrations of 200 µg ml−1, leaving transcription by RNA polymerase I unaffected. As seen in Fig. 3B, the inclusion of α-amanitin (+ α-ama) inhibited all transcription except for that of the rRNA in the nucleolus, as well as transcription of the active VSG expression site located in the expression site body (ESB), which is seen as a small discrete spot [34]. There was no discernible difference in transcription of RNA Pol I after blocking VSG synthesis for 24 hours. Therefore the total block in protein synthesis was not caused by a general arrest in transcription.
Induction of a VSG synthesis block results in disassociation of ribosomes from the ER and a reduction in polysomes
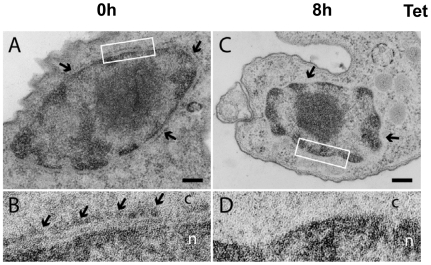
If the observed translation arrest was operating at the level of translation elongation, one would expect to see unaffected distribution of ribosomes on the ER. In order to investigate this, we determined the distribution of ribosomes on the endoplasmic reticulum (ER) using transmission electron microscopy (TEM) and a nonstandard TEM fixing and staining technique. Normally ribosomes are distributed uniformly on the outer membrane of the nuclear envelope, which is contiguous with the rough ER (indicated with arrows in Fig. 4A and B). After induction of VSG221 RNAi in T. brucei 221VG1.1 cells for 8 hours, the nuclear envelope appeared to be devoid of significant numbers of bound ribosomes (Fig. 4C and D). This disassociation of ribosomes from the ER coincided with the previously observed decrease in protein synthesis. This result is consistent with the observed translation arrest being a consequence of disassociation of polysomes in the stalled cells rather than a scenario whereby polysomes remain intact but translation elongation is blocked.
Figure 4. Ultrastructural evidence for disassociation of ribosomes from the endoplasmic reticulum after blocking VSG synthesis.
A) In non-induced T. brucei 221VG1.1 cells (0 h), the outer membrane of the nuclear envelope is studded with ribosomes (arrows). Membranes are not contrasted because cells are not treated with osmium tetroxide. Scale bars represent 200 nm. B) A region of the nuclear envelope indicated with a white box in (A) shown at 4x higher magnification. An array of membrane-associated ribosomes is indicated with arrows. c = cytoplasm and n = nucleoplasm. C) After induction of VSG RNAi for 8 hours, large stretches of nuclear envelope are devoid of ribosomes (arrows). D) A region of the nuclear envelope indicated with a white box in (C) shown at 4x higher magnification. No membrane-associated ribosomes are observed.
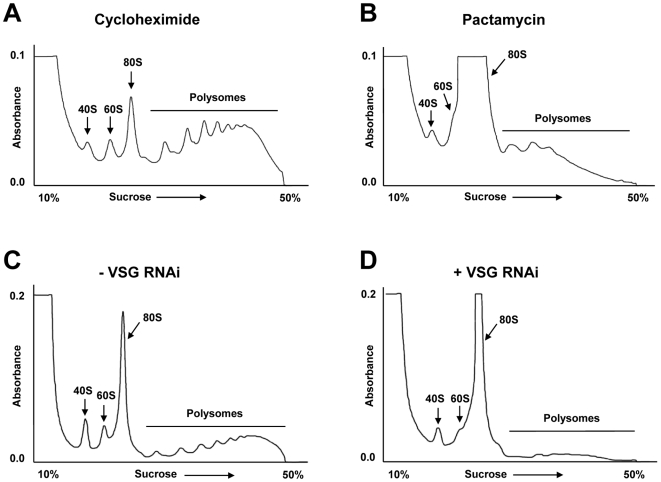
Polysomes are large complexes composed of translating ribosomes associated with mRNA transcripts which fractionate at the bottom of sucrose gradients. In contrast, free ribosomes fractionate at the top of the gradient. Polysome profiles from normal cells and cells where a VSG RNAi mediated block of VSG synthesis had been induced were determined by separating lysed T. brucei over sucrose gradients (Fig. 5). We first determined the polysome profile of T. brucei incubated with cycloheximide (50 µg ml−1), which blocks translation elongation [35]. The ribosomes as monitored by absorbance at 254 nm were primarily found in polysomes, which contain transcripts associated with the translating ribosomes (Fig. 5A). In contrast, if T. brucei is incubated with pactamycin (200 ng ml−1) which blocks translation initiation [35], [36], ribosomes disassociated from the transcripts, and there was a dramatic increase in free ribosomes (80S)(Fig. 5B).
Figure 5. Polysome profile analysis indicates that the VSG221 RNAi induced global translation arrest operates at the level of translation initiation.
A) Polysome profile of T. brucei 221VG1.1 cells which have been incubated with cycloheximide (50 µg ml−1) to block translation elongation. The percentage of sucrose from top to bottom of the gradient is indicated on the X-axis, with the absorbance at 254 nm indicated on the Y-axis. The free ribosomes (80S) and ribosomal subunits (40S and 60S) are indicated with arrows. The polysomes (translating ribosomes bound to transcript) are indicated with a bar. B) Polysome profile of T. brucei 221VG1.1 cells which have been incubated with pactamycin (200 ng ml−1) to block translation initiation. C) Polysome profile of T. brucei 221VG1.1 in the absence (−) of VSG RNAi. D) Polysome profile of T. brucei 221VG1.1 where VSG221 RNAi has been induced for 24 hours (+).
In a similar fashion, polysome profiles were determined for T. brucei 221VG1.1 in the presence or absence of a VSG synthesis block. In the absence of VSG221 RNAi (Fig. 5C), most of the ribosomes were associated with polysomes. However, after the induction of VSG221 RNAi for 24 hours, there was a drastic decrease in polysomes and an increase in free ribosomes (80S) (Fig. 5D). These experiments demonstrate that the global translation arrest observed after the induction of VSG221 RNAi was operating through a block in translation initiation rather than translation elongation.
We have not found evidence for striking changes in levels of a range of different T. brucei translation factors (eIF4A, eIF4E1-4) [37](M. Narayanan, O. Neto, M.C. and G.R. unpublished) after the induction of a VSG RNAi induced cell-cycle arrest. Phosphorylation of translation initiation factor eIF2α is an important potential mechanism for blocking global protein synthesis at the level of translation initiation [38], [39]. However, we have not found evidence for changes in levels or phosphorylation state of T. brucei eIF2α in cells stalled after the induction of VSG RNAi for up to 24 hours (M. Narayanan and G. R. unpublished results). Similar to what we describe here, there is a comparable reduction in polysome abundance after the induction of a heat-shock response in trypanosomes, yet no evidence for changes in the phosphorylation state of eIF2α [40]. This could indicate significant differences in translation control in trypanosomes compared with other eukaryotes.
A concern with a global translation arrest is that it is a consequence of dying cells. We know that levels of transcription, lipid synthesis and endocytosis of FITC-tomato lectin are not obviously affected in the arrested cells (Fig. 3, [13]), indicating that they are metabolically active. In order to investigate protein composition in these stalled cells, we used proteomic two dimensional Difference Gel Electrophoresis (2D-DIGE analysis) [41]. We did not see any significant up- or down-regulation of the levels of any individual protein in cells stalled after the induction of VSG RNAi for 8 hours, when the cell-cycle arrest is already maximal (Sup. Fig. 6). Although this type of proteomic analysis would only be expected to sample the most abundant proteins (in our case 1486 different spots), this result indicates that the arrested cells do not have a drastically different protein composition from normal. This would support the observation that the cells arrested immediately after the induction of the cell-cycle arrest are not an obviously dying system that is degrading. Because blocking VSG synthesis could be expected to result in aberrant expression of other surface proteins in the cell as a compensatory effect, we looked specifically at the procyclic specific cell surface protein procyclin. However, we did not observe that blocking the major cell surface protein (VSG) resulted in significant upregulation of procyclin, either at the RNA or the protein level (Fig. S7).
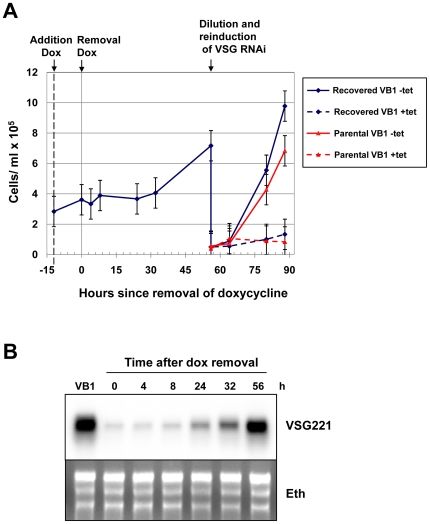
Figure 6. The cell-cycle arrest in T. brucei 221VB1.1 triggered by blocking VSG221 synthesis is reversible after the removal of doxycycline.
A) Doxycycline was added to a culture of T. brucei 221VB1.1 (VB1) for 12 hours (indicated with an arrow labeled addition dox) to induce VSG221 RNAi and a subsequent cell-cycle arrest. The cells were then washed to remove the doxycycline (removal dox) and growth was monitored. After an initial stalled period, the trypanosomes started to grow. In order to establish that these growing trypanosomes were still responsive to VSG221 RNAi, these growing trypanosomes were diluted and VSG221 RNAi was induced with 1 µg ml−1 tetracycline (blue dashed line). As a control, VSG221 RNAi was also induced in the parental T. brucei 221VB1.1 cells (red lines). The standard deviation of counts in triplicate is indicated with error bars. B) Northern blot analysis of RNA from T. brucei 221VB1.1 cells following removal of doxycycline. RNA from the noninduced T. brucei 221VB1.1 (VB1) is compared with RNA from T. brucei 221VB1.1 cells where doxycycline has been removed from the cultures for the time in hours (h) indicated above. The blot was hybridised with a probe for VSG221. An ethidium stain (Eth) of the blot is indicated below.
The VSG RNAi mediated cell-cycle arrest is reversible
As a global translation arrest is an extreme phenotype, we next determined if the cell-cycle arrest triggered by the induction of VSG RNAi is reversible. T. brucei 221VB1.1 was incubated with the water soluble tetracycline analogue doxycycline for 12 hours to induce VSG221 RNAi and induce a cell-cycle arrest (Fig. 6A). Doxycycline was then removed by washing the cells and transferring them to fresh medium. VSG RNAi was induced using 300 pg ml−1 doxycycline, as this concentration reproducibly triggered a maximum cell cycle arrest, yet was low enough to facilitate the washing step (Fig. S8). Comparable cell cycle arrest results were obtained using 2 ng ml−1 tetracycline. However using tetracycline resulted in greater experimental variability compared with doxycycline, possibly due to differences in its solubility or cell permeability.
After an initial lag of about 24 hours following doxycycline removal, the cells resumed growth, presumably due to the translation arrest impeding rapid recovery. Northern blot analysis showed that VSG221 transcript levels returned back to normal levels 56 hours after the stalled cells were removed from doxycycline (Fig. 6B). These recovered cells stalled abruptly if VSG221 RNAi was induced again, showing that they were still fully responsive to VSG RNAi. To determine the cloning efficiency of this experimental procedure, serial dilutions were plated out in microtitre dishes. The observed rescued trypanosomes arose reliably in about 15% of the wells when less than 80 stalled cells were allowed to recover, which is about 30x less efficient than that of cloning the parental T. brucei 221VB1.1 line (Fig. S9). This relatively low cloning efficiency is presumably due to deleterious aspects of the VSG RNAi induction, including the generation of internalised flagella in some of the cells [13]. However, our recovered cells arise more rapidly than would be expected from generation by background mutations, as the mutation rate in T. brucei has been estimated to be about 10−9 per bp per cell generation [42]. These results argue that the cell-cycle arrest triggered by blocking VSG synthesis is reversible. After a brief arrested state, some of the cells re-enter the cell-cycle and resume dividing. The reversibility of the precise precytokinesis cell-cycle arrest suggests that this phenomenon is not necessarily a dead end pathway. It could possibly have relevance in allowing the trypanosome to react to fluctuations in either VSG synthesis and/or deposition of VSG on the cell surface that are less drastic than those described here, where essentially all VSG synthesis is blocked for a significant period of time.
Discussion
We show that although the induction of VSG221 RNAi normally induces a precise precytokinesis cell-cycle arrest in VSG221 expressing trypanosomes, cells did not stall in the cell-cycle if VSG117 (which is not recognised by the VSG221 RNAi) was also expressed from the active VSG221 expression site. This argues that the cell-cycle arrest observed after the induction of VSG221 RNAi is a consequence of lack of newly synthesised VSG rather than toxicity of the VSG221 siRNA. Surprisingly, an extreme and global block in protein synthesis was induced in the stalled cells, whereby total translation was reduced to 1–4% normal levels after 24 hours induction of VSG221 RNAi. No major changes in transcription or transcript levels were observed that explain this protein synthesis block. However, after 8 hours induction of VSG RNAi ribosomes appeared to have disassociated from the ER. Polysome analysis of the stalled cells showed that the translation block was operating at the level of translation initiation rather than translation elongation. Despite the striking changes in the arrested cells, particularly with regards to the global arrest in protein synthesis, the cell-cycle arrest was reversible suggesting that VSG synthesis and/or deposition on the cell-surface is possibly being monitored as part of a normal cell-cycle checkpoint.
Earlier, it has been shown that T. brucei can be genetically modified to express two VSGs from the telomere of the active VSG expression site [43]. We show that a second VSG could also be efficiently expressed if it was inserted immediately downstream of the promoter of the active VSG expression site rather than in its usual telomeric location. The invariably telomeric location of VSGs within VSG expression sites therefore presumably facilitates VSG recombinogenicity rather than being essential for expression.
Surprisingly, the precise precytokinesis arrest observed after blocking VSG synthesis coincided with an extreme and global block in protein synthesis down to less than 1–4% normal levels. No major decrease in transcription or steady state transcript levels was observed that explains this protein synthesis block. Although there was some reduction in transcripts encoding structural proteins including the flagellar protein PFR2 and tubulin, the observed reduction of just a few transcripts does not explain the global protein synthesis arrest phenotype. Similarly, Northern blots probed with spliced leader RNA specific oligonucleotides showed no drastic reduction in total mRNA (S.T. and G.R. unpublished results). These results are compatible with our observation that transcription was not detectably disrupted in these stalled cells. These data agree with previously reported results [44], which show that the T. brucei transcriptome remains largely unchanged in trypanosomes when confronted with a variety of stresses. This indicates that T. brucei does not respond to environmental challenges through transcriptional regulation, and in the absence of protein synthesis, still transcribes and then degrades large amounts of excess transcript.
In addition, there were no abrupt decreases in intracellular amino acid pools which could explain this block in protein synthesis. Incorporation of methionine and serine into protein decreased drastically. However, phospholipid synthesis was not greatly altered in these stalled cells (T.K.S. unpublished data), allowing serine uptake (at about 20% normal levels) and incorporation into phospholipids to continue. This continued uptake of serine indicates that the protein synthesis block was not a consequence of disruption in general amino acid uptake, possibly through minor perturbations in the VSG coat negatively affecting amino acid transporters. Also if this were the case, then the stalled cells would not be able to escape the cell-cycle arrest.
Polysome analysis of the stalled cells showed that the translation block was operating at the level of translation initiation rather than translation elongation. Compatible with this, after 8 hours induction of VSG RNAi ribosomes appeared to have disassociated from the ER. The signal sequence on nascent peptides targets the translating ribosome to the ER membrane, facilitating anchoring of the ribosome to the ER through the binding of the signal recognition particle (SRP) to SRP receptors on the ER surface [45], [46]. As VSG is the major secreted protein in bloodstream form T. brucei, blocking its synthesis would result in a drastic reduction in nascent peptides with signal sequences. Ribosomal subunits in mammalian cells remain membrane-bound after pharmacological inhibition of protein synthesis [45]. Even if ribosomal subunits initially remained membrane-bound upon termination of VSG translation, ongoing translation of cytosolic transcripts would result in a clearance of ribosomes from membranes [35]. This probably occurs early after induction of VSG RNAi, and is followed by the observed global translation arrest.
Why is there a global translation arrest in trypanosomes stalled precytokinesis after the induction of a VSG synthesis block? The first possibility is that the translation arrest is triggered as part of a general stress response, perhaps as a consequence of lack of VSG synthesis or deposition on the cell surface. In our experimental system the ER is being depleted of newly synthesised VSG, which accounts for 10% of the total protein, and which could generate a form of stress. In some eukaryotes a global translation block can be observed after the induction of the unfolded protein response (UPR) [reviewed in: [47]]. UPR is triggered by ER stress, including the accumulation of unfolded protein within the ER. If this ER stress continues for a prolonged period, translation arrests in order to stop the stream of additional protein into the overburdened ER [38], [48]. If the situation persists, the cell undergoes apoptosis [47]. Our VSG RNAi induced arrest does not appear to be the same as that induced by UPR, as there was no significant upregulation of the chaperone BiP [13], which is characteristic of UPR [49]. In addition, in the absence of VSG synthesis the cells stall and remain metabolically active rather than entering apoptosis. It is possible that trypanosomes have a novel stress response pathway different to UPR, which is triggered by the stress induced by a restriction operating on the deposition of VSG on the cell surface. In this regard it is striking that knock-down of actin, which would be expected to be involved in VSG recycling, also results in a striking arrest in translation (Fig. S3).
Another stress response in trypanosomes is the spliced leader RNA silencing response (SLS), which occurs after the induction of stress in the ER by pH stress, or by blocking synthesis of the signal recognition particle (SRP) receptor, which facilitates the translocation of secretory proteins across the ER [32]. The SLS response results in the down-regulation of the spliced leader RNA genes, resulting in drastic reduction in total mRNA and subsequent cell death [32]. The translation arrest observed after the induction of VSG RNAi is not coupled to an SLS response, as there was no reduction in spliced leader transcript after induction of VSG RNAi for up to 24 hours (Fig. S5). In addition, there was no significant reduction in total trans-spliced mRNA transcripts (another expected consequence of induction of an SLS response) after the induction of VSG RNAi for up to 24 hours (S.T. and G.R unpublished results; [32]). Therefore the ER stress caused by blocking VSG synthesis appears to use different pathways than those that trigger the spliced leader RNA silencing response in African trypanosomes.
A second possible explanation for the observed translation arrest, is that trypanosomes always transiently arrest protein synthesis when they are in the precytokinesis stage of the cell-cycle. Earlier, it has been shown in mammalian cells that protein synthesis slows down when cells are in the G2/M stage of the cell-cycle [50]. However in our case, this is unlikely to provide the explanation. After the induction of VSG RNAi, the stalled population of trypanosomes is up to 60% enriched for precytokinesis stage cells, but still contains cells in other stages of the cell-cycle which normally would be expected to be translationally active [13]. This scenario is not compatible with our observed block in protein synthesis down to less than 1–4% normal levels.
A third possibility is that the observed translation arrest is a general feature of RNAi phenotypes that result in growth inhibition and eventual death. This is not the case, as cells arrested as a consequence of another lethal phenotype (induction of a tubulin synthesis block) do not show reduced levels of translation (Fig. S3). In addition, we have no evidence that our cells are an obviously dying system within the 24 hour period used in these analyses. Our arrested cells are viable for relatively long periods, with virtually all cells intact and metabolically active (other than protein synthesis) after 24 hours [13]. Endocytosis of FITC-tomato lectin appears unimpaired [13], and as shown here, there was no significant drop in transcription. In addition, the proteomic analysis did not detect changes in the most abundant proteins even in fully stalled cells (Fig. S6), confirming that we are not looking at a system that is obviously falling apart. Last, arrested cells were able to reenter the cell-cycle when doxycycline was removed.
We favour an explanation whereby induction of a VSG synthesis block triggers a stress response resulting in a global translation arrest. Although it is unclear what exactly is being “sensed” in this stress response pathway, our data is most compatible with a scenario whereby the cell senses the restriction occurring from the lack of deposition of VSG on the cell surface. The fact that downregulation of actin (which is necessary for vesicular trafficking of VSG) also triggers a severe translation arrest is compatible with this possibility.
Does this arrest operate in vivo during an infection? Our experiments here document an extreme situation whereby virtually all VSG synthesis is rapidly blocked by the induction of VSG RNAi. It is unclear if anything this extreme occurs within a natural trypanosome infection. VSG synthesis also halts in “stumpy form” T. brucei where transcription of the active VSG expression site stops [51]. There are some superficial similarities between our cells arrested after the induction of a VSG synthesis block and stumpy form T. brucei. Similar to stumpy cells, our arrested cells are shorter and broader than normal, arguing that this morphological change could be a consequence of a restriction operating on VSG [13]. In addition, stumpy form T. brucei also show reduced levels of protein synthesis and a reduction in polysomes [52]. However, stumpy form T. brucei are stalled in G0 rather than precytokinesis, and have undergone other morphological changes that facilitate differentiation into procyclic form trypanosomes that are not seen in our stalled cells. It is unlikely that VSG synthesis is ever drastically reduced in bloodstream form T. brucei to the extent seen after the induction of VSG RNAi. However, possibly the stress response documented here has evolved to allow the trypanosome to reduce cell growth in response to fluctuations in amount of synthesised VSG rather than complete ablation of VSG transcript. An important feature of this stress response is reversibility, which could argue that the triggered cell-cycle arrest has biological relevance, rather than being an irreversible end-point from which the trypanosome is unable to escape. The challenge for the future will be in unravelling exactly which pathways are involved in these novel cell-cycle checkpoints.
Materials and Methods
Trypanosome strains, culturing and genetic modification
Bloodstream form T. brucei 427 was cultured in HMI-9 medium with 10% SerumPlus (SAFC Biosciences) and 10% fetal calf serum (Gibco). All trypanosomes used were bloodstream form variants derived from T. brucei 427 90-13 expressing VSG221 [53]. The T. brucei 221VB1.1 and T. brucei 221VG1.1 cell lines are described in [13]. The T. brucei 221VP117 transformants have the p221-purVSG117UTR construct inserted into the T. brucei 221VB1.1 cell line. The construct is integrated immediately behind the VSG221 expression site promoter using the target fragments of the 221GP1 construct described in [54]. This construct contains the VSG117 gene from pXS5:117VSG (gift of Jay Bangs)[55] flanked downstream by the VSG221 UTR and polyadenylation sequences. The VSG117 cassette is inserted downstream of the puromycin drug resistance gene, which is flanked up and downstream by α-tubulin gene RNA processing signals [54]. Homogeneity of the VSG coat expressed was ensured by maintaining trypanosome transfectants with drug markers in the active VSG expression site on the appropriate drug selection pressure to prevent switches to different VSG expression sites. Tet-system approved fetal calf serum (Clontech) was used for some experiments requiring tetracycline free conditions.
Nucleic acid and polysome analysis
For Northern blot analysis, total RNA was purified using RNeasy RNA isolation kits (Qiagen). Approximately 1.5 µg total RNA was loaded per lane on 1.5% formaldehyde agarose gels, electrophoresed and blotted using standard protocols. Northern blots were hybridized with random primed probes radiolabeled with [32P]-dCTP using Amersham Ready-To-Go DNA labeling Beads (-dCTP) (GE Healthcare). Details on the exact sequence of the different Northern probes can be obtained from the authors. Blots were imaged with a Bio-Rad Personal Molecular Imager FX. Quantitation of signal was performed using Quantity One software, and radioactive signals were plotted in arbitrary units.
Polysome analysis was performed according to [36] and [40], whereby 400 ml cultures of T. brucei 221VG1.1 cells were grown to a density of about 7.5×105 cells ml−1. VSG RNAi was induced by incubating the cells with 1 µg ml−1 tetracycline for 24 hours. Pactamycin treated trypanosomes were incubated with 200 ng ml−1 pactamycin (gift of Pfizer Global Research & Development, Groton, Connecticut, USA) for 20 minutes at 37°C, and cycloheximide-treated trypanosomes were incubated with 50 µg ml−1 cycloheximide for 10 minutes at 37°C prior to the start of polysome purification procedure. Cells were harvested by centrifugation at 4°C and washed with HMI-9 without serum (containing either pactamycin or cycloheximide where appropriate). Cells were resuspended in polysome buffer (120 mM KCl, 2 mM MgCl2, 20 mM Tris pH 7.5, 1 mM DTT) containing either cycloheximide or pactamycin where appropriate. Cells were lysed using the detergent n-octylglycoside (which does not absorb at 254 nm), loaded on top of 10%–50% sucrose gradients and centrifuged for 2 hours at 36,000 rpm at 4°C in a Beckman ultracentrifuge using a SW41 rotor. Gradients were then harvested using a peristaltic pump and analysed using an absorbance reader at 254 nm.
RNA analysis to determine the presence or absence of an SL response was performed using end-labeled 9091 oligonucleotide to detect the spliced leader and U3 oligonucleotide (1×105 cpm pmol−1) according to [56]. After annealing at 60°C for 15 minutes, the sample was kept on ice for one minute. Next, one unit of reverse transcriptase (Expand RT, Roche Molecular Biochemicals) and one unit of RNase inhibitor (Promega) were added, and extension was performed at 42°C for 90 minutes. The reaction was analysed on a 6% polyacrylamide denaturing gel.
Microscopy
Labeling of nascent RNA using BrUTP was performed essentially according to [34], only the Saponin (Sigma) incubation was performed at 4°C. After the transcription reaction, cells were washed and fixed in 2% paraformaldehyde and BrUTP labeled transcripts were detected with a monoclonal anti-BrdUTP antibody (Roche). For transmission electron microscopy analysis of membrane bound ribosomes, cells were fixed by addition of 2.5% glutaraldehyde directly to the culture medium followed by fixation in 2.5% glutaraldehyde in 0.1 M phosphate buffer pH 7.0 for 2 hours at room temperature without osmium tetroxide post-fixation. Cells were dehydrated in a graded series of ethanol and embedded in Epon resin. Thin sections were stained 5 minutes with 5% aqueous uranyl acetate and 1 minute with 0.1% lead citrate and examined in a FEI Tecnai 12 transmission electron microscope.
Cell-cycle arrest reversibility and cell volume measurements
T. brucei 221VB1.1 was cultured with a starting cell density of about 3×105 cells ml−1. Doxycycline was added to a final concentration of 300 pg ml−1 (minimum effective concentration) and incubated for 12 hours in order to induce VSG221 RNAi. Cells were centrifuged, washed with 20 ml HMI-9, and then resuspended in HMI-9 with the appropriate drugs but without doxycycline. Total RNA was isolated at different time points and analysed using Northern blots as described above.
Recovery efficiency of the cells that resume growth after removal of doxycycline was determined by plating out serial dilutions (10x, 100x, 1000x and 10,000x) of the culture (at about 3×105 cells ml−1) in fresh HMI-9 medium with appropriate drugs but without doxycycline. Each dilution was aliquotted over 48 wells of 96-well plates. The cloning efficiency using this method was also established using the parental T. brucei 221VB1.1 cell line in the absence of induction of VSG221 RNAi.
In order to determine the cell volume, the parental T. brucei 221VB1.1 cell line and the T. brucei double expressers 221VP117.1 and 221VP117.2 were cultured to a concentration of 8–10×105 cells ml−1. Fifty-fold dilutions of each culture were prepared in CASY® ton dilution liquid and the cell sizes were determined by using the Coulter size exclusion method on a CASY® Cell Counter and Analyser System model TT (Schärfe, Reutlingen, Germany) according to the manufacturer's instructions. The cell volumes were determined as an average of the measurements from five independent experiments.
Protein analysis and in vivo metabolic labeling
For Western blot analysis, T. brucei 221VP117 cells were cultured and VSG221 RNAi was induced by incubating the cultures with 1 µg ml−1 tetracycline for up to 96 hours. Cells were maintained at a concentration of about 7–10×105 cells ml−1 throughout the experiment. Western blot analysis was performed by analysing lysate from 4×105 cells per lane using 10% polyacrylamide gels, and transferred to nitrocellulose membrane using standard protocols. Blots were probed with antibodies raised against the N-termini of VSG117 or VSG221 (gift of Piet Borst) or BiP (gift of Jay Bangs). Anti-EP procyclin antibody is a mouse monoclonal antibody obtained from Cedarlane Laboratories (CLP0001A). Blots were imaged using Western Lightning® Detection Kit (Perkin Elmer) and a BioRad Fluor-S MAX imager.
For the metabolic labeling experiments, mid-logarithmic growth T. brucei 221VG1.1 cells induced with tetracycline (1 µg ml−1) for different times were centrifuged (800 g 10 minutes) and washed in minimal essential media minus either methionine or serine, before resuspension in the same media at 1×107 cells ml−1. Cells were labeled for 1 hour at 37°C with either 5 µCi ml−1 [35S]-methionine (MP Biomedicals, 1175 Ci mmol−1) or 50 µCi ml−1 of [3H]-serine (20 Ci mmol−1, ARC) in a shaking water bath. The cells were collected by centrifugation and washed briefly in TDB buffer (25 mM KCl, 400 mM NaCl, 5 mM MgSO4, 100 mM Na2HPO4, NaH2PO4, 100 mM glucose) prior to samples taken for either protein or lipid analysis as previously described [26]. Proteins were separated on a 10% SDS-PAGE gel (4×106 cell equivalents per lane) and visualised by Coomassie blue staining. To detect radiolabeled proteins the destained gel was soaked in En3hance™ (NEN) for 30 minutes, washed with water twice, soaked in 10% glycerol and dried and exposed to XAR-5 film at −70°C.
For the in vivo radiolabel incorporation into total, protein or lipid fractions T. brucei 221VG1.1 cells were induced with tetracycline (1 µg ml−1) for different times, and then labeled with [35S]-methionine or [3H]-serine for one hour as described above. Cells were split into three equal volumes and processed as follows: for total incorporation, cells were spun down through tetrachlorophenyl-modified silicone oil (Medford) and washed with TDB, prior to lysis with 1% SDS. Total uptake of radiolabel was quantified by scintillation counting. For incorporation into protein, cells were centrifuged and washed with TDB, followed by TCA protein precipitation and washing of the protein pellet, followed by reconstitution with 1% SDS and total radiolabel incorporation into protein quantified by scintillation counting. For the measurement of incorporation into lipids, cells were spun down and washed with TDB followed by extraction of lipids/glycolipids with CHCl3∶MeOH and CHCl3∶MeOH∶H2O (10∶10∶3). These extracts were desalted and the incorporation of radiolabel into the lipid fraction was quantified by scintillation counting. Values are means with standard deviations of three separate labeling experiments. The values at time zero are normalised to 100% whereby the deviation of total incorporation between different experiments was no greater than 10%.
Amino acid analysis
For the amino acid analysis, triple aliquots of noninduced (30 ml of cells at a density of 1.5×106) or cells where VSG RNAi had been induced with tetracycline for 24 hours (30 ml of cells at a density of 1.8×106) were collected by centrifugation (850 g for 5 minutes at room temperature), washed twice briefly in ice-cold MEM (1 ml) and suspended in ice-cold MEM (250 µl, containing norleucine [2 µl of 1.5 mM] as internal standard). This cell suspension was immediately added to 1 ml 50°C ethanol to lyse the cells and cooled on ice for 10 minutes. After centrifugation at 14,000 rpm at 4°C for 30 minutes the supernatant was transferred to a new Eppendorf tube and dried under vacuum. An aliquot (10 µl) of a freshly prepared mixture (ethanol: sodium acetate [1 M]: triethylamine, [2∶2∶1 v/v]) was added, vortexed and thoroughly dried under vacuum. A 20 µl aliquot of a freshly prepared derivatising agent (water: triethylamine: and PITC phenylisothiocyanate [2∶2∶1 v/v]) was added to the dry samples, vortexed, centrifuged at full speed for one minute and allowed to stand at room temperature for 20 minutes, prior to drying under vacuum. The derivatised amino acids were dissolved in 100 µl of starting buffer and analysed using a PICO-TAG HPLC system (Waters). Triplicate samples of both a standard mix of amino acids containing 2 µl of 0.1–1.5 mM of each amino acid and an equivalent volume (250 µl) of MEM was used in order to correct the values obtained from the cell samples to obtain intracellular amino acid pools.
Supporting Information
The cell volume of T. brucei expressing only VSG221 on its surface is not significantly different to that of T. brucei “double-expressors” expressing both VSG117 and VSG221 on their surface. The cell volumes were determined as pseudo-spheres using a CASY® Cell Counter. The parental T. brucei VB1.1 cell line is compared with that of the double-expressers T. brucei 221VP117.1 and 221VP117.2. A) The graph at the top shows data from a representative experiment whereby the percentage of cells with a respective cell diameter as pseudo-spheres is indicated. B) The values shown are the average of five independent experiments with the standard deviation indicated. The average peak volume is calculated from the projected cell diameter if the cells are represented as pseudo-spheres. C) The values shown below are the average of five independent experiments with the standard deviation indicated with error bars.
(0.24 MB TIF)
Global translation arrest after the induction of VSG RNAi monitored using [3H]-serine labeling of cells. A) T. brucei 221VG1.1 cells had VSG221 RNAi induced with tetracycline for the time in hours (h) indicated above, prior to labeling with [3H]-serine for 1 hour. Proteins were separated by SDS-PAGE and visualised by Coomassie staining (Coom.). B) Triplicate aliquots of the [3H]-serine labeled cells were processed to determine the mean rate of [3H]-serine incorporation into total protein versus time. The standard deviation is indicated with error bars.
(1.06 MB TIF)
Induction of a lethal RNAi mediated phenotype does not always trigger a global translation arrest in T. brucei. A) Various T. brucei cell lines with RNAi constructs allowing the tetracycline inducible knock-down of clathrin, PFR2, tubulin, actin or VSG221 were analysed. The levels of total protein synthesis were investigated after the induction of RNAi with tetracycline for 0 or 24 hours prior to labeling with [35S]-methionine for one hour. After this period cells were fully arrested. Total protein was separated on an SDS-PAGE gel. The top panel shows [35S]-labeled proteins detected by fluorography. Bottom panel is the corresponding Coomassie stained gel. B) Quantitation of triplicate samples of the of [35S]-methionine labeled cells after the induction of RNAi for 0 or 24 hours were processed to determine the mean rate (+/− the standard deviation) of [35S]-methionine incorporation into total protein after induction of the indicated knock-down.
(0.87 MB TIF)
Quantitation of the radioactive signal from the Northern blot analysis of transcripts from T. brucei 221VG1.1 cells where VSG221 RNAi has been induced for the respective time in hours (Fig. 3A). The signal is indicated as arbitrary units of radioactivity after quantitation was performed using a BioRad PhosphorImager and Quantity One software.
(0.18 MB TIF)
Blocking VSG synthesis by the induction of VSG RNAi does not result in downregulation of SL RNA, indicating that there is no induction of an SL response. RNA analysed was isolated from trypanosomes where VSG RNAi had been induced with tetracycline (Tet) for the time in hours (h) indicated above. Primer extension reactions to detect either the spliced leader (SL) RNA or the control U3 RNA were performed using radiolabeled oligonucleotides as described in the Experimental Procedures. The reaction products were electrophoresed on a polyacrylamide gel. The U3 or SL RNAs are indicated on the right.
(0.93 MB TIF)
A) 2D-DIGE comparison of cells grown in the presence or absence of VSG221 RNAi. T. brucei 221VB1.1 cells were either grown in the absence of tetracycline, or in the presence of tetracycline (Tet) for 8 hours to induce VSG221 RNAi. Three replicate sample pairs consisting of lysates from induced or noninduced cells were compared by 2D-DIGE. 2D-DIGE is a proteomic technique that allows the direct comparison of two sample types on a single gel. Each sample to be compared is pre-labeled with one of three CyDye DIGE fluor dyes (GE Healthcare). Here, one sample from each pair was labeled with CyDye5 and the other sample with CyDye3, such that in two of the pairs the noninduced sample was labeled with CyDye5 while in the other pair the noninduced sample was labeled with CyDye3. A standard sample consisting of an equal proportion of each of the six samples was generated and labeled with CyDye2. For each of the three replicates, the induced and noninduced samples, together with one third of the standard sample were combined and subjected to two dimensional electrophoresis. The standard sample is therefore present on all gels, and allows normalisation of protein abundance within each gel and statistical analysis across all gels. Proteins were separated in the first dimension on pH 3–11 NL IPG strips (GE Healthcare) and in the second dimension by SDS-PAGE-10% acrylamide: bisacrylamide 37.5∶1. On the 2D gels shown, molecular weight (Mw) decreases from top to bottom, and pH increases from left to right. Spots were visualised on a Typhoon scanner (GE Healthcare) and gel images were analysed and matched by reference to the standard sample using the DeCyder software suite (GE Healthcare). A total of 1486 spots were matched between at least two of the replicate gels and average ratios between induced and noninduced time points were obtained. B) No significant changes in protein levels were observed after the induction of VSG221 RNAi for 8 hours. Log10 average ratios of spots from induced and noninduced lysates were calculated for each of the 1486 spots analysed. The frequency of log10 ratios was calculated with a resolution of 0.025 and plotted as a percentage of the total number of spots analysed. Over 95% of the proteins showed a less than 25% change after the induction of VSG221 RNAi (area between the grey vertical bars), and no changes greater than 37% were observed.
(1.48 MB TIF)
There is no evidence for upregulation of procyclin after the induction of a VSG221 RNAi induced cell-cycle arrest in T. brucei VB1.1. A) Northern blot analysis does not show evidence for significant upregulation of procyclin after the induction of a VSG221 RNAi mediated cell-cycle arrest. RNA from the procyclic T. brucei 29-13 cell line (PF) was compared with RNA from the T. brucei 221VB1.1 cell line in which VSG221 RNAi had been induced with tetracycline for the time in hours (h) indicated above. The blot was hybridised with a probe for procyclin (CPT4) from [57], VSG221 to show the degree of VSG221 transcript knock-down, or actin as a loading control. Quantitation of the radioactive signal from the Northern blot analysis is indicated in arbitrary units of radioactivity, and was performed using a BioRad PhosphorImager with QuantityOne software. B) Western blot analysis of T. brucei VB1.1 stalled by the induction of VSG221 RNAi does not show evidence for the upregulation of EP procyclin. Protein lysates from bloodstream form T. brucei HNI(V02) [58] (BF), procyclic T. brucei 29-13 cell line (PF), or T. brucei 221VB1.1 where VSG221 RNAi had been induced for the time in hours (h) indicated above. The blot was probed with an antibody for EP procyclin or BiP as a loading control.
(0.32 MB TIF)
Titration of the minimum concentration of tetracycline or doxycycline which induces a maximal VSG221 RNAi mediated cell-cycle arrest. The T. brucei 221VB1.1 cell line was incubated with the indicated amount of tetracycline (Tet) or doxycycline (Dox) for the time in hours indicated below. The density of trypanosomes is indicated per ml x 105. The average of triplicate counts is shown, with the standard deviation indicated with error bars.
(0.28 MB TIF)
A) Cloning of recovered T. brucei 221VB1.1 cells after induction of VSG221 RNAi. Cells were induced using doxycycline for 12 hours, and then subsequently washed to remove the doxycycline. Ten-fold serial dilutions of washed cells were made and plated out over 48 wells of a 96 well plate with the indicated number of cells per well. The percentage of positive wells is indicated. Results are the average of three independent experiments with the standard deviation indicated with error bars. B) Cloning the parental T. brucei 221VB1.1 cells without the induction of VSG221 RNAi. Each serial dilution of the washed cells was plated out in 48 wells of a 96 well plate with the indicated number of cells per well. The percentage of positive wells is indicated. Results are the average of two independent experiments.
(0.19 MB TIF)
Acknowledgments
We are very grateful to Elisabetta Ullu (Yale University) for discussions and advice concerning the polysome analysis, Beatriz Castilho and Osvaldo Neto (University of São Paulo, Brazil) for advice and antibodies for analysis of T. brucei translation factors and translation factor kinases, and Mani Narayanan for analyses of T. brucei translation factors. We are grateful to Viola Denninger for making different T. brucei RNAi cell lines. We thank Helen Banks for help with the cell volume analysis and Mike Shaw for help with electron microscopy. We are grateful to Jay Bangs (University of Wisconsin) for VSG117 containing constructs and antibodies, and to James Minchin for performing initial analyses of double-expressing trypanosomes. We are very grateful to Pfizer Global Research & Development (Groton, Connecticut, USA) for the generous gift of the translation initiation inhibitor pactamycin. We acknowledge the lab of Peter Cook (University of Oxford) for technical advice. We would like to thank Peter Taylor (University of Dundee) for use of his PICO-TAG HPLC system. We thank Megan Lindsay, Tara Stanne, Viola Denninger, Manish Kushwaha, Mani Narayanan and Alexander Fullbrook for comments on the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was funded by the Wellcome Trust. T.K.S. and G.R. are Wellcome Trust Senior Research Fellows in the Basic Biomedical Sciences. K.G. is a Wellcome Trust Principal Research Fellow. Work in the lab of M.C. is funded by the Wellcome Trust. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cross GA. Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology. 1975;71:393–417. doi: 10.1017/s003118200004717x. [DOI] [PubMed] [Google Scholar]
- 2.Magez S, Schwegmann A, Atkinson R, Claes F, Drennan M, et al. The role of B-cells and IgM antibodies in parasitemia, anemia, and VSG switching in Trypanosoma brucei-infected mice. PLoS Pathog. 2008;4:e1000122. doi: 10.1371/journal.ppat.1000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pays E, Vanhamme L, Perez-Morga D. Antigenic variation in Trypanosoma brucei: facts, challenges and mysteries. Curr Opin Microbiol. 2004;7:369–374. doi: 10.1016/j.mib.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Horn D, Barry JD. The central roles of telomeres and subtelomeres in antigenic variation in African trypanosomes. Chromosome Res. 2005;13:525–533. doi: 10.1007/s10577-005-0991-8. [DOI] [PubMed] [Google Scholar]
- 5.Taylor JE, Rudenko G. Switching trypanosome coats: what's in the wardrobe? Trends Genet. 2006;22:614–620. doi: 10.1016/j.tig.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Barry JD, Marcello L, Morrison LJ, Read AF, Lythgoe K, et al. What the genome sequence is revealing about trypanosome antigenic variation. Biochem Soc Trans. 2005;33:986–989. doi: 10.1042/BST20050986. [DOI] [PubMed] [Google Scholar]
- 7.Marcello L, Barry JD. Analysis of the VSG gene silent archive in Trypanosoma brucei reveals that mosaic gene expression is prominent in antigenic variation and is favored by archive substructure. Genome Res. 2007;17:1344–1352. doi: 10.1101/gr.6421207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freymann D, Down J, Carrington M, Roditi I, Turner M, et al. 2.9 A resolution structure of the N-terminal domain of a variant surface glycoprotein from Trypanosoma brucei. JMolBiol. 1990;216:141–160. doi: 10.1016/S0022-2836(05)80066-X. [DOI] [PubMed] [Google Scholar]
- 9.Blum ML, Down JA, Gurnett AM, Carrington M, Turner MJ, et al. A structural motif in the variant surface glycoproteins of Trypanosoma brucei. Nature. 1993;362:603–609. doi: 10.1038/362603a0. [DOI] [PubMed] [Google Scholar]
- 10.Masterson WJ, Doering TL, Hart GW, Englund PT. A novel pathway for glycan assembly: biosynthesis of the glycosyl-phosphatidylinositol anchor of the trypanosome variant surface glycoprotein. Cell. 1989;56:793–800. doi: 10.1016/0092-8674(89)90684-3. [DOI] [PubMed] [Google Scholar]
- 11.Engstler M, Thilo L, Weise F, Grunfelder CG, Schwarz H, et al. Kinetics of endocytosis and recycling of the GPI-anchored variant surface glycoprotein in Trypanosoma brucei. J Cell Sci. 2004;117:1105–1115. doi: 10.1242/jcs.00938. [DOI] [PubMed] [Google Scholar]
- 12.Engstler M, Pfohl T, Herminghaus S, Boshart M, Wiegertjes G, et al. Hydrodynamic flow-mediated protein sorting on the cell surface of trypanosomes. Cell. 2007;131:505–515. doi: 10.1016/j.cell.2007.08.046. [DOI] [PubMed] [Google Scholar]
- 13.Sheader K, Vaughan S, Minchin J, Hughes K, Gull K, et al. Variant surface glycoprotein RNA interference triggers a precytokinesis cell cycle arrest in African trypanosomes. Proc Natl Acad Sci U S A. 2005;102:8716–8721. doi: 10.1073/pnas.0501886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jetton N, Rothberg KG, Hubbard JG, Wise J, Li Y, et al. The cell cycle as a therapeutic target against Trypanosoma brucei: Hesperadin inhibits Aurora kinase-1 and blocks mitotic progression in bloodstream forms. Mol Microbiol. 2009;72:442–458. doi: 10.1111/j.1365-2958.2009.06657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z, Umeyama T, Wang CC. The chromosomal passenger complex and a mitotic kinesin interact with the Tousled-like kinase in trypanosomes to regulate mitosis and cytokinesis. PLoS ONE. 2008;3:e3814. doi: 10.1371/journal.pone.0003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Wang CC. KMP-11, a basal body and flagellar protein, is required for cell division in Trypanosoma brucei. Eukaryot Cell. 2008;7:1941–1950. doi: 10.1128/EC.00249-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benz C, Clayton CE. The F-box protein CFB2 is required for cytokinesis of bloodstream-form Trypanosoma brucei. Mol Biochem Parasitol. 2007;156:217–224. doi: 10.1016/j.molbiopara.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Broadhead R, Dawe HR, Farr H, Griffiths S, Hart SR, et al. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature. 2006;440:224–227. doi: 10.1038/nature04541. [DOI] [PubMed] [Google Scholar]
- 19.Lillico S, Field MC, Blundell P, Coombs GH, Mottram JC. Essential Roles for GPI-anchored Proteins in African Trypanosomes Revealed Using Mutants Deficient in GPI8. Mol Biol Cell. 2003;14:1182–1194. doi: 10.1091/mbc.E02-03-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Figueiredo LM, Janzen CJ, Cross GA. A histone methyltransferase modulates antigenic variation in African trypanosomes. PLoS Biol. 2008;6:e161. doi: 10.1371/journal.pbio.0060161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen CL, Goulding D, Field MC. Clathrin-mediated endocytosis is essential in Trypanosoma brucei. EMBO J. 2003;22:4991–5002. doi: 10.1093/emboj/cdg481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ngo H, Tschudi C, Gull K, Ullu E. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Natl Acad Sci U S A. 1998;95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Salcedo JA, Perez-Morga D, Gijon P, Dilbeck V, Pays E, et al. A differential role for actin during the life cycle of Trypanosoma brucei. EMBO J. 2004;23:780–789. doi: 10.1038/sj.emboj.7600094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nolan DP, Garcia-Salcedo JA. Loss of actin does not affect export of newly synthesized proteins to the surface of Trypanosoma brucei. Mol Biochem Parasitol. 2008;157:233–235. doi: 10.1016/j.molbiopara.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Martin KL, Smith TK. Phosphatidylinositol synthesis is essential in bloodstream form Trypanosoma brucei. Biochem J. 2006;396:287–295. doi: 10.1042/BJ20051825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin KL, Smith TK. The glycosylphosphatidylinositol (GPI) biosynthetic pathway of bloodstream-form Trypanosoma brucei is dependent on the de novo synthesis of inositol. Mol Microbiol. 2006;61:89–105. doi: 10.1111/j.1365-2958.2006.05216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibellini F, Hunter WN, Smith TK. The ethanolamine branch of the Kennedy pathway is essential in the bloodstream form of Trypanosoma brucei. Mol Microbiol. 2009;73:826–843. doi: 10.1111/j.1365-2958.2009.06764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erondu NE, Donelson JE. Differential expression of two mRNAs from a single gene encoding an HMG1-like DNA binding protein of African trypanosomes. Mol Biochem Parasitol. 1992;51:111–118. doi: 10.1016/0166-6851(92)90206-y. [DOI] [PubMed] [Google Scholar]
- 29.Hughes K, Wand M, Foulston L, Young R, Harley K, et al. A novel ISWI is involved in VSG expression site downregulation in African trypanosomes. EMBO J. 2007;26:2400–2410. doi: 10.1038/sj.emboj.7601678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rout MP, Field MC. Isolation and characterization of subnuclear compartments from Trypanosoma brucei. Identification of a major repetitive nuclear lamina component. J Biol Chem. 2001;276:38261–38271. doi: 10.1074/jbc.M104024200. [DOI] [PubMed] [Google Scholar]
- 31.Bastin P, Sherwin T, Gull K. Paraflagellar rod is vital for trypanosome motility. Nature. 1998;391:548. doi: 10.1038/35300. [DOI] [PubMed] [Google Scholar]
- 32.Lustig Y, Sheiner L, Vagima Y, Goldshmidt H, Das A, et al. Spliced-leader RNA silencing: a novel stress-induced mechanism in Trypanosoma brucei. EMBO Rep. 2007;8:408–413. doi: 10.1038/sj.embor.7400930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pombo A, Jackson DA, Hollinshead M, Wang Z, Roeder RG, et al. Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. Embo J. 1999;18:2241–2253. doi: 10.1093/emboj/18.8.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seiser RM, Nicchitta CV. The fate of membrane-bound ribosomes following the termination of protein synthesis. J Biol Chem. 2000;275:33820–33827. doi: 10.1074/jbc.M004462200. [DOI] [PubMed] [Google Scholar]
- 36.Djikeng A, Shi H, Tschudi C, Shen S, Ullu E. An siRNA ribonucleoprotein is found associated with polyribosomes in Trypanosoma brucei. RNA. 2003;9:802–808. doi: 10.1261/rna.5270203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhalia R, Marinsek N, Reis CR, Katz R, Muniz JR, et al. The two eIF4A helicases in Trypanosoma brucei are functionally distinct. Nucleic Acids Res. 2006;34:2495–2507. doi: 10.1093/nar/gkl290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamanaka RB, Bennett BS, Cullinan SB, Diehl JA. PERK and GCN2 contribute to eIF2alpha phosphorylation and cell cycle arrest after activation of the unfolded protein response pathway. Mol Biol Cell. 2005;16:5493–5501. doi: 10.1091/mbc.E05-03-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moraes MC, Jesus TC, Hashimoto NN, Dey M, Schwartz KJ, et al. Novel membrane-bound eIF2alpha kinase in the flagellar pocket of Trypanosoma brucei. Eukaryot Cell. 2007;6:1979–1991. doi: 10.1128/EC.00249-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kramer S, Queiroz R, Ellis L, Webb H, Hoheisel JD, et al. Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2(alpha) phosphorylation at Thr169. J Cell Sci. 2008;121:3002–3014. doi: 10.1242/jcs.031823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westermeier R, Scheibe B. Difference gel electrophoresis based on lys/cys tagging. Methods Mol Biol. 2008;424:73–85. doi: 10.1007/978-1-60327-064-9_7. [DOI] [PubMed] [Google Scholar]
- 42.Valdes J, Taylor MC, Cross MA, Ligtenberg MJ, Rudenko G, et al. The viral thymidine kinase gene as a tool for the study of mutagenesis in Trypanosoma brucei. NucleicAcidsRes. 1996;24:1809–1815. doi: 10.1093/nar/24.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munoz-Jordan JL, Davies KP, Cross GA. Stable expression of mosaic coats of variant surface glycoproteins in Trypanosoma brucei. Science. 1996;272:1795–1797. doi: 10.1126/science.272.5269.1795. [DOI] [PubMed] [Google Scholar]
- 44.Koumandou VL, Natesan SK, Sergeenko T, Field MC. The trypanosome transcriptome is remodelled during differentiation but displays limited responsiveness within life stages. BMC Genomics. 2008;9:298. doi: 10.1186/1471-2164-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Potter MD, Seiser RM, Nicchitta CV. Ribosome exchange revisited: a mechanism for translation-coupled ribosome detachment from the ER membrane. Trends Cell Biol. 2001;11:112–115. doi: 10.1016/s0962-8924(00)01905-x. [DOI] [PubMed] [Google Scholar]
- 46.Halic M, Beckmann R. The signal recognition particle and its interactions during protein targeting. Curr Opin Struct Biol. 2005;15:116–125. doi: 10.1016/j.sbi.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 47.Bernales S, Papa FR, Walter P. Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- 48.Wek RC, Cavener DR. Translational control and the unfolded protein response. Antioxid Redox Signal. 2007;9:2357–2371. doi: 10.1089/ars.2007.1764. [DOI] [PubMed] [Google Scholar]
- 49.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 50.Sivan G, Kedersha N, Elroy-Stein O. Ribosomal slowdown mediates translational arrest during cellular division. Mol Cell Biol. 2007;27:6639–6646. doi: 10.1128/MCB.00798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amiguet-Vercher A, Perez-Morga D, Pays A, Poelvoorde P, Van Xong H, et al. Loss of the mono-allelic control of the VSG expression sites during the development of Trypanosoma brucei in the bloodstream. Mol Microbiol. 2004;51:1577–1588. doi: 10.1111/j.1365-2958.2003.03937.x. [DOI] [PubMed] [Google Scholar]
- 52.Brecht M, Parsons M. Changes in polysome profiles accompany trypanosome development. Mol Biochem Parasitol. 1998;97:189–198. doi: 10.1016/s0166-6851(98)00151-0. [DOI] [PubMed] [Google Scholar]
- 53.Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 54.Sheader K, te Vruchte D, Rudenko G. Bloodstream form-specific up-regulation of silent vsg expression sites and procyclin in Trypanosoma brucei after inhibition of DNA synthesis or DNA damage. J Biol Chem. 2004;279:13363–13374. doi: 10.1074/jbc.M312307200. [DOI] [PubMed] [Google Scholar]
- 55.Bangs JD, Ransom DM, McDowell MA, Brouch EM. Expression of bloodstream variant surface glycoproteins in procyclic stage Trypanosoma brucei: role of GPI anchors in secretion. EMBO J. 1997;16:4285–4294. doi: 10.1093/emboj/16.14.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tkacz ID, Lustig Y, Stern MZ, Biton M, Salmon-Divon M, et al. Identification of novel snRNA-specific Sm proteins that bind selectively to U2 and U4 snRNAs in Trypanosoma brucei. RNA. 2007;13:30–43. doi: 10.1261/rna.174307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rudenko G, Bishop D, Gottesdiener K, Van der Ploeg LH. Alpha-amanitin resistant transcription of protein coding genes in insect and bloodstream form Trypanosoma brucei. EMBO J. 1989;8:4259–4263. doi: 10.1002/j.1460-2075.1989.tb08611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rudenko G, Chaves I, Dirks-Mulder A, Borst P. Selection for activation of a new variant surface glycoprotein gene expression site in Trypanosoma brucei can result in deletion of the old one. MolBiochemParasitol. 1998;95:97–109. doi: 10.1016/s0166-6851(98)00099-1. [DOI] [PubMed] [Google Scholar]
- 59.Ariyanayagam MR, Oza SL, Guther ML, Fairlamb AH. Phenotypic analysis of trypanothione synthetase knockdown in the African trypanosome. Biochem J. 2005;391:425–432. doi: 10.1042/BJ20050911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The cell volume of T. brucei expressing only VSG221 on its surface is not significantly different to that of T. brucei “double-expressors” expressing both VSG117 and VSG221 on their surface. The cell volumes were determined as pseudo-spheres using a CASY® Cell Counter. The parental T. brucei VB1.1 cell line is compared with that of the double-expressers T. brucei 221VP117.1 and 221VP117.2. A) The graph at the top shows data from a representative experiment whereby the percentage of cells with a respective cell diameter as pseudo-spheres is indicated. B) The values shown are the average of five independent experiments with the standard deviation indicated. The average peak volume is calculated from the projected cell diameter if the cells are represented as pseudo-spheres. C) The values shown below are the average of five independent experiments with the standard deviation indicated with error bars.
(0.24 MB TIF)
Global translation arrest after the induction of VSG RNAi monitored using [3H]-serine labeling of cells. A) T. brucei 221VG1.1 cells had VSG221 RNAi induced with tetracycline for the time in hours (h) indicated above, prior to labeling with [3H]-serine for 1 hour. Proteins were separated by SDS-PAGE and visualised by Coomassie staining (Coom.). B) Triplicate aliquots of the [3H]-serine labeled cells were processed to determine the mean rate of [3H]-serine incorporation into total protein versus time. The standard deviation is indicated with error bars.
(1.06 MB TIF)
Induction of a lethal RNAi mediated phenotype does not always trigger a global translation arrest in T. brucei. A) Various T. brucei cell lines with RNAi constructs allowing the tetracycline inducible knock-down of clathrin, PFR2, tubulin, actin or VSG221 were analysed. The levels of total protein synthesis were investigated after the induction of RNAi with tetracycline for 0 or 24 hours prior to labeling with [35S]-methionine for one hour. After this period cells were fully arrested. Total protein was separated on an SDS-PAGE gel. The top panel shows [35S]-labeled proteins detected by fluorography. Bottom panel is the corresponding Coomassie stained gel. B) Quantitation of triplicate samples of the of [35S]-methionine labeled cells after the induction of RNAi for 0 or 24 hours were processed to determine the mean rate (+/− the standard deviation) of [35S]-methionine incorporation into total protein after induction of the indicated knock-down.
(0.87 MB TIF)
Quantitation of the radioactive signal from the Northern blot analysis of transcripts from T. brucei 221VG1.1 cells where VSG221 RNAi has been induced for the respective time in hours (Fig. 3A). The signal is indicated as arbitrary units of radioactivity after quantitation was performed using a BioRad PhosphorImager and Quantity One software.
(0.18 MB TIF)
Blocking VSG synthesis by the induction of VSG RNAi does not result in downregulation of SL RNA, indicating that there is no induction of an SL response. RNA analysed was isolated from trypanosomes where VSG RNAi had been induced with tetracycline (Tet) for the time in hours (h) indicated above. Primer extension reactions to detect either the spliced leader (SL) RNA or the control U3 RNA were performed using radiolabeled oligonucleotides as described in the Experimental Procedures. The reaction products were electrophoresed on a polyacrylamide gel. The U3 or SL RNAs are indicated on the right.
(0.93 MB TIF)
A) 2D-DIGE comparison of cells grown in the presence or absence of VSG221 RNAi. T. brucei 221VB1.1 cells were either grown in the absence of tetracycline, or in the presence of tetracycline (Tet) for 8 hours to induce VSG221 RNAi. Three replicate sample pairs consisting of lysates from induced or noninduced cells were compared by 2D-DIGE. 2D-DIGE is a proteomic technique that allows the direct comparison of two sample types on a single gel. Each sample to be compared is pre-labeled with one of three CyDye DIGE fluor dyes (GE Healthcare). Here, one sample from each pair was labeled with CyDye5 and the other sample with CyDye3, such that in two of the pairs the noninduced sample was labeled with CyDye5 while in the other pair the noninduced sample was labeled with CyDye3. A standard sample consisting of an equal proportion of each of the six samples was generated and labeled with CyDye2. For each of the three replicates, the induced and noninduced samples, together with one third of the standard sample were combined and subjected to two dimensional electrophoresis. The standard sample is therefore present on all gels, and allows normalisation of protein abundance within each gel and statistical analysis across all gels. Proteins were separated in the first dimension on pH 3–11 NL IPG strips (GE Healthcare) and in the second dimension by SDS-PAGE-10% acrylamide: bisacrylamide 37.5∶1. On the 2D gels shown, molecular weight (Mw) decreases from top to bottom, and pH increases from left to right. Spots were visualised on a Typhoon scanner (GE Healthcare) and gel images were analysed and matched by reference to the standard sample using the DeCyder software suite (GE Healthcare). A total of 1486 spots were matched between at least two of the replicate gels and average ratios between induced and noninduced time points were obtained. B) No significant changes in protein levels were observed after the induction of VSG221 RNAi for 8 hours. Log10 average ratios of spots from induced and noninduced lysates were calculated for each of the 1486 spots analysed. The frequency of log10 ratios was calculated with a resolution of 0.025 and plotted as a percentage of the total number of spots analysed. Over 95% of the proteins showed a less than 25% change after the induction of VSG221 RNAi (area between the grey vertical bars), and no changes greater than 37% were observed.
(1.48 MB TIF)
There is no evidence for upregulation of procyclin after the induction of a VSG221 RNAi induced cell-cycle arrest in T. brucei VB1.1. A) Northern blot analysis does not show evidence for significant upregulation of procyclin after the induction of a VSG221 RNAi mediated cell-cycle arrest. RNA from the procyclic T. brucei 29-13 cell line (PF) was compared with RNA from the T. brucei 221VB1.1 cell line in which VSG221 RNAi had been induced with tetracycline for the time in hours (h) indicated above. The blot was hybridised with a probe for procyclin (CPT4) from [57], VSG221 to show the degree of VSG221 transcript knock-down, or actin as a loading control. Quantitation of the radioactive signal from the Northern blot analysis is indicated in arbitrary units of radioactivity, and was performed using a BioRad PhosphorImager with QuantityOne software. B) Western blot analysis of T. brucei VB1.1 stalled by the induction of VSG221 RNAi does not show evidence for the upregulation of EP procyclin. Protein lysates from bloodstream form T. brucei HNI(V02) [58] (BF), procyclic T. brucei 29-13 cell line (PF), or T. brucei 221VB1.1 where VSG221 RNAi had been induced for the time in hours (h) indicated above. The blot was probed with an antibody for EP procyclin or BiP as a loading control.
(0.32 MB TIF)
Titration of the minimum concentration of tetracycline or doxycycline which induces a maximal VSG221 RNAi mediated cell-cycle arrest. The T. brucei 221VB1.1 cell line was incubated with the indicated amount of tetracycline (Tet) or doxycycline (Dox) for the time in hours indicated below. The density of trypanosomes is indicated per ml x 105. The average of triplicate counts is shown, with the standard deviation indicated with error bars.
(0.28 MB TIF)
A) Cloning of recovered T. brucei 221VB1.1 cells after induction of VSG221 RNAi. Cells were induced using doxycycline for 12 hours, and then subsequently washed to remove the doxycycline. Ten-fold serial dilutions of washed cells were made and plated out over 48 wells of a 96 well plate with the indicated number of cells per well. The percentage of positive wells is indicated. Results are the average of three independent experiments with the standard deviation indicated with error bars. B) Cloning the parental T. brucei 221VB1.1 cells without the induction of VSG221 RNAi. Each serial dilution of the washed cells was plated out in 48 wells of a 96 well plate with the indicated number of cells per well. The percentage of positive wells is indicated. Results are the average of two independent experiments.
(0.19 MB TIF)