Abstract
Dimorphism or morphogenic conversion is exploited by several pathogenic fungi and is required for tissue invasion and/or survival in the host. We have identified a homolog of a master regulator of this morphological switch in the plant pathogenic fungus Fusarium oxysporum f. sp. lycopersici. This non-dimorphic fungus causes vascular wilt disease in tomato by penetrating the plant roots and colonizing the vascular tissue. Gene knock-out and complementation studies established that the gene for this putative regulator, SGE1 (SIX Gene Expression 1), is essential for pathogenicity. In addition, microscopic analysis using fluorescent proteins revealed that Sge1 is localized in the nucleus, is not required for root colonization and penetration, but is required for parasitic growth. Furthermore, Sge1 is required for expression of genes encoding effectors that are secreted during infection. We propose that Sge1 is required in F. oxysporum and other non-dimorphic (plant) pathogenic fungi for parasitic growth.
Author Summary
Plant pathogenic fungi have evolved many ways to infect their hosts and can have devastating effects on commercial crop production. Dissecting their infection strategies and understanding the molecular pathways involved in pathogenesis have been and continue to be the subject of intensive research. New insights gained may help to develop disease controlling strategies. Fusarium oxysporum has become a model for root invading, pathogenic fungi. This fungus attacks a wide range of plant species worldwide and the only effective disease control strategies in the field are crop rotation and usage of resistant plant varieties, if at all available. In the last decade, many genes have been identified that play a role during pathogenesis, many of which are linked to general strain fitness. Only few genes have been identified that are required for pathogenesis but do not affect vegetative growth. This paper describes the characterization of such a gene. Its protein product, Sge1, is conserved in the fungal kingdom and represents a new class of transcriptional regulators involved in morphological switching in dimorphic fungal pathogens. In F. oxysporum, the Sge1 protein is required for parasitic growth and is associated with expression of parasitic phase-specific genes. We suggest that the function of Sge1 is conserved in (plant) pathogenic fungi.
Introduction
The fungus Fusarium oxysporum is found in both agricultural and non-cultivated soils throughout the world. The species consists of non-pathogenic and pathogenic isolates, both known as efficient colonizers of the root rhizosphere. The pathogenic isolates, grouped into formae specialis depending on their host range [1],[2], cause wilt or rot disease in important agricultural and ornamental plant species, such as tomato, banana, cotton and tulip bulbs, thereby causing serious problems in commercial crop production [3],[4]. Recently, F. oxysporum has also been reported as an emerging human pathogen, causing opportunistic mycoses [5]–[7].
In the absence of plant roots F. oxysporum survives in the soil either as dormant propagules (chlamydospores) or by growing saprophytically on organic matter [1],[8]. When growing on roots of a suitable host F. oxysporum appears to switch from a saprophyte into a pathogen. As a pathogen F. oxysporum needs to overcome host defence responses and sustain growth within the host in order to establish disease. To do so, F. oxysporum likely undergoes reprogramming of gene expression. In the last decade, genes have been identified that do not seem to be required for saprophytic growth, but are involved in or required for pathogenicity and/or are specifically expressed during in planta growth. Examples are SIX1, encoding a small secreted protein, and FOW2 and FTF1, both encoding Zn(II)2Cys6-type transcriptional regulators [9]–[12].
In an insertional mutagenesis screen aimed at identification of pathogenicity factors of Fusarium oxysporum f. sp. lycopercisi (Fol), a gene now called SGE1 (SIX Gene Expression 1) was identified that shows homology to the transcriptional regulators Candida albicans WOR1 and Histoplasma capsulatum RYP1 [9]. These transcription factors have been identified as major regulators of morphological switching in these human pathogens: from a filamentous to a yeast form in H. capsulatum and from a white to opaque cell type in C. albicans [10]–[13]. In both fungi, these morphological transitions are correlated with the ability to cause disease. Targeted deletion of RYP1 in H. capsulatum or WOR1 in C. albicans locks the fungus in its filamentous form or white cell type, respectively.
In this work we characterize SGE1 and show that it shares many characteristics with WOR1 and RYP1. In addition, we show that expression of effector genes is lost in the SGE1 deletion mutant. We conclude that Sge1 plays a major role during parasitic growth, defined as extensive in planta growth leading to wilt symptoms, in F. oxysporum f. sp. lycopersici.
Results
Isolation and characterization of SGE1 and FoPAC2
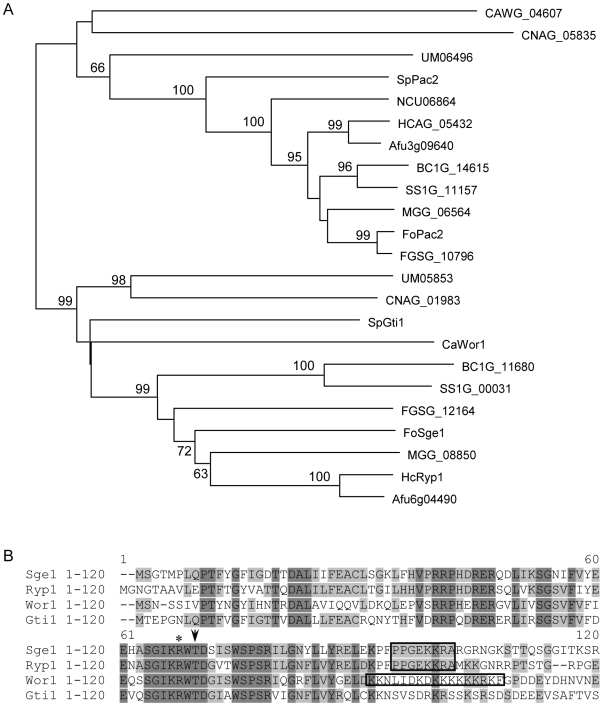
In an insertional mutagenesis screen aimed at identifying genes involved in pathogenicity a non-pathogenic mutant (5G2) and one severely reduced in pathogenicity (101E1) were identified that both carried a single T-DNA insertion into the ORF of FOXG_10510 [9], hereafter called SGE1 (SIX Gene Expression 1). The SGE1 ORF contains no introns and encodes a protein of 330 amino acids (http://www.broad.mit.edu/annotation/genome/fusarium_group/MultiHome.html). Sequence analysis revealed that the N-terminus (amino acids 1–120) contains a TOS9 (COG5037) and a Gti1_Pac2 family domain (Pfam09729) and is conserved in the fungal kingdom; all fungi of which the genome sequence was examined, including ascomycetes, basidiomycetes and zygomycetes, contain related genes that divide in two groups based on sequence similarity of the predicted proteins. Most ascomycetes have one member in each group, except Neurospora crassa which lacks a member of the SGE1 group (Figure 1A). The basidiomycete Coprinus cinereus and the zygomycete Rhizopus oryzae contain more than two members, still with at least one member in each group (data not shown). The branching order within the two groups does not always follow species phylogeny, making orthology questionable. Examples are the placement of FoSge1 and FGSG_12164 basal to the Magnaporthe grisea homolog (MGG_00850), the placement of CAWG_04607 of C. albicans basal to Pac2 of Schizosaccharomyces pombe (and closer to basidiomycete homologs) and of NCU06864 of N. crassa basal to homologs of other filamentous fungi (pezizomycotina) (Figure 1A). Sge1 is in the same group as Histoplasma capsulatum Ryp1 and Candida albicans Wor1, both identified as regulators for morphological switching [10]–[13], and Schizosaccharomyces pombe Gti1, which plays a role in gluconate uptake upon glucose starvation [14]. A potential protein kinase A phosphorylation site (KRWTDS/G) is conserved between these proteins (Figure 1B). In addition, a nuclear localization motif is present in Sge1 (+93 to +100) that is shared with Ryp1 (Figure 1B). The F. oxysporum protein related to Sge1, encoded by FOXG_12728, shows high similarity to S. pombe Pac2, a protein controlling the onset of sexual development [15]. Interestingly, in the same insertional mutagenesis screen mentioned above, a Fol mutant with reduced pathogenicity (30C11) was identified in which one of two T-DNA insertions resides in the FOXG_12728 ORF [9], hereafter called FoPAC2.
Figure 1. Representation of Sge1 homologs in fungi.
A) Phylogenetic (neighbour joining, mid-point rooted) tree of Sge1 and FoPac2 with homologs from C. albicans (Wor1 and CAWG_04607), S. pombe (SpGti1 and SpPac2), H. capsulatum (HcRyp1 and HCAG_05432), Magnaporthe grisea (MGG_08850 and MGG_06564), Aspergillus fumigatus (Afu6g04490 and Afu3g09640), Fusarium graminearum (FGSG_12164 and FGSG_10796), Botrytis cinerea (BC1G_11680 and BC1G_14615), Sclerotinia sclerotiorum (SS1G_00031 and SS1G_11157), Cryptococcus neoformans (CNAG_01983 and CNAG_05835), Ustilago maydis (UM05853 and UM06496) and Neurospora crassa (NCU06864), constructed using MacVector software. Only fully aligned parts of the multiple sequence alignment were used (manual curation). Bootstrap percentages are provided only for branches receiving 60% or more support (1000 replications). Branch length reflects the extent of sequence divergence. B) Protein sequence alignment of the Sge1 N-terminal region with H. capsulatum Ryp1, C. albicans Wor1 and S. pombe Gti1. Conserved residues are shaded black, similar residues are shaded grey. The arrow head indicates the conserved threonine residue within the potential protein kinase A phosphorylation site and the asterisk indicates the mutated residue in Sge1R66S. Predicted nucleur localization signals are boxed. The protein sequence alignment were created using VectorNTI software.
SGE1 is strictly required for pathogenicity
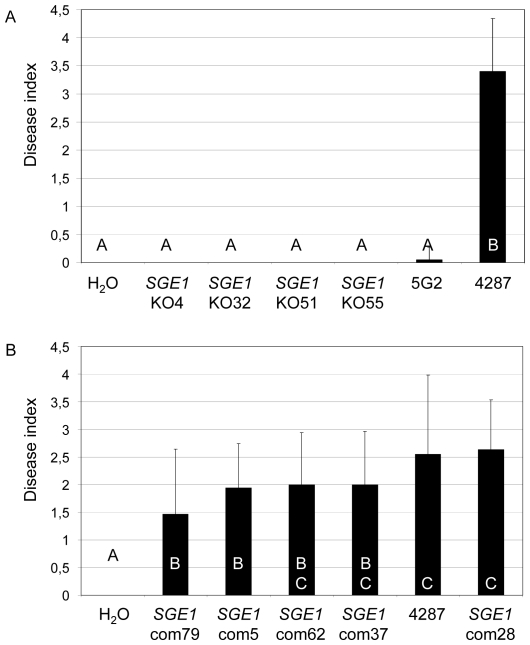
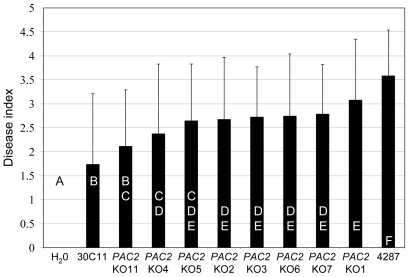
To assess the involvement of SGE1 and FoPAC2 in pathogenicity, gene knock-out mutants were generated by homologous recombination. Four independent SGE1 and eight independent FoPAC2 knock-out mutants were obtained, with the deletions confirmed by PCR and Southern analysis (Figure S1, S3 and S4). The SGE1 deletion mutants were non-pathogenic on tomato in a root dip bioassay and corroborated the severely reduced to non-pathogenic phenotype of the insertional mutagenesis mutants (Figure 2A). Re-introduction of the SGE1 gene in locus by homologous recombination in a knock-out mutant (Figure S2 and S3) restored pathogenicity (Figure 2B), confirming that the loss of pathogenicity was due to loss of SGE1. Deletion of FoPAC2 only had a minor effect on pathogenicity. All knock-out mutants were significantly different from the wild type in disease causing ability, but seven out of the eight mutants were also significantly different from the original insertion mutant (30C11) (Figure 3). This indicates that FoPAC2 plays at most a minor role during infection and that most probably additional defects in the 30C11 mutant added to the reduced pathogenicity phenotype. Since the loss of pathogenicity was complete upon deletion of SGE1, we focussed on this gene for further analysis.
Figure 2. Sge1 is required for full pathogenicity.
Nine to eleven days old tomato seedlings were inoculated with fungal spore suspensions using root-dip inoculation and the disease index (ranging from 0, healthy plant to 4, severely diseased plant or dead plant) was scored after three weeks. Error bars indicate standard deviation and capitals define statistically different groups (ANOVA, p = 0.95). A) Average disease index of 20 plants three weeks after mock inoculation (H2O) or inoculation with four independent SGE1 deletion mutants (SGE1KO4, 32, 51, and 55), insertional mutant 5G2 or wild type (4287). B) Average disease index of 20 plants three weeks after mock inoculation (H20) or inoculated with five independent SGE1 complementation strains (SGE1com5, 37, 62, and 79) or wild type (4287).
Figure 3. FoPac2 only plays a minor role in pathogenicity.
Nine to eleven days old tomato seedlings were inoculated with fungal spore suspensions using root-dip inoculation [50] and the disease index (ranging from 0, healthy plant to 4, severely diseased plant or dead plant) was scored after three weeks. Error bars indicate standard deviation and capitals define statistically different groups (ANOVA, p = 0.95). Average disease index of 20 plants three weeks after mock inoculation (H2O) or inoculation with eight independent FoPac2 deletion mutants (FoPac2KO1, 2, 3, 4, 5, 6, 7, and 11), insertional mutant 30C11 or wild type (4287).
SGE1 is not required for vegetative growth, but quantitatively affects conidiation
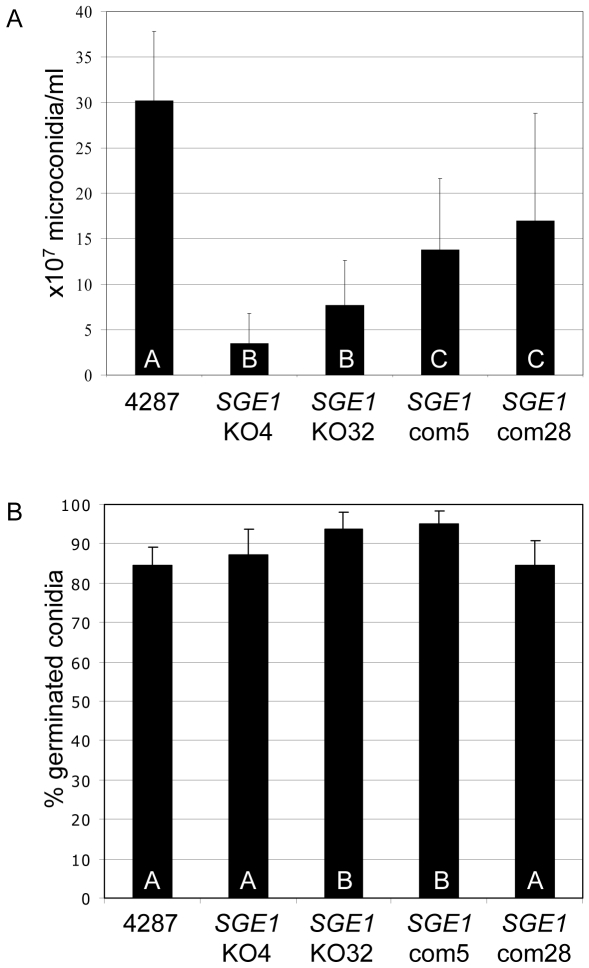
Previously, we reported that vegetative growth of the insertional mutants 5G2 and 101E1 are indistinguishable from that of the wild type on various carbon sources [9]. To more fully analyze potential metabolic defects of the sge1 mutant, we made use of BIOLOG FF MicroPlates, in which each well contains a different carbon source [16]. Also in this assay no reproducible differences were observed between growth of the wild type and the SGE1 deletion mutant on 95 different carbon sources (Figure S5). Microconidia and macroconidia generated in minimal or CMC liquid medium were phenotypically indistinguishable from wild type (Figure S6). However, the SGE1 deletion mutants produced about 6-fold less microconidia compared to the wild type in both media, and this phenotype was only partially restored in the SGE1 complementation mutants (Figure 4A). The conidial germination rates were comparable to wild type (Figure 4B), indicating that, although less microconidia are formed, they are fully viable. These observations indicate that SGE1 is quantitatively involved in conidiogenesis, but is not required for conidial fitness, overall (colony) morphology, vegetative growth or carbon source utilization.
Figure 4. Sge1 is involved in conidiogenesis.
Microconidia production was determined after five days of growth in minimal medium and counted in a Bürker-Türk haemocytometer. A) The average number of microconidia of five independent experiments each with five replicates. B) The average germination rate of the isolated microconidia on PDA medium after six hours of incubation at 25°C. Error bars indicate standard deviation and capitals define statistically different groups (ANOVA, p = 0.95).
SGE1 expression is upregulated during infection of tomato roots
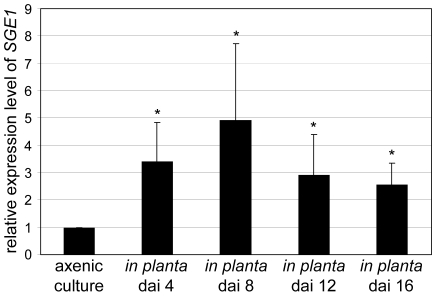
C. albicans WOR1 and H. capsulatum RYP1 are 45-fold and 4-fold upregulated upon transition from white to opaque cells in C. albicans and from filamentous growth to yeast cells in H. capsulatum, respectively [11],[17]. To determine the relative expression levels of SGE1 during saprophytic and parasitic growth quantitative PCR was performed. Expression levels of SGE1 were determined both in axenic culture and during tomato root infection at different time points after inoculation and compared to the level of the constitutively expressed elongation factor 1 alpha gene (EF-1α). We found that SGE1 expression is upregulated 2- to 5-fold during infection with maximal expression eight days after inoculation (Figure 5).
Figure 5. SGE1 is upregulated during infection of tomato roots.
Nine to eleven days old tomato seedlings were inoculated with either water or wild type Fol using root-dip inoculation. Roots were harvested 4, 8, 12, and 16 days after inoculation followed by RNA isolation and cDNA synthesis. Quantitative PCR was used to determine the expression levels of the constitutively expressed gene EF-1α and SGE1 in axenic culture (minimal medium) and during infection. Relative expression levels were subsequently calculated using the comparative C(t) method. *, significantly different from the SGE1 expression level in axenic culture at a 95% confidence interval.
SGE1 is not essential for colonization or penetration of the root surface
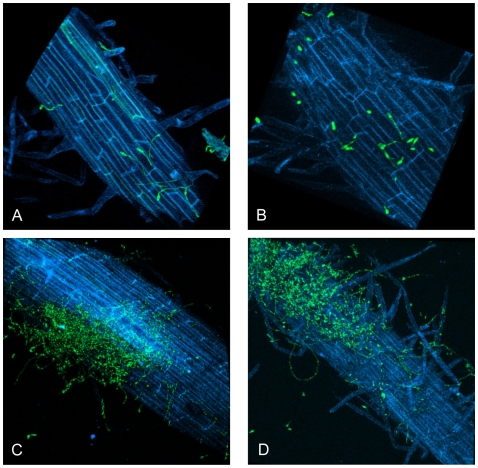
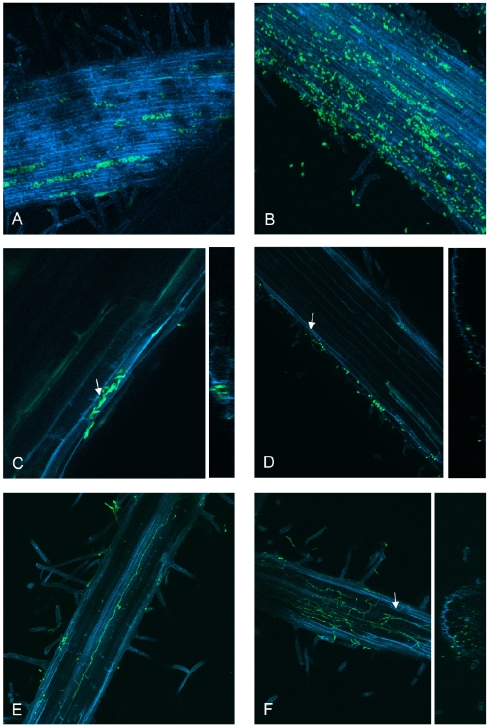
To determine at which stage during infection the SGE1 deletion mutant is halted, tomato root colonization by the mutant was visualized using fluorescent binocular and confocal laser scanning microscopy. Tomato seedlings were infected with a GFP expressing wild type strain or SGE1 deletion strain and colonization was followed over time. After two days, patches of colonization on the roots were observed for both strains (Figure S7), indicating that the SGE1 deletion mutant is not impaired in root surface colonization. This observation was confirmed by confocal laser scanning microscopy. For both strains, germinated spores were observed on the root surface 24 hours after infection (Figure 6A and 6B) and mycelial mass increased in time leading to patches of colonization as described above (Figure 6C and 6D). No difference in colonization was observed between the wild type and the sge1 mutant in the first 2 days after inoculation. However, after three days, plant cells filled with spores and mycelium were observed in roots infected with the wild type strain (Figure 7A), indicating that penetration of the root surface had occurred (Figure 7C). A mixture of germinated spores and mycelium was also observed on plant roots infected with the sge1 mutant, however, growth was dispersed over the entire root (Figure 7B) and was not confined to single plant cells as observed for the wild type. Occasionally, the sge1 mutant was observed within the root cortex (Figure 7D), indicating that the sge1 mutant is capable of penetrating the surface. This is in line with our observation that the sge1 mutant is able to penetrate a cellophane sheet (Figure S8), an assay used to assess the ability to form penetration hyphae [18]. Four days after infection fungal growth within the roots was observed. In roots infected with the wild type, growth within the xylem vessel was observed once (Figure 7E). Also in one case, extensive growth of the sge1 mutant was observed within the root (Figure 7F); however, this growth was mainly intercellular. In both cases the fungal growth extended from a point where the root was broken. Since these cases were too rare for a proper quantification, we can only conclude that both the wild type and the sge1 mutant are capable of in planta growth.
Figure 6. The SGE1 deletion mutant is not impaired in root colonization.
The infection behaviour of the SGE1 deletion mutant was determined by confocal microscopy. Nine to eleven days old tomato seedlings were inoculated with SGE1 deletion mutant spore suspension and root colonization was determined after one to two days after inoculation. Germinated spores were observed on a tomato root infected with a GFP-expressing virulent strain (A) or with the SGE1 deletion mutant (B). Patches of colonization were found on a tomato root infected with a GFP-expressing virulent strain (C) or with the SGE1 deletion mutant (D).
Figure 7. The SGE1 deletion mutant is not impaired in penetration.
The infection behaviour of the SGE1 deletion mutant was determined by confocal microscopy. Nine to eleven days old tomato seedlings were inoculated with SGE1 deletion mutant spore suspension and root colonization was determined after three to four days after inoculation. Fungal growth within single cell compartments was observed in a tomato root infected with a GFP-expressing virulent strain (A). More dispersed fungal growth was observed in a tomato root infected with the SGE1 deletion mutant (B). Fungal growth within cells was observed in a tomato root infected with a GFP-expressing virulent strain (C) or, less frequently, with the SGE1 deletion mutant (D). Fungal growth within the xylem vessel of a tomato root was seen upon infection with a GFP-expressing virulent strain (E) and intercellular growth was seen upon infection with the SGE1 deletion mutant (F). The arrows indicate the position in the YZ projection.
To determine whether the sge1 mutant is able to colonize xylem vessels as extensively as the wild type, tomato seedlings were inoculated with the wild type, SGE1 knock-out or the SGE1 complementation strain and potted according to the bioassay procedure (i.e. with damaged roots to allow relatively easy entry into vessels). One week after inoculation the hypocotyl was cut in slices of several millimeters and put on rich (PDA) medium. After two days F. oxysporum outgrowth was observed from hypocotyl pieces previously inoculated with the wild type and the SGE1 complementation strain, but not from the hypocotyl pieces previously inoculated with the SGE1 knock-out mutant (Figure S9). Based on the above described observations, it can be concluded that Sge1 is not required for early pathogenesis-related functions, such as root colonization and penetration, but appears to be required for extensive growth within plant cells and the xylem.
Sge1 is localized in the nucleus
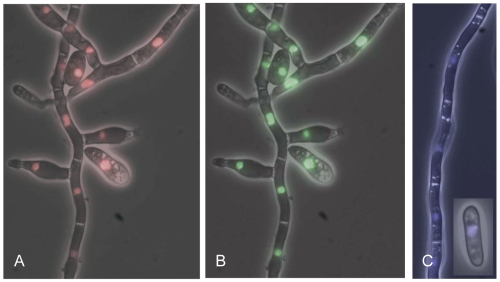
The homologs of Sge1 in C. albicans and H. capsulatum are nuclear proteins [10]–[13]. To determine whether this is also the case for Sge1 its subcellular localization was investigated. For this purpose, Sge1 was fused C-terminally to the fluorescent proteins CFP or RFP. Constructs expressing these fusion proteins were introduced in the SGE1 deletion mutant by homologous recombination at the sge1 locus (verified by Southern analysis (Figure S3)). The functionality of the fusion proteins was determined in a bioassay. SGE1::CFP restored pathogenicity to almost wild type levels (Figure S10). Disease symptoms were also observed upon infection with strains expressing SGE1::RFP, albeit severely reduced compared to the wild type infection (Figure S10). Subcellular localization for both fusion proteins was similar in that they are localized in the nucleus in both spores and hyphae (Figure 8). Nuclear localization was verified by introduction of a construct encoding histone H2B::GFP [19] into the SGE1::RFP strain. Both fusion proteins localized to the same compartment (Figure 8). These observations support the potential role of Sge1 as a transcriptional regulator in F. oxysporum.
Figure 8. Sge1 is localized in the nucleus.
The subcellular localization of the fluorescent proteins Sge1::RFP and Sge1::CFP was determined after five days of growth in minimal medium using a fluorescence microscope. A) Subcellular localization of Sge1::RFP. B) Subcellular localization of H2B::GFP expressed in the Sge1::RFP background. C) Subcellular localization of Sge1::CFP.
The putative Pka phosphorylation site is required for Sge1 function
As described above, the Sge1 protein contains a potential Pka phosphorylation site and this site is conserved in all Sge1 homologs (Figure 1B). Replacement of the conserved threonine residue by an alanine impaired S. pombe Gti1 function [14]. In an attempt to identify Sge1 interacting partners using a yeast two-hybrid screen, the SGE1 gene, fused to the portion of GAL4 encoding the Gal4 DNA binding domain, was introduced in Saccharamyces cerevisiae. To our surprise, after numerous transformation attempts only one colony was obtained. After re-isolation of the plasmid from this colony followed by sequencing it turned out that a point mutation in the SGE1 sequence (C196A) had occurred resulting in an amino acid change from an arginine into a serine at position 66, altering the Pka phosphorylation site (Figure 1). In contrast to wild type SGE1, this mutant form of SGE1 could be easily re-transformed to yeast. We concluded that SGE1 is normally toxic in S. cerevisiae and that the R>S mutation leads to tolerance in this yeast. To determine the effect of this mutation on the activity of Sge1 in Fol, a construct expressing Sge1R66S was introduced in the SGE1 deletion mutant. Correct gene replacement was again confirmed by PCR (Figure S11). All strains encoding the Sge1R66S protein were non-pathogenic (Figure 9), suggesting that an intact Pka phosphorylation site is required for Sge1 to function properly.
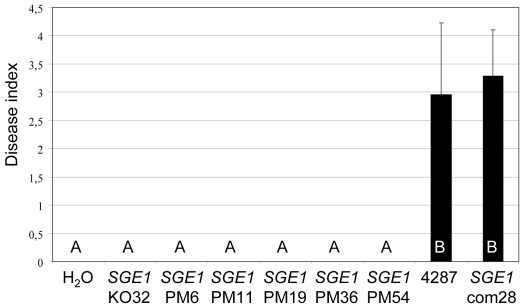
Figure 9. An intact Pka phosphorylation site in Sge1 is essential for pathogenicity.
Nine to eleven days old tomato seedlings were inoculated with fungal spore suspensions using root-dip inoculation and the disease index (ranging from 0, healthy plant to 4, severely diseased plant or dead plant) was scored after three weeks. Error bars indicate standard deviation and capitals define statistically different groups (ANOVA, p = 0.95). Average disease index of 20 plants three weeks after mock inoculation (H2O) or inoculation with five independent Sge1R66S complementation mutants (SGEPM6, 11, 19, 36 and 54), an SGE1 complementation mutant (SGE1com28) or wild type (4287).
SGE1 regulates expression of effector genes
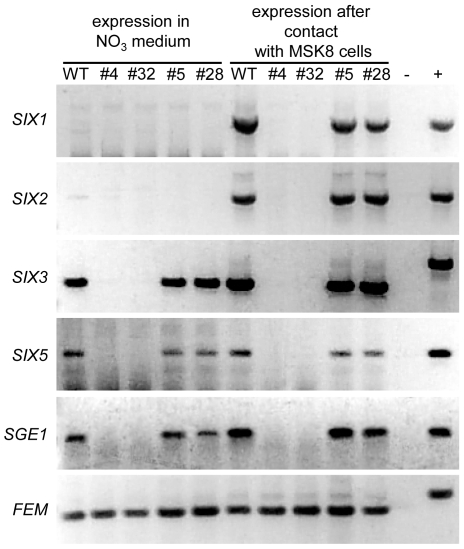
The transcriptional regulators Ryp1 and Wor1 regulate expression of phase specific genes [11],[20]. Since Sge1 shares many features with Ryp1 and Wor1, we hypothesized that expression of F. oxysporum genes specifically expressed during infection could be altered. Examples of such genes are those encoding effectors, small in planta secreted proteins, called Six (Secreted in xylem) proteins in Fol. Recently, it was shown that F. oxysporum secretes numerous Six proteins during infection [21] (Houterman and Rep, unpublished). SIX1, encoding Avr3, is strongly induced upon penetration of the root cortex and plays a role during pathogenicity [22],[23]. In addition, Six3 (Avr2) and Six4 (Avr1) have been shown to play a role in virulence as well as resistance protein recognition [24],[25]. Since the SGE1 deletion mutant does not show extensive in planta growth, precluding assessment of SIX gene expression during root infection, we incubated the mutant together with tomato cells in culture, a situation known to induce SIX1 expression [22]. MSK8 tomato cells were inoculated with a conidial suspension of the wild type or the SGE1 deletion mutant. After 24 h the cultures were harvested and the presence of SIX gene transcripts was determined by RT-PCR and compared to transcript levels in axenic cultures. As expected, SIX1 gene expression was very low when the wild type strain was grown in axenic culture and expression was strongly upregulated upon incubation with MSK8 cells. A similar expression pattern was observed for SIX2 (Figure 10). In contrast, the SIX3 and SIX5 genes were expressed in the wild type strain in axenic culture as well as in the presence of MSK8 cells (Figure 10). Interestingly, in the sge1 mutant expression of all four SIX genes was lost both in axenic culture and in the presence of MSK8 cells. Expression was restored in the complemented strain (Figure 10). These results show that expression of at least four SIX genes tested is dependent on the presence of SGE1.
Figure 10. SGE1 regulates expression of effector genes.
Expression levels of SIX1, SIX2, SIX3, SIX5, SGE1 and the constitutively expressed gene FEM1 were determined in the SGE1 deletion (#4 and #32) and complementation mutants (#5 and #28) and in the wild type (WT) grown in vitro and under in planta-mimicking conditions. Total RNA was isolated from mycelium harvested after five days of growth in minimal medium (in vitro) and from mycelium grown for 24 h in BY-medium in the presence of one week old MSK8 cells (in planta-mimicking) followed by RT-PCR. −, negative control. +, positive (genomic DNA) control.
The SGE1 deletion mutant exhibits biocontrol activity
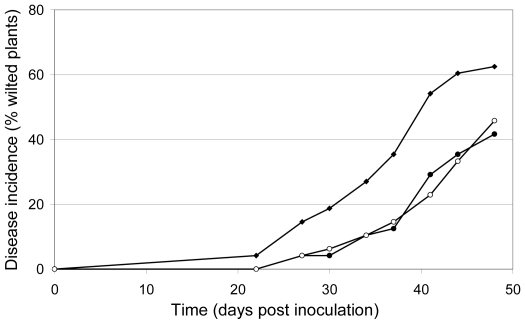
Biocontrol activity has been reported to be dependent on colonization of superficial cell layers, a capacity shared between pathogenic and protective strains and has been reported to be independent of competition for putative penetration sites or nutrients [26]. Fusarium oxysporum f. sp. lycopersici exerts biocontrol activity on flax when added in a 100-fold excess relative to a F. oxysporum strain pathogenic towards flax. To determine whether the sge1 mutant is still able to protect flax against wilt disease like its parental strain, flax cv. viking was treated with either a pathogenic isolate of F. oxysporum f. sp. lini (Foln3) alone or in a 1∶100 ratio with either the sge1 mutant or the wild type strain 4287. The first wilt symptoms were observed 22 days after inoculation in the treatment with Foln3 (Figure 11). In the Foln3/sge1 mutant and Foln3/wild type combination treatments the first disease symptoms were delayed by 3 days. Disease symptoms were always reduced in the Foln3/sge1 mutant and Foln3/wild type treatments compared to the Foln3 treatment: 48 days post inoculation disease symptoms were reduced by 27 and 33%, by the sge1 mutant and the wild type, respectively (Figure 11). The ANOVA performed on AUDPCs indicated that this difference was significant at the probability of 91%. We conclude that the sge1 mutant is able to protect flax against Fusarium wilt as well as the wild type strain, suggesting that it can colonize roots efficiently.
Figure 11. The sge1 mutant and its parental strain exhibit biocontrol activity on flax.
The protective capacity of the sge1 mutant and its parental strain were determined on flax. Flax cv. viking was inoculated with the pathogenic isolate F. oxysporum f. sp. lini (104 conidia/ml) alone or in combination with the sge1 mutant or the 4287 wild type strain (106 conidia/ml). Fusarium wilt incidence is expressed as percentage of wilted plants. Black diamond, F. oxysporum f. sp. lini. Open circle, F. oxysporum f. sp. lini and sge1 mutant. Black circle, F. oxysporum f. sp. lini and the wild type strain 4287.
Discussion
Fungi have various ways to adapt their morphology to the environment, one example being dimorphism. Dimorphism is a strategy frequently employed by fungal pathogens where this developmental transition is correlated with the ability to cause disease (reviewed by [27],[28]). In this study, we have characterized Sge1, the F. oxysporum homolog of the master regulator of morphological switching Wor1 and Ryp1. Although F. oxysporum does not display an immediately obvious morphological switch like C. albicans or H. capsulatum, Sge1 was found to be required for colonization of the xylem system and disease development.
Sge1, together with its homolog FoPac2, described in this work, belong to a class of conserved fungal proteins. Both proteins have apparent orthologs across the fungal kingdom and the N-terminal domain of these proteins is always more conserved than the C-terminus, which is generally rich in glutamine residues. Despite their similarity, the SGE1 and FoPAC2 deletion mutants generated in this study display different phenotypes, indicating that the functions of these proteins are not redundant. The FoPac2 and Sge1 homologs in S. pombe, Pac2 and Gti1, respectively, also have a different function although both proteins are involved in regulation of transition processes. Pac2 is a negative regulator of sexual development, which is induced under nitrogen starvation [15] and Gti1 is a positive regulator of metabolic reprogramming by inducing gluconate uptake under glucose starvation [14]. The best characterized members of the class to which Sge1 belongs are Wor1 in C. albicans and Ryp1 in H. capsulatum [10]–[13].
Ryp1 and Sge1 share a conserved and apparently functional nuclear localization signal. In addition, all three proteins contain a putative protein kinase A (Pka) phosphorylation site. Another common feature of these proteins is increased expression of their genes upon transition. WOR1 and RYP1 are both under tight transcriptional regulation in that hardly any expression of these genes can be detected in the white and the filamentous growth phase, respectively. Their expression is upregulated 45- and 4-fold during and after the transition to the opaque or the yeast growth phase [10],[11],[13],[17]. Although SGE1 expression can easily be detected in the saprophytic growth phase, it was found to be upregulated 2- to 5-fold during in planta growth. WOR1 is preceded by an exceptionally large intergenic region of 10.3 kb (average length in C. albicans ∼0.9 kb (http://www.broad.mit.edu/annotation/genome/candida_albicans/GeneStatsSummary.html) and it is able to bind to several positions in its own upstream region, although the protein does not contain an annotated DNA binding domain [13]. Also Ryp1, which is preceded by an intergenic region of about 2 kb, is able to bind to several positions in its own upstream region [11]. The intergenic region preceding SGE1 is larger than the average in the F. oxysporum genome: 4.3 kb versus 2.0 kb (http://www.broad.mit.edu/annotation/genome/fusarium_group/GeneStatsSummary.html), but whether Sge1 is able to activate its own transcription by binding to its promoter remains to be elucidated. Nevertheless, given the nuclear localization of Sge1, the homology to the transcriptional regulators Wor1 and Ryp1, and the effect of deletion of SGE1 on expression of SIX genes, we propose that also Sge1 also acts as a transcriptional regulator.
The requirement of Sge1 for expression of effector genes may explain the non-pathogenic phenotype of the SGE1 deletion mutant. Based on the apparent absence of xylem colonization as observed with confocal microscopy and the absence of the sge1 mutant in hypocotyls from infected plants, we conclude that although the mutant is still able to colonize roots and penetrate the root surface, it is not able to grow extensively in living cells or the xylem system. The ability of the sge1 mutant to colonize roots superficially is supported by the biocontrol activity of the sge1 mutant. The only additional phenotype of the mutant that we found was a 6-fold lower microconidia production compared to the wild type strain. An aberrant conidiogenesis behaviour was also observed for the H. capsulatum ryp1 mutant [11], indicating a conserved role for these proteins in conidiogenesis. In C. albicans, the regulatory network that drives white-opaque switching, to which Wor1 belongs, is beginning to be understood. A gene counteracting WOR1 is EFG1, which is necessary for maintaining the white state [29]. Wor1 is able to bind to the upstream region of EFG1, thereby regulating its expression [20]. Interestingly, the Fusarium homolog of Efg1 is FoStuA, a protein required for conidiogenesis in F. oxysporum [30]. However, STUA expression levels in the wild type and in the SGE1 deletion mutant were comparable (data not shown). How Sge1 affects conidiogenesis remains, therefore, to be elucidated.
As mentioned above, a common feature of Sge1 and its homologs is the presence of a potential protein kinase A phosphorylation site. The functionality of this phosphorylation site has been investigated in S. pombe. It was shown that gluconate import under repressing conditions (high glucose concentration) occurred in a protein kinase A deletion mutant. Therefore, it was speculated that Pka1 inhibits Gti1 protein function. However, alteration of the Pka phosphorylation site by replacement of the conserved threonine residue by an alanine did not result in activation of the Gti1 protein and the expected gluconate import was not observed, indicating that either Pka1 does not inhibit Gti1 activity by phosphorylation or that the mutation impairs Gti1 function or its stability [14]. In Sge1, the same Pka site appears to be required for activity. A stable transformant of SGE1 in S. cerevisiae was obtained only after a spontaneous mutation of an arginine into a serine in the potential phosphorylation site, suggesting that expression of wild type SGE1 in yeast inhibits growth. S. cerevisiae contains a SGE1 homolog, YEL007W, and this gene has been implicated to play a role in regulation of smooth ER, cell adhesion and budding [31]–[33]. It could be that expression of both genes is lethal to yeast. Introduction of the mutated SGE1 gene in the SGE1 deletion mutant of Fol failed to restore the pathogenicity defect, confirming that the R66S mutation impairs Sge1 function, possibly due to an effect on phosphorylation. Since there is only a minor increase in SGE1 expression levels upon in planta growth, post-translational modifications such as phosphorylation could play a key role in activation of Sge1. Unfortunately, initial attempts to demonstrate phosphorylation by Pka1 of purified Sge1 and Sge1R66S in vitro remained inconclusive (data not shown) and will be subject of further investigation.
The loss of effector (SIX) gene expression in the sge1 mutant supports a role of Sge1 as a transcriptional activator controlling the onset of parasitic growth. In Fol, these effector genes encode small cysteine rich proteins secreted during colonization of xylem vessels, designated Secreted in xylem (Six) proteins [21],[34]. SIX1 is only expressed during in planta growth and encodes Avr3, as it is required for I-3 mediated resistance [22],[34]. Six3 is Avr2 and is required for I-2 mediated resistance [25]. Both Six1/Avr3 and Six3/Avr2 are required for full virulence [23],[25]. No function has yet been assigned to Six2 and Six5. Here, we show that, like Six1, Six2 is highly expressed during in planta-mimicking growth conditions (co-cultivation with tomato cells), but SIX3 and SIX5, divergently transcribed from the same promoter region, are expressed in synthetic medium. It could be that Sge1 activity under axenic growth conditions is too low for induction of SIX1 and SIX2, but high enough to induce SIX3 and SIX5 expression. The increased SGE1 expression levels during in planta growth may then lead to expression of all SIX genes. Currently, a SGE1 over-expression mutant is being generated in order to determine whether increased SGE1 expression alone can lead to expression of all SIX genes under axenic growth conditions. At the moment, it is not known whether Sge1 influences the expression of the SIX genes directly or indirectly, for instance through involvement of other (transcription) factors, nor how SGE1 expression itself is regulated. Preliminary promoter analysis of the SIX genes revealed a potential common motif (unpublished data). The role of this motif in SIX gene expression and the question whether Sge1 is able to bind DNA and the SIX gene promoter regions in particular remain to be elucidated.
All the genes tested in the SGE1 deletion mutant (SIX1, SIX2, SIX3, and SIX5) were found to be dependent on Sge1 for their expression, even during growth in synthetic medium. Although deletion of individual SIX genes (SIX1 or SIX3) only leads to a minor reduction in pathogenicity, the loss of expression of all SIX genes in the sge1 mutant may be the primary cause of the complete non-pathogenic phenotype, due to the inability to suppress host defence responses. Still, loss of pathogenicity upon deletion of SGE1 may not be due to loss of production of effector proteins only. The majority of pathogenicity genes found in F. oxysporum have pleiotropic functions [35],[36]. It is therefore unlikely that expression of these genes is altered in the sge1 mutant, since this mutant did not display growth defects other than a minor effect in conidiogenesis. One described F. oxysporum mutant that is only disturbed in pathogenicity is the fow2 mutant [37]. Expression of FOW2 nor that of two other pathogenicity genes, the protein kinase gene SNF1 [38] and velvet homolog gene FOXG_00016 [39], is altered in the SGE1 deletion mutant (unpublished data). However, preliminary results suggest that the expression of the FTF1 transcription factor gene, implicated to be involved in pathogenicity [40], seems to be altered in the sge1 mutant (unpublished data). In addition, secondary metabolite profiling revealed that the sge1 mutant is reduced in the production of fusaric acid, beauvericin and a number of unknown metabolites compared to the wild type (U. Thrane and C.B. Michielse, unpublished data). High concentrations of fusaric acid may contribute to pathogenicity by reducing host resistance [41],[42]. Thus, processes implicated in pathogenesis other than SIX gene expression are also altered in the sge1 mutant, supporting the hypothesis that Sge1 plays a central role during parasitic growth. Deletion of the Sge1 homolog in Botrytis cinerea, BC1G_11680, also leads to a severely reduced pathogenicity phenotype (C.B. Michielse and P. Tudzynski, unpublished data), indicating that the role of Sge1-like proteins in (plant) pathogenic fungi might be conserved.
The regulation of expression of phase specific genes (SIX1, SIX2), and genes involved in virulence (SIX1, SIX3) by Sge1 resembles the functions of Wor1 in C. albicans and Ryp1 in H. capsulatum. Wor1 has been shown to bind directly to the upstream region of target genes [20]. Interestingly, these target genes all have a large upstream region, indicating that large upstream regions might be a prerequisite for Wor1 binding. Whether this is a common feature shared between Wor1 and Sge1 is still unknown. Future efforts include the identification of additional genes regulated by Sge1 and, eventually, to unravel the network of transcriptional regulators involved in activating the pathogenicity program in F. oxysporum. This will also reveal whether Sge1 indeed plays a master role in this network, like its homologs in dimorphic fungi. Given the striking conservation of Sge1 across fungi, we expect that many features of this network will turn out to be similar in pathogens of both plants and animals. Obviously, since Sge1 homologs are also present in non-pathogenic fungi, this regulatory network must serve a fundamental morphological or physiological switch function and pathogens would have adopted this network for adapting to the host environment.
Materials and Methods
Strains, plant material and culture conditions
Fusarium oxysporum f. sp. lycopersici strain 4287 (race 2; FGSC9935) was used as the parent strain for fungal transformation. It was stored as a monoconidial culture at −80°C and revitalized on potato dextrose agar (PDA, Difco) at 25°C. Agrobacterium tumefaciens EHA105 [43] was used for Agrobacterium-mediated transformation of F. oxysporum and was grown in 2YT medium [44] containing 20 µg/ml rifampicin at 28°C. Introduction of the plasmids into the Agrobacterium strain was performed as described by Mattanovich et al [45]. Escherichia coli DH5 alpha (Invitrogen) was used for construction, propagation, and amplification of the plasmids and was grown in LB medium at 37°C containing either 100 µg/ml ampicillin or 50 µg/ml kanamycin depending on the resistance marker of the plasmid used. Plant line Moneymaker ss590 (Gebr. Eveleens b.v., The Netherlands) was used to assess pathogenicity of F. oxysporum strains and transformants. Biocontrol assays were performed on flax (Linum usitatissimum) cv. viking using F. oxysporum f. sp. lini (Foln3, MYA-1201) isolated from a diseased flax plant in French Britany as pathogenic strain.
Construction of gene replacement and complementation constructs
To generate the SGE1 disruption construct, pSGE1KO, PCR was performed using a BAC clone containing the SGE1 gene as a template with primer combination FP878–FP879 and FP880–FP881 in order to amplify a 901 bp HindIII fragment corresponding to the upstream region and a 1032 bp KnpI fragment corresponding to the downstream region, respectively (Table S1). The fragments were sequentially cloned into pPK2hphgfp [9] and proper orientation of the inserts was checked by PCR.
The SGE1 complementation construct, pSGE1com, was generated by cloning a 2234 bp HindIII fragment containing the SGE1 ORF including 901 bp upstream and 341 bp downstream sequences into pUC19, resulting in pUC19SGE1. The HindIII SGE1 fragment was subsequently transferred from pUC19SGE1 into pRW1p [24], resulting in pSGE1com. The SGE1 complementation construct carrying a point mutation in the ORF of SGE1, pSGE1comPM, was generated by replacing a 465 bp SacII/BglII SGE1 fragment in pSGE1com by a 465 bp SacII/BglII fragment isolated from the yeast-two-hybrid bait vector pASSGE1PM. This vector was re-isolated from Saccharomyces cerevisiae after previously being transformed with pASSGE1. The latter vector was generated by cloning an NcoI-EcoRI 1011 bp PCR product obtained with the primers FP1484 and FP1485 corresponding to the SGE1 ORF in pAS2.1 (Clontech).
To generate the FoPAC2 gene disruption construct, a 1037 bp upstream fragment and a 749 bp downstream fragment was amplified from genomic DNA by PCR with the primer pairs FP1796–FP1797 and FP1798–FP1799, respectively. The PCR products were cloned into pGEM-T Easy (Promega) and, subsequently, the KpnI/PacI upstream and the AscI/HindIII downstream fragment were sequentially cloned in pRW2h [24].
The Sge1-fluorescent protein fusion constructs pSGE1::CFP and pSGE1::RFP were generated in a multi-step approach. First, pUC19SGE1 was amplified by PCR with the primers FP1120 and FP1121 flanked at the 5′ end by an ApaI and a SpeI restriction site, respectively. The amplified product was digested with ApaI and religated, resulting in pUC19SGE1as, containing the SGE1 ORF with the ApaI and SpeI preceding the SGE1 stop codon. Secondly, a CFP [46] and a mRFP [47] fragment were generated by PCR using primer pair FP1122–FP1123 and FP1120–FP1121, respectively. The CFP and RFP PCR products were digested with ApaI and SpeI and directionally cloned in pUC19SGE1as, resulting in pUCSGE1::CFP and pUCSGE1::RFP, respectively. Finally, a HindIII fragment corresponding to each fusion cassette was cloned into HindIII digested pRW1p and proper orientation of the fragments was checked by PCR. The gpdAH2B::GFP expression cassette was isolated as a XbaI/NheI fragment from pH2B::GFP (kind gift from Dr. Ram, Leiden, The Netherlands) and cloned into XbaI/NheI digested pRW2h [24] to generate pRWH2B::GFP.
Fungal transformation and disease assay
Agrobacterium-mediated transformation of F. oxysporum f. sp. lycopersici was performed as described by Mullins and Chen [48] with minor adjustments [49]. Depending on the selection marker used, transformants were selected on Czapek Dox agar (CDA, Oxoid) containing 100 µg/ml Hygromycin (Duchefa) or on CDA containing 0.1 M TrisHCl pH 8 and 100 µg/ml Zeocin (InvivoGen).
Plant infection was performed using 9 to 11 days old seedlings (Moneymaker ss590), following the root-dip inoculation method [50]. Disease index was scored and statistical analysis performed as described earlier [9].
Analysis of conidiogenesis, conidia germination, carbon source utilization and cellophane penetration
For quantization of microconidia production, five independent experiments were performed, each with five replicates. Microconidia were harvested after five days of growth in 50 ml minimal medium (3% sucrose, 10 mM KNO3 and 0.17% yeast nitrogen base without amino acids and ammonia) and 106 spores were used to inoculate 100 ml minimal medium. After five days the microconidia produced were harvested and counted in a Bürker-Türk haemocytometer. The isolated conidia were also used to determine germination rates. To this end 600 spores were added into a 6-wells plate with each well containing 250 µl PDA and incubated overnight at 4°C, then transferred to 25°C and germinated conidia were counted after six hours of incubation. Macroconidia development was analyzed in liquid carboxymethyl cellulose medium as described by Ohara and Tsuge [30]. Conidia were fixed in 0.4% p-formaldehyde and stained with Hoechst 33342 (250 µg/ml) and calcofluor white (25 µg/ml) to visualize nuclei and cell walls, respectively. Stained cells were observed with a BX50 fluorescence microscope and a U-MWU filter (Olympus).
Carbon-source utilization was tested in a BIOLOG FF MicroPlate (BIOLOG). A conidial suspension (104 conidia in 150 µl) of each strain was inoculated in each well of the plate and incubated at 25°C. The absorbance of each well at 600 nm was measured with a microtiter plate reader (Packerd Spectra Count) after 1, 2, 3 and 4 days of incubation.
The cellophane penetration assay was performed as described [18], with minor adjustments. CDA was used as basal medium in the assay and was inoculated with 105 conidia. After incubation of 5 days at 25°C, the cellophane was removed and after a subsequent incubation of 2 days at 25°C fungal growth was scored.
Southern analysis
Genomic DNA was isolated as described by Kolar et al [51] with minor adjustments [9]. For Southern analysis, 10 µg genomic DNA of each transformant was digested with Acc65I or BglII for SGE1 or FoPAC2 transformants, respectively, and incubated overnight at 37°C. The samples were loaded on a 1% 0.5× Tris-borate/EDTA gel and run for 18 hours at 45 V. The digested DNA was transferred to Hybond-N+ (Amersham Pharmacia) as described by Sambrook et al [44]. The probes used for Southern analysis are: (1) a 432 bp SGE1 upstream fragment obtained by PCR with primers FP842 and FP1174 (Table S1) and (2) a 488 bp FoPAC2 downstream fragment obtained by PCR with primers FP2198 and FP 2199. The DecaLabel™ DNA Labelling Kit (Fermentas) was used to label probes with [α-32P]dATP. Hybridization was done overnight at 65°C in 0.5 M sodium phosphate buffer, pH 7.2, containing 7% SDS and 1 mM EDTA. Blots were washed with 0.2× SSC, 0.1% SDS. Hybridization signals were visualized by phosphorimaging (Molecular Dynamics).
RT and quantitative PCR
Samples for expression analysis were obtained by adding 0,5 ml of 107 conidia/ml of wild type, SGE1 deletion or SGE1 complementation strain to 4,5 ml of a one week old MSK8 cell culture grown in BY-medium [52]. After 24 hours of incubation at 22°C, the material was harvested, washed two times with sterile water and freeze-dried. Total RNA was isolated using Trizol (Invitrogen) and prior to cDNA synthesis the RNA was treated with DnaseI (Fermentas). First strand cDNA was synthesized with M-MuLV reverse transcriptase following the instruction of the manufacturer (Fermentas). Supertaq (SphaeroQ), 1 µl of the cDNA reaction and primers FP1999/FP2000, FP998/FP1001, FP962/FP963, FP1993/FP1994, FP2131/FP2132 and FP157/FP158 (Table S1) to detect SIX1 (CAE55879), SIX2 (CAE55868), SIX3 (CAJ83999), SIX5 (ACN87967), SGE1 (FOXG_10510) and FEM1 (AAL47843) transcripts, respectively, were used in the RT-PCR.
Quantitative PCR (qPCR) was performed with Platinum SYBR Green qPCR (Invitrogen) using a 7500 Realtime PCR System (Applied Biosystems). To quantify mRNA levels of SGE1 and of the constitutively expressed EF-1α gene, we used primers FP2131/FP2132 and FP2029/FP2030, respectively (Table S1). EF-1α was used to calculate the relative expression levels of SGE1 in axenic and MSK8 cultures infected with F. oxysporum wild type, SGE1 deletion or SGE1 complementation mutants. Real time PCR primer efficiencies were calculated using LinRegPCR [53] and relative expression levels were calculated according to the comparative C(t) method [54].
Microscopic analysis
Sge1::mRFP, Sge1::CFP and H2B::GFP fusion proteins were visualized using a BX50 fluorescence microscope with the appropriate excitation and emission filters for CFP, mRFP and GFP (Olympus). For this purpose, 10 µl of five days old minimal medium cultures was spotted onto glass slides.
A Nikon A1 microscope was used to monitor tomato root infection by the wild type and SGE1 deletion mutant. Excitation was provided for the GFP signal with an Argon (488 nm) laser (emission: 405–455 nm) and for the root auto-fluorescence with an UV diode (405 nm) laser (emission: 420–470 nm). Images were line-sequential scanned (2 µm slices, 1024×1024 pixels) using water objective plan fluor 20× Imm, 0.75 NA. Pictures were analyzed with the Nikon NIS and ImageJ software. For preparation of the slides, 9 days old tomato seedlings were carefully removed from the potting soil, rinsed with water and intact roots were inoculated with 20 ml tap water containing 107 conidia/ml and incubated for 4 days at room temperature in a petridish. The roots were rinsed with water, cut from the hypocotyl, placed in a drop of water on a glass slide and covered with a cover glass. A bridge mounted on the glass slide prevented squashing of the root material.
Inoculum preparation and biocontrol assay
Inocula were prepared as described [55], except that minimal liquid medium [56] in which sucrose was replaced by glucose (5 g/l) and sodium nitrate by ammonium tartrate (1 g/l) was used instead of Malt medium. A heat treated (100°C for 1 h) silty-loam soil from Dijon (35.1% clay, 47% loam, 15.1% sand, and 1.22% organic C [pH = 6.9]) was added to module trays containing 96 wells each of 50 ml. To prevent contamination between treatments, only every second row of wells was filled with soil. The soil in each well was inoculated with 5 mL of conidia suspension. The concentration of the conidia suspensions were adjusted to obtain the following inoculum densities: 1×104 conidia ml−1 of soil for the pathogenic strain Foln3 and 1×106 conidia ml−1 of soil for the two others strains. The soil surface was covered with a thin layer of calcinated clay granules (Oil Dri Chem-Sorb, Brenntag Bourgogne, Montchanin, France) and three seeds of flax, cv. viking, were sown in each pot. A thin layer of Chem-Sorb was used to cover the seeds. Plants were grown in a growth chamber in the first 2 weeks; the growing conditions were 8 h 15°C N/16 h 17°C D, with a light intensity of 33 µE m−2 s−1. The plants were thinned to one plant per pot, and from week 3 the temperature was kept at 22°C N/25°C D. There were three replicates of 16 individual plants per treatment, randomly arranged, and the experiment was replicated. Plants were watered every day, and once a week water was replaced by a 500-fold dilution of a commercial nutrient stock solution (“Hydrodrokani AO”, Hydro Agri, Nanterre, France). Plants showing characteristic symptoms of yellowing were recorded twice a week and removed. To compare the ability of strains to induce disease or, on the contrary, to protect the plant against wilt, ANOVA was performed on Area under the Disease Progress Curves followed by Newman and Keuls test at the probability of 95%.
Supporting Information
Primers used in this study.
(0.01 MB PDF)
Analysis of transformants deleted for SGE1. A knock-out construct containing a hygromycin-GFP expression cassette flanked by 901 bp up- and 1032 bp downstream sequences of SGE1 was introduced in the wild type strain by Agrobacterium-mediated transformation. A) Schematic representation of the knock-out strategy for SGE1 drawn to scale. The arrow heads indicate the positions of the original T-DNA insertions. The small arrows represent the primers used to check homologous recombination (a and b) and the absence of the open reading frame (c and d). B) Verification of homologous recombination by PCR using primers a and b. C) Verification of the absence of the SGE1 open reading frame by PCR using primers c and d. The 1 kb DNA ladder of Fermentas (www.fermentas.com) is used as a marker. −, negative control. +, positive control (genomic DNA). The figures are composed from different parts of an ethidium bromide gel, which results in minor colour differences.
(0.41 MB PDF)
Analysis of transformants complemented with SGE1. A complementation construct containing a phleomycin expression cassette and the SGE1 gene including 901 bp up- and 341 bp downstream sequences was introduced in the SGE1 knock-out mutant #32 by Agrobacterium-mediated transformation. A) Schematic representation of the complementation strategy for SGE1 drawn to scale. The small arrows represent the primers used to check the presence of the open reading frame (c and d). B) Verification of the presence of the SGE1 ORF by PCR using primers c and d. The 1 kb DNA ladder of Fermentas (www.fermentas.com) is used as a marker. −, negative control.
(0.09 MB PDF)
Southern analysis of the SGE1 deletion and complementation mutants. Southern analysis was performed to verify correct homologous recombination at the SGE1 locus in the SGE1 disruptants and complemented strains. To this end, chromosomal DNA of the various mutants was digested with Acc65I, blotted and hybridized with a 432 bp probe corresponding to the SGE1 promoter. The SGE1 locus of the wild type strain is visible as a 1.6 kb fragment corresponding to the SGE1 upstream region and to the 5′ part of the SGE1 ORF. In the 5G2 insertional mutagenesis mutant, the SGE1 ORF is disrupted due to a T-DNA insertion (see Figure S1A). As a result the 1.6 kb fragment observed in a wild type situation is replaced by a 1.8 kb fragment corresponding to a part of the SGE1 promoter region and the hygromycin expression cassette present on the T-DNA. In the SGE1 disruption mutants introduction of the gene replacement cassette by homologous recombination led to the expected replacement of the 1.6 kb fragment with a fragment containing part of the SGE1 upstream region and the gene disruption cassette which should be larger than 4.9 kb. Introduction of the SGE1 complementation cassette, including the SGE1::FP fusion protein complementation cassettes, by homologous recombination restored the wild type SGE1 locus. WT, wild type. 5G2, insertional mutagenesis mutant 5G2. KO, SGE1 knock-out mutants. com, SGE1 complementation mutants. CC, SGE1::CFP complementation mutants. RC, SGE1::RFP complementation mutants.
(0.05 MB PDF)
Analysis of transformants deleted for FoPAC2. A knock-out construct containing a hygromycin expression cassette flanked by 1037 bp up- and 749 bp downstream sequences of FoPAC2 was introduced in the wild type strain by Agrobacterium-mediated transformation. A) Schematic representation of the knock-out strategy for FoPAC2 drawn to scale. The arrow head indicates the position of the original T-DNA insertion. The small arrows represents the primers used to check homologous recombination (a and b) and the absence of the open reading frame (c and d). B) Verification of the absence of the FoPAC2 open reading frame by PCR using primers c and d. C) Verification of homologous recombination by PCR using primers a and b. D) Southern analysis of the FoPAC2 deletion mutants. Chromosomal DNA was digested with BglII, blotted and hybridized with a 488 bp probe corresponding to upstream region. The FoPAC2 locus of the wild type strain is visible as a 7.0 kb fragment. In the 30C11 insertional mutagenesis mutant, the FoPAC2 ORF is disrupted due to a T-DNA insertion (see Figure S4A). As a result the 7.0 kb fragment observed in a wild type situation is replaced by a 12.3 kb fragment corresponding to a part of the FoPAC2 upstream region and the hygromycin expression cassette present on the T-DNA. In the FoPAC2 disruption mutants introduction of the gene replacement cassette by homologous recombination led to the expected replacement of the 7.0 kb fragment with a 11.4 kb fragment containing part of the FoPAC2 upstream region and the gene disruption cassette. The 1 kb DNA ladder of Fermentas (www.fermentas.com) is used as a marker. −, negative control. +, positive control (genomic DNA). Panels B and C are composed of different parts of an ethidium bromide gel, which results in minor colour differences.
(0.15 MB PDF)
The SGE1 deletion mutant is not impaired in carbon source utilization. To analyze carbon utilization of the sge1 mutant, BIOLOG FF MicroPlates containing in each well a different carbon source were used. A conidial suspension (104 conidia in 150 µl) of the wild type or the SGE1 deletion stain was inoculated in each well and incubated at 25°C. The absorbance of each well at 600 nm was measured with a microtiter plate reader (Packerd Spectra Count) after 4 days of incubation. The ratios were calculated by dividing the measured values of the mutant strain by those of the wild type strain. Only values higher than 1.5 or lower than 0.5 were marked (boxed) as carbon sources on which the sge1 mutant appeared to display a different growth rate than the wild type strain in this experiment. An additional plate assay containing these carbon sources showed that these differences were not reproducible (data not shown).
(0.40 MB TIF)
Micro- and macroconidia of the SGE1 deletion mutant are morphologically indistinguishable from wild type. Micro- and macroconidia development was analyzed in liquid carboxymethyl cellulose medium. Conidia were fixed in 0.4% p-formaldehyde and stained with Hoechst 33342 (250 µg/ml) and calcofluor white (25 µg/ml) to visualize nuclei and cell walls, respectively. The left panel depicts a phase contrast recording and the right panel depicts a UV-exposed recording of conidia of the sge1 mutant.
(1.38 MB PDF)
SGE1 is not essential for superficial root colonization. The root colonization behaviour of the SGE1 deletion mutant was determined by binocular microscopy. Nine to eleven days old tomato seedlings were inoculated with wild type or an SGE1 deletion mutant spore suspension and root colonization was determined after two to five days after inoculation. Patches of colonization were already visible after three days. The left panel depicts phase contrast recordings of a GFP-expressing virulent strain (38H3) and of the SGE1 deletion mutant. The right panel depicts UV-exposed recordings.
(0.82 MB PDF)
The SGE1 mutant is not impaired in cellophane penetration. The capacity of the SGE1 deletion strain to penetrate cellophane was determined using a cellophane penetration assay. CDA medium covered by a cellophane sheet was inoculated with a drop containing 105 conidia. After incubation of 5 days at 25°C (left panel), the cellophane was removed and after a subsequent incubation of 2 days at 25°C fungal growth of both the wild type and the SGE1 deletion mutant was clearly observed (right panel).
(0.25 MB PDF)
The sge1 mutant is impaired in extensive in planta growth. To determine whether the SGE1 deletion mutant is able to growth within xylem vessels, tomato seedlings were inoculated with the wild type, SGE1 knock-out or the SGE1 complementation strain and potted into soil according to the bioassay procedure. One week after inoculation the hypocotyl was cut in slices of several millimeters which were placed on rich (PDA) medium. F. oxysporum outgrowth was observed from the hypocotyl pieces previously inoculated with the wild type and the SGE1 complementation strain, but not from the hypocotyl pieces previously inoculated with the SGE1 knock-out mutant.
(0.26 MB PDF)
Pathogenicity is partially restored in SGE1::FP complementation strains. Nine to eleven days old tomato seedlings were inoculated with fungal spore suspensions following the root-dip inoculation method and the disease index (ranging from 0 healthy plant to 4 severely diseased plant/dead plant) was scored after three weeks. Average disease index of 20 plants three weeks after mock inoculation (H2O) or inoculation with a SGE1 deletion mutant (SGE1KO32), SGE1 (SGE1com28), SGE1::CFP (SGE1comCC4, 5, and 10) and SGE1::RFP (SGEcomRC2, 6, and 11) complementation mutants or wild type (4287). Error bars indicate standard deviation and capitals define statistically different groups (ANOVA, p = 0.95).
(0.03 MB PDF)
Analysis of transformants complemented with SGE1PM. A complementation construct containing a phleomycin expression cassette and the SGE1PM gene, encoding Sge1R66S, including 901 bp up- and 341 bp downstream sequences, was introduced in the SGE1 knock-out mutant #32 by Agrobacterium-mediated transformation. A) Verification of the presence of the SGE1 ORF by PCR using primers c and d (see Figure S1A). B) Verification of homologous recombination by PCR using primers a and b (see Figure S2). The 1 kb DNA ladder of Fermentas (www.fermentas.com) is used as a marker. −, negative control. Deletion mutant SGE1KO32 and complementation mutant SGE1com28 were used as a negative and positive control, respectively.
(0.34 MB PDF)
Acknowledgments
We thank Antonio Di Pietro for providing F. oxysporum strain 4287, Arthur Ram for the vector pH2B::GFP, Wilfried Jonkers for technical assistance, Ben Cornelissen for critically reading this manuscript and Ulf Thrane for metabolic profile analysis. Furthermore, we thank Harold Lemereis, Thijs Hendrix, and Ludek Tikovsky for managing the plant growth facilities and their assistance with bioassays.
Footnotes
The authors have declared that no competing interests exist.
Most of this work was funded by the Utopa Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gordon TR, Martyn RD. The evolutionary biology of Fusarium oxysporum. Ann Rev Phytopathol. 1997;35:111–128. doi: 10.1146/annurev.phyto.35.1.111. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong GM, Armstrong JK. Formae speciales and races of Fusarium oxysporum and Alternaria alternata. In: Cook R, editor. Fusarium: Diseases, Biology and Taxonomy. Penn State University Press; 1981. pp. 391–399. [Google Scholar]
- 3.Brayford D. Fusarium oxysporum f. sp. radicis-lycopersici, IMI descriptions of fungi and bacteria no. 1270. Mycopathologia. 1996;133:61–63. [Google Scholar]
- 4.Ploetz RC. Popolation biology of Fusarium oxysporum f. sp. cubense. In: Ploetz RC, editor. Fusarium wilt of banana. St. Paul, Minnesota: APS Press; 1990. pp. 63–76. [Google Scholar]
- 5.Albisetti M, Lauener RP, Gungor T, Schar G, Niggli FK, et al. Disseminated Fusarium oxysporum infection in hemophagocytic lymphohistiocytosis. Infection. 2004;32:364–366. doi: 10.1007/s15010-004-3135-8. [DOI] [PubMed] [Google Scholar]
- 6.Anaissie EJ, Kuchar RT, Rex JH, Francesconi A, Kasai M, et al. Fusariosis associated with pathogenic Fusarium species colonization of a hospital water system: a new paradigm for the epidemiology of opportunistic mold infections. Clin Infect Dis. 2001;33:1871–1878. doi: 10.1086/324501. [DOI] [PubMed] [Google Scholar]
- 7.Romano C, Miracco C, Difonzo EM. Skin and nail infections due to Fusarium oxysporum in Tuscany, Italy. Mycoses. 1998;41:433–437. doi: 10.1111/j.1439-0507.1998.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 8.Agrios GN. Plant Pathology. San Diego, California: Academic Press; 1997. [Google Scholar]
- 9.Michielse CB, van Wijk R, Reijnen L, Cornelissen BJ, Rep M. Insight into the molecular requirements for pathogenicity of Fusarium oxysporum f. sp. lycopersici through large-scale insertional mutagenesis. Genome Biol. 2009;10:R4. doi: 10.1186/gb-2009-10-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang G, Wang H, Chou S, Nie X, Chen J, et al. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci U S A. 2006;103:12813–12818. doi: 10.1073/pnas.0605270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen VQ, Sil A. Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc Natl Acad Sci U S A. 2008;105:4880–4885. doi: 10.1073/pnas.0710448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srikantha T, Borneman AR, Daniels KJ, Pujol C, Wu W, et al. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot Cell. 2006;5:1674–1687. doi: 10.1128/EC.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zordan RE, Galgoczy DJ, Johnson AD. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci U S A. 2006;103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caspari T. Onset of gluconate-H+ symport in Schizosaccharomyces pombe is regulated by the kinases Wis1 and Pka1, and requires the gti1+ gene product. J Cell Sci. 1997;110:2599–2608. doi: 10.1242/jcs.110.20.2599. [DOI] [PubMed] [Google Scholar]
- 15.Kunitomo H, Sugimoto A, Wilkinson CR, Yamamoto M. Schizosaccharomyces pombe pac2+ controls the onset of sexual development via a pathway independent of the cAMP cascade. Curr Genet. 1995;28:32–38. doi: 10.1007/BF00311879. [DOI] [PubMed] [Google Scholar]
- 16.Preston-Mafham J, Boddy L, Randerson PF. Analysis of microbial community functional diversity using sole-carbon-source utilisation profiles - a critique. FEMS Microbiol Ecol. 2002;42:1–14. doi: 10.1111/j.1574-6941.2002.tb00990.x. [DOI] [PubMed] [Google Scholar]
- 17.Tsong AE, Miller MG, Raisner RM, Johnson AD. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell. 2003;115:389–399. doi: 10.1016/s0092-8674(03)00885-7. [DOI] [PubMed] [Google Scholar]
- 18.Prados Rosales RC, Di Pietro A. Vegetative hyphal fusion is not essential for plant infection by Fusarium oxysporum. Eukaryot Cell. 2008;7:162–171. doi: 10.1128/EC.00258-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maruyama J, Nakajima H, Kitamoto K. Visualization of nuclei in Aspergillus oryzae with EGFP and analysis of the number of nuclei in each conidium by FACS. Biosci Biotechnol Biochem. 2001;65:1504–1510. doi: 10.1271/bbb.65.1504. [DOI] [PubMed] [Google Scholar]
- 20.Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007;5:e256. doi: 10.1371/journal.pbio.0050256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houterman PM, Speijer D, Dekker HL, De Koster CG, Cornelissen BJC, et al. The mixed xylem sap proteome of Fusarium oxysporum-infected tomato plants. Mol Plant Pathol. 2007;8:215–221. doi: 10.1111/j.1364-3703.2007.00384.x. [DOI] [PubMed] [Google Scholar]
- 22.van der Does HC, Duyvesteijn RG, Goltstein PM, van Schie CC, Manders EM, et al. Expression of effector gene SIX1 of Fusarium oxysporum requires living plant cells. Fungal Genet Biol. 2008;45:1257–1264. doi: 10.1016/j.fgb.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Rep M, Meijer M, Houterman PM, van der Does HC, Cornelissen BJ. Fusarium oxysporum evades I-3-mediated resistance without altering the matching avirulence gene. Mol Plant Microbe Interact. 2005;18:15–23. doi: 10.1094/MPMI-18-0015. [DOI] [PubMed] [Google Scholar]
- 24.Houterman PM, Cornelissen BJ, Rep M. Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathog. 2008;4:e1000061. doi: 10.1371/journal.ppat.1000061. doi: 10.1371/journal.ppat.1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Houterman PM, Ma L, van Ooijen G, de Vroomen MJ, Cornelissen BJ, et al. The effector protein Avr2 of the xylem colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J. 2009;58:970–978. doi: 10.1111/j.1365-313X.2009.03838.x. [DOI] [PubMed] [Google Scholar]
- 26.Alabouvette C, Olivain C, Migheli Q, Steinberg C. Microbiological control of soil-borne diseases: capacities of non pathogenic strains of Fusarium oxysporum to control Fusarium wilts. New Phytol. 2009 doi: 10.1111/j.1469-8137.2009.03014.x. in press. [DOI] [PubMed] [Google Scholar]
- 27.Klein BS, Tebbets B. Dimorphism and virulence in fungi. Curr Opin Microbiol. 2007;10:314–319. doi: 10.1016/j.mib.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nadal M, Garcia-Pedrajas MD, Gold SE. Dimorphism in fungal plant pathogens. FEMS Microbiol Lett. 2008;284:127–134. doi: 10.1111/j.1574-6968.2008.01173.x. [DOI] [PubMed] [Google Scholar]
- 29.Sonneborn A, Tebarth B, Ernst JF. Control of white-opaque phenotypic switching in Candida albicans by the Efg1p morphogenetic regulator. Infect Immun. 1999;67:4655–4660. doi: 10.1128/iai.67.9.4655-4660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohara T, Tsuge T. FoSTUA, encoding a basic helix-loop-helix protein, differentially regulates development of three kinds of asexual spores, macroconidia, microconidia, and chlamydospores, in the fungal plant pathogen Fusarium oxysporum. Eukaryot Cell. 2004;3:1412–1422. doi: 10.1128/EC.3.6.1412-1422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li F, Palecek SP. Identification of Candida albicans genes that induce Saccharomyces cerevisiae cell adhesion and morphogenesis. Biotechnol Prog. 2005;21:1601–1609. doi: 10.1021/bp050236c. [DOI] [PubMed] [Google Scholar]
- 32.Wright R, Parrish ML, Cadera E, Larson L, Matson CK, et al. Parallel analysis of tagged deletion mutants efficiently identifies genes involved in endoplasmic reticulum biogenesis. Yeast. 2003;20:881–892. doi: 10.1002/yea.994. [DOI] [PubMed] [Google Scholar]
- 33.Zettel MF, Garza LR, Cass AM, Myhre RA, Haizlip LA, et al. The budding index of Saccharomyces cerevisiae deletion strains identifies genes important for cell cycle progression. FEMS Microbiol Lett. 2003;223:253–258. doi: 10.1016/S0378-1097(03)00384-7. [DOI] [PubMed] [Google Scholar]
- 34.Rep M, van der Does HC, Meijer M, van Wijk R, Houterman PM, et al. A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol Microbiol. 2004;53:1373–1383. doi: 10.1111/j.1365-2958.2004.04177.x. [DOI] [PubMed] [Google Scholar]
- 35.Di Pietro A, Madrid MP, Caracuel Z, Delgado-Jarana J, Roncero MI. Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol Plant Pathol. 2003;4:315–325. doi: 10.1046/j.1364-3703.2003.00180.x. [DOI] [PubMed] [Google Scholar]
- 36.Michielse CB, Rep M. Pathogene profile update: Fusarium oxysporum. Mol Plant Pathol. 2009;10:311–324. doi: 10.1111/j.1364-3703.2009.00538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Imazaki I, Kurahashi M, Iida Y, Tsuge T. Fow2, a Zn(II)2Cys6-type transcription regulator, controls plant infection of the vascular wilt fungus Fusarium oxysporum. Mol Microbiol. 2007;63:737–753. doi: 10.1111/j.1365-2958.2006.05554.x. [DOI] [PubMed] [Google Scholar]
- 38.Ospina-Giraldo MD, Mullins E, Kang S. Loss of function of the Fusarium oxysporum SNF1 gene reduces virulence on cabbage and Arabidopsis. Curr Genet. 2003;44:49–57. doi: 10.1007/s00294-003-0419-y. [DOI] [PubMed] [Google Scholar]
- 39.Lopez-Berges MS, A DIP, Daboussi MJ, Wahab HA, Vasnier C, et al. Identification of virulence genes in Fusarium oxysporum f. sp. lycopersici by large-scale transposon tagging. Mol Plant Pathol. 2009;10:95–107. doi: 10.1111/j.1364-3703.2008.00512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramos B, Alves-Santos FM, Garcia-Sanchez MA, Martin-Rodrigues N, Eslava AP, et al. The gene coding for a new transcription factor (ftf1) of Fusarium oxysporum is only expressed during infection of common bean. Fungal Genet Biol. 2007;44:864–876. doi: 10.1016/j.fgb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 41.Bouizgarne B, El-Maarouf-Bouteau H, Frankart C, Reboutier D, Madiona K, et al. Early physiological responses of Arabidopsis thaliana cells to fusaric acid: toxic and signalling effects. New Phytol. 2006;169:209–218. doi: 10.1111/j.1469-8137.2005.01561.x. [DOI] [PubMed] [Google Scholar]
- 42.Gaumann E. Fusaric acid as a wilt toxin. Phytopathol. 1957;47:342–357. [Google Scholar]
- 43.Hood EE, Gelvin SB, Melchers S, Hoekema A. New Agrobacterium helper plasmids for gene transfer to plants (EHA105). Trans Res. 1993;2:208–218. [Google Scholar]
- 44.Sambrook J, Russel DW. Molecular Cloning: A laboratory manual. New York: Cold Spring Harbor laboratory press; 2001. [Google Scholar]
- 45.Mattanovich D, Ruker F, Machado AC, Laimer M, Regner F, et al. Efficient transformation of Agrobacterium spp. by electroporation. Nucleic Acids Res. 1989;17:6747. doi: 10.1093/nar/17.16.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolwerk A, Lagopodi AL, Lugtenberg BJ, Bloemberg GV. Visualization of interactions between a pathogenic and a beneficial Fusarium strain during biocontrol of tomato foot and root rot. Mol Plant Microbe Interact. 2005;18:710–721. doi: 10.1094/MPMI-18-0710. [DOI] [PubMed] [Google Scholar]
- 47.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, et al. A monomeric red fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mullins ED, Chen X, Romaine P, Raina r, Geiser DM, et al. Agrobacterium-mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathol. 2001;91:173–180. doi: 10.1094/PHYTO.2001.91.2.173. [DOI] [PubMed] [Google Scholar]
- 49.Takken FL, Van Wijk R, Michielse CB, Houterman PM, Ram AF, et al. A one-step method to convert vectors into binary vectors suited for Agrobacterium-mediated transformation. Curr Genet. 2004;45:242–248. doi: 10.1007/s00294-003-0481-5. [DOI] [PubMed] [Google Scholar]
- 50.Wellman FL. A technique for studying host resistance and pathogenicity in tomato Fusarium wilt. Phytopathol. 1939;29:945–956. [Google Scholar]
- 51.Kolar M, Punt PJ, van den Hondel CA, Schwab H. Transformation of Penicillium chrysogenum using dominant selection markers and expression of an Escherichia coli lacZ fusion gene. Gene. 1988;62:127–134. doi: 10.1016/0378-1119(88)90586-0. [DOI] [PubMed] [Google Scholar]
- 52.Felix G, Grosskopf DG, Regenass M, Basse CW, Boller T. Elicitor-Induced Ethylene Biosynthesis in Tomato Cells: Characterization and Use as a Bioassay for Elicitor Action. Plant Physiol. 1991;97:19–25. doi: 10.1104/pp.97.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 54.Livak KJ. ABI Prism 7700 Sequence Detection System. 2001. User bulletin 2, Applied Biosystems.
- 55.Trouvelot S, Olivain C, Recorbet G, Migheli Q, Alabouvette C. Recovery of Fusarium oxysporum Fo47 Mutants affected in their biocontrol activity after transposition of the Fot1 element. Phytopathol. 2002;92:936–945. doi: 10.1094/PHYTO.2002.92.9.936. [DOI] [PubMed] [Google Scholar]
- 56.Correll JC, Klittich CJR, Leslie JF. Nitrate non-utilizing mutants of Fusarium oxysporum and their use in vegetative compatibility tests. Phytopathol. 1987;77:1640–1646. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in this study.
(0.01 MB PDF)
Analysis of transformants deleted for SGE1. A knock-out construct containing a hygromycin-GFP expression cassette flanked by 901 bp up- and 1032 bp downstream sequences of SGE1 was introduced in the wild type strain by Agrobacterium-mediated transformation. A) Schematic representation of the knock-out strategy for SGE1 drawn to scale. The arrow heads indicate the positions of the original T-DNA insertions. The small arrows represent the primers used to check homologous recombination (a and b) and the absence of the open reading frame (c and d). B) Verification of homologous recombination by PCR using primers a and b. C) Verification of the absence of the SGE1 open reading frame by PCR using primers c and d. The 1 kb DNA ladder of Fermentas (www.fermentas.com) is used as a marker. −, negative control. +, positive control (genomic DNA). The figures are composed from different parts of an ethidium bromide gel, which results in minor colour differences.
(0.41 MB PDF)
Analysis of transformants complemented with SGE1. A complementation construct containing a phleomycin expression cassette and the SGE1 gene including 901 bp up- and 341 bp downstream sequences was introduced in the SGE1 knock-out mutant #32 by Agrobacterium-mediated transformation. A) Schematic representation of the complementation strategy for SGE1 drawn to scale. The small arrows represent the primers used to check the presence of the open reading frame (c and d). B) Verification of the presence of the SGE1 ORF by PCR using primers c and d. The 1 kb DNA ladder of Fermentas (www.fermentas.com) is used as a marker. −, negative control.
(0.09 MB PDF)
Southern analysis of the SGE1 deletion and complementation mutants. Southern analysis was performed to verify correct homologous recombination at the SGE1 locus in the SGE1 disruptants and complemented strains. To this end, chromosomal DNA of the various mutants was digested with Acc65I, blotted and hybridized with a 432 bp probe corresponding to the SGE1 promoter. The SGE1 locus of the wild type strain is visible as a 1.6 kb fragment corresponding to the SGE1 upstream region and to the 5′ part of the SGE1 ORF. In the 5G2 insertional mutagenesis mutant, the SGE1 ORF is disrupted due to a T-DNA insertion (see Figure S1A). As a result the 1.6 kb fragment observed in a wild type situation is replaced by a 1.8 kb fragment corresponding to a part of the SGE1 promoter region and the hygromycin expression cassette present on the T-DNA. In the SGE1 disruption mutants introduction of the gene replacement cassette by homologous recombination led to the expected replacement of the 1.6 kb fragment with a fragment containing part of the SGE1 upstream region and the gene disruption cassette which should be larger than 4.9 kb. Introduction of the SGE1 complementation cassette, including the SGE1::FP fusion protein complementation cassettes, by homologous recombination restored the wild type SGE1 locus. WT, wild type. 5G2, insertional mutagenesis mutant 5G2. KO, SGE1 knock-out mutants. com, SGE1 complementation mutants. CC, SGE1::CFP complementation mutants. RC, SGE1::RFP complementation mutants.
(0.05 MB PDF)
Analysis of transformants deleted for FoPAC2. A knock-out construct containing a hygromycin expression cassette flanked by 1037 bp up- and 749 bp downstream sequences of FoPAC2 was introduced in the wild type strain by Agrobacterium-mediated transformation. A) Schematic representation of the knock-out strategy for FoPAC2 drawn to scale. The arrow head indicates the position of the original T-DNA insertion. The small arrows represents the primers used to check homologous recombination (a and b) and the absence of the open reading frame (c and d). B) Verification of the absence of the FoPAC2 open reading frame by PCR using primers c and d. C) Verification of homologous recombination by PCR using primers a and b. D) Southern analysis of the FoPAC2 deletion mutants. Chromosomal DNA was digested with BglII, blotted and hybridized with a 488 bp probe corresponding to upstream region. The FoPAC2 locus of the wild type strain is visible as a 7.0 kb fragment. In the 30C11 insertional mutagenesis mutant, the FoPAC2 ORF is disrupted due to a T-DNA insertion (see Figure S4A). As a result the 7.0 kb fragment observed in a wild type situation is replaced by a 12.3 kb fragment corresponding to a part of the FoPAC2 upstream region and the hygromycin expression cassette present on the T-DNA. In the FoPAC2 disruption mutants introduction of the gene replacement cassette by homologous recombination led to the expected replacement of the 7.0 kb fragment with a 11.4 kb fragment containing part of the FoPAC2 upstream region and the gene disruption cassette. The 1 kb DNA ladder of Fermentas (www.fermentas.com) is used as a marker. −, negative control. +, positive control (genomic DNA). Panels B and C are composed of different parts of an ethidium bromide gel, which results in minor colour differences.
(0.15 MB PDF)
The SGE1 deletion mutant is not impaired in carbon source utilization. To analyze carbon utilization of the sge1 mutant, BIOLOG FF MicroPlates containing in each well a different carbon source were used. A conidial suspension (104 conidia in 150 µl) of the wild type or the SGE1 deletion stain was inoculated in each well and incubated at 25°C. The absorbance of each well at 600 nm was measured with a microtiter plate reader (Packerd Spectra Count) after 4 days of incubation. The ratios were calculated by dividing the measured values of the mutant strain by those of the wild type strain. Only values higher than 1.5 or lower than 0.5 were marked (boxed) as carbon sources on which the sge1 mutant appeared to display a different growth rate than the wild type strain in this experiment. An additional plate assay containing these carbon sources showed that these differences were not reproducible (data not shown).
(0.40 MB TIF)
Micro- and macroconidia of the SGE1 deletion mutant are morphologically indistinguishable from wild type. Micro- and macroconidia development was analyzed in liquid carboxymethyl cellulose medium. Conidia were fixed in 0.4% p-formaldehyde and stained with Hoechst 33342 (250 µg/ml) and calcofluor white (25 µg/ml) to visualize nuclei and cell walls, respectively. The left panel depicts a phase contrast recording and the right panel depicts a UV-exposed recording of conidia of the sge1 mutant.
(1.38 MB PDF)
SGE1 is not essential for superficial root colonization. The root colonization behaviour of the SGE1 deletion mutant was determined by binocular microscopy. Nine to eleven days old tomato seedlings were inoculated with wild type or an SGE1 deletion mutant spore suspension and root colonization was determined after two to five days after inoculation. Patches of colonization were already visible after three days. The left panel depicts phase contrast recordings of a GFP-expressing virulent strain (38H3) and of the SGE1 deletion mutant. The right panel depicts UV-exposed recordings.
(0.82 MB PDF)
The SGE1 mutant is not impaired in cellophane penetration. The capacity of the SGE1 deletion strain to penetrate cellophane was determined using a cellophane penetration assay. CDA medium covered by a cellophane sheet was inoculated with a drop containing 105 conidia. After incubation of 5 days at 25°C (left panel), the cellophane was removed and after a subsequent incubation of 2 days at 25°C fungal growth of both the wild type and the SGE1 deletion mutant was clearly observed (right panel).
(0.25 MB PDF)
The sge1 mutant is impaired in extensive in planta growth. To determine whether the SGE1 deletion mutant is able to growth within xylem vessels, tomato seedlings were inoculated with the wild type, SGE1 knock-out or the SGE1 complementation strain and potted into soil according to the bioassay procedure. One week after inoculation the hypocotyl was cut in slices of several millimeters which were placed on rich (PDA) medium. F. oxysporum outgrowth was observed from the hypocotyl pieces previously inoculated with the wild type and the SGE1 complementation strain, but not from the hypocotyl pieces previously inoculated with the SGE1 knock-out mutant.
(0.26 MB PDF)
Pathogenicity is partially restored in SGE1::FP complementation strains. Nine to eleven days old tomato seedlings were inoculated with fungal spore suspensions following the root-dip inoculation method and the disease index (ranging from 0 healthy plant to 4 severely diseased plant/dead plant) was scored after three weeks. Average disease index of 20 plants three weeks after mock inoculation (H2O) or inoculation with a SGE1 deletion mutant (SGE1KO32), SGE1 (SGE1com28), SGE1::CFP (SGE1comCC4, 5, and 10) and SGE1::RFP (SGEcomRC2, 6, and 11) complementation mutants or wild type (4287). Error bars indicate standard deviation and capitals define statistically different groups (ANOVA, p = 0.95).
(0.03 MB PDF)
Analysis of transformants complemented with SGE1PM. A complementation construct containing a phleomycin expression cassette and the SGE1PM gene, encoding Sge1R66S, including 901 bp up- and 341 bp downstream sequences, was introduced in the SGE1 knock-out mutant #32 by Agrobacterium-mediated transformation. A) Verification of the presence of the SGE1 ORF by PCR using primers c and d (see Figure S1A). B) Verification of homologous recombination by PCR using primers a and b (see Figure S2). The 1 kb DNA ladder of Fermentas (www.fermentas.com) is used as a marker. −, negative control. Deletion mutant SGE1KO32 and complementation mutant SGE1com28 were used as a negative and positive control, respectively.
(0.34 MB PDF)