A combination of experimentation and modeling reveal that multiple myosin VI molecules coordinately transport cargo over the actin filament network.
Abstract
Unconventional myosins interact with the dense cortical actin network during processes such as membrane trafficking, cell migration, and mechanotransduction. Our understanding of unconventional myosin function is derived largely from assays that examine the interaction of a single myosin with a single actin filament. In this study, we have developed a model system to study the interaction between multiple tethered unconventional myosins and a model F-actin cortex, namely the lamellipodium of a migrating fish epidermal keratocyte. Using myosin VI, which moves toward the pointed end of actin filaments, we directly determine the polarity of the extracted keratocyte lamellipodium from the cell periphery to the cell nucleus. We use a combination of experimentation and simulation to demonstrate that multiple myosin VI molecules can coordinate to efficiently transport vesicle-size cargo over 10 µm of the dense interlaced actin network. Furthermore, several molecules of monomeric myosin VI, which are nonprocessive in single molecule assays, can coordinate to transport cargo with similar speeds as dimers.
Introduction
Several unconventional myosins, including myosin I, V, and VI, have been implicated in membrane traffic in the actin cortex (Eichler et al., 2006; Buss and Kendrick-Jones, 2008; Kim and Flavell, 2008), which is generally found to be polarized, with the actin filament barbed ends being attached to the plasma membrane. Their role in these processes has been inferred from loss-of-function experiments in vivo (Buss et al., 2001; Varadi et al., 2005; Krendel et al., 2007). Myosin VI has been implicated in moving vesicles from the cell surface to the interior of cells, through the cortical actin meshwork (Buss et al., 2001), which is in keeping with its ability to move toward the pointed ends of actin filaments (Wells et al., 1999).
Cortical F-actin is generally organized as a thin (< 0.5 µm) dense network below the plasma membrane (Mooseker, 1985). There are many examples of using an exposed cellular cytoskeleton for in vitro experiments (Sawada and Sheetz, 2002; Sivaramakrishnan et al., 2008a; Brawley and Rock, 2009). The choice of cell type that is used to obtain a native actin cytoskeleton is critical, depending on the type of information on myosin function one is interested in obtaining. Microdissection of Nitella axillaris to expose the polarized actin cytoskeleton was used to study movement of 1-µm-sized microspheres coated with muscle myosin II (Sheetz and Spudich, 1983). The uniform polarity of the long actin bundles in N. axillaris enabled the establishment of the first quantitative in vitro motility assay with muscle myosin speeds comparable to those seen in muscle (Sheetz and Spudich, 1983). Detergent extraction is another way to reveal and maintain an intact cytoskeleton (Svitkina et al., 1995). Brawley and Rock (2009) recently examined the movement of single dimeric forms of myosin V, VI, and X on detergent-extracted Drosophila S2 and mammalian COS-7 and U2OS cells. They found that myosin V selectively moves toward the cell periphery, that myosin VI moves toward the cell center, and that myosin X is recruited to filopodia. These cell types have an actin cortical region that extends in toward the cell interior by <1 µm. The goal of our study was to define quantitatively the behavior, on a cellular F-actin interlaced network, of nanospheres coated with multiple copies of myosin VI in both monomeric and dimeric states, and to compare and contrast them. Thus, we needed a cell type with an extensive lamellipodial actin cortex. We chose the keratocyte lamellipodium, which has been characterized using light (Schaub et al., 2007) and electron microscopy (Svitkina and Borisy, 1998). It spans ∼30 × 10 µm, providing a large surface for observing interaction of unconventional myosins. The actin polarity is thought to be uniform (Small et al., 1995), with the pointed ends of the actin filaments pointing inward toward the nucleus. This widely held hypothesis is supported by the findings of our study.
Myosin VI is necessary for timely transport of endocytic cargo such as transferrin to early endosomes (Hasson, 2003). Because actin filaments in the terminal web are polarized with their pointed ends away from the cell edge, myosin VI–based movement of early endosomes along actin filaments has been inferred to facilitate endocytosis (Mooseker and Tilney, 1975). However, individual myosin VI molecules isolated from cells are monomeric and therefore are nonprocessive (Lister et al., 2004). Indeed, several unconventional myosins, including myosin I, VI, and VII, have been shown to be nonprocessive when isolated from cells, leading to questions about whether or how they can transport vesicles/organelles in vivo (Loubéry and Coudrier, 2008). Myosin VI, although monomeric as a pure protein, binds to endosomes through adaptor proteins such as GIPC and Dab2, which can dimerize on endocytic vesicles (Lou et al., 2001; Xu et al., 1998); this suggests that myosin VI may function as a dimer in vivo. This suggestion is supported by anisotropy Förster resonance energy transfer measurements in vivo that suggest that myosin VI molecules are precisely positioned in close proximity on endosomes (Altman et al., 2007). However, whether dimerization is required for processive movement of cargo is unclear.
Complicating the issue, myosin VI and V spend most of their time bound to F-actin (high duty ratio), whereas myosin I transitions from a low to high duty ratio motor under load (De La Cruz et al., 2001; Moore et al., 2001; Laakso et al., 2008). Multiple myosin heads with high duty ratio could interfere with each other, preventing the movement of cargo bound to multiple motors along the cortical actin mesh.
In this study, we examine the coordinated interaction of multiple copies of myosin VI molecules with the isolated keratocyte actin network. We address an outstanding question in the unconventional myosin field, namely the ability of multiple myosin molecules to interact, regardless of their oligomerization state, in order to bring about the transport of cargo.
Results and discussion
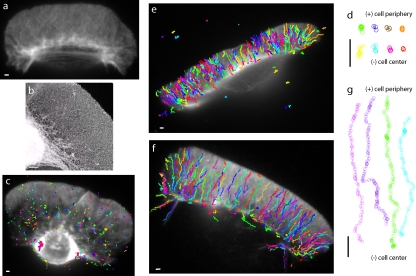
Fig. 1 a shows a fluorescence image of the keratocyte actin cytoskeleton after extraction, displaying the characteristic densely packed interlaced actin network (Schaub et al., 2007). High-resolution images obtained using the platinum replica technique (Fig. 1 b) confirm that the dense interlaced actin network after detergent extraction is maintained in the absence of fixatives. Although endocytosis may or may not occur in the keratocyte lamellipodium, this dense actin meshwork is likely to be similar to the smaller actin cortex found in other cells. To model the movement of endocytic vesicles on such a network, we use myosin VI coupled to 200-nm-diameter fluorescent nanospheres.
Figure 1.
Interaction of myosin VI artificial dimer-coated nanospheres with extracted keratocyte lamellipodial F-actin networks. (a) Fluorescence micrograph of a fish epidermal keratocyte after extraction. (b) Platinum replica electron micrograph of an extracted fish epidermal keratocyte showing the dense, interlaced actin network in the keratocyte lamellipodium. (c) Trajectories of movement of 200-nm-diameter nanospheres coated with a single myosin VI dimer on the extracted keratocyte. (d) Sample trajectories from panel c aligned such that the cell periphery is at the top and the cell center is at the bottom. (e and f) Trajectories of movement of nanospheres coated with ∼10 myosin VI dimers, such that approximately two dimers interact with the keratocyte lamellipodium. All nanospheres, without exception, move toward the cell center. A nanosphere landing near the cell edge can traverse the entire length of the keratocyte lamellipodium. (g) Sample trajectories from panel f aligned such that the cell periphery is at the top and cell center is at the bottom. Myosin VI is a pointed end–directed motor; hence, the polarity of F-actin, inferred from the observed movement, is indicated by barbed end (+) and pointed end (−) in panels d and g. Bars, 1 µm.
Nanospheres coated with a single myosin VI artificial dimer land on the keratocyte network, move a short distance (<1 µm), then detach and diffuse away (Fig. 1 c). Myosin VI artificial dimers were created by the insertion of a GCN4 leucine zipper coiled-coil motif in its tail domain (De La Cruz et al., 2001). Insertion of GCN4 ensures dimerization, as has been observed for artificial dimers in single molecule experiments (Rock et al., 2001; Okten et al., 2004). Fig. 1 d shows trajectories of these nanospheres, starting with their landing on (top) and ending with their detachment from (bottom) the keratocyte actin network. The short run lengths (<1 µm) are consistent with previous measurements (Sweeney et al., 2007; Brawley and Rock, 2009). Myosin VI heads release from the actin filament when bound to ATP. Occasionally, when both heads bind an ATP, the dimer releases from the actin and the nanosphere diffuses into solution, terminating the run.
When multiple myosin VI dimers are coupled together through a nanosphere, the nanosphere will release from the actin network only when all the heads are bound to ATP. The frequency of this depends on the rate at which both heads of a dimer are simultaneously bound to ATP (<1 s−1) relative to the rate at which the myosin transitions back to a state when it strongly binds to F-actin (∼40 s−1; Sweeney et al., 2007). Thus, coupling multiple myosin VI motors is likely to keep a myosin VI–coated nanosphere in contact with the actin network longer before it diffuses away. However, the presence of multiple dimers may also interfere with movement, a result of one dimer trying to step while others are bound with both heads to F-actin. Interference between dimers could prevent the nanosphere from moving along the network.
Therefore, we studied the movement of myosin VI–coated nanospheres with multiple dimers to resolve whether they can coordinate movement along the actin network. We found that nanospheres coated with ∼10 artificial dimers, such that ∼2 dimers are able to interact with the keratocyte actin mesh (Fig. S1), move across the entire keratocyte lamellipodium (Fig. 1, e and f). They move in an almost linear pattern, starting from the cell edge and proceeding toward the cell center (Fig. 1 g). All movement, without exception, is directed away from the cell edge toward the cell center (Fig. 1 g and Video 1, which is a sample video of Fig. 1 f).
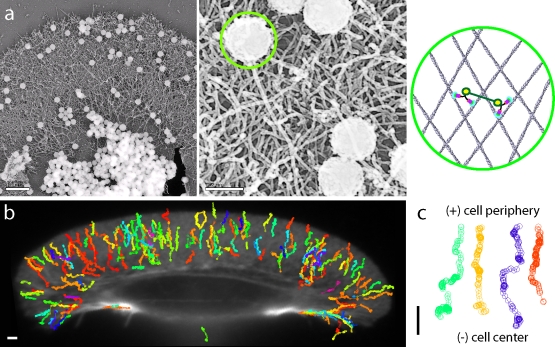
Platinum replica electron micrographs of the keratocyte lamellipodium acquired after movement of the nanospheres with ∼10 dimers do not indicate reorganization of or damage to the actin cytoskeleton (Fig. 2 a). This suggests that the movement is brought about by the cooperative interaction between multiple dimers and the actin network and not by reorganization or reorientation of the lamellipodium, as could be possible with multiple myosin heads. This finding was confirmed by cross-linking the actin cytoskeleton using a fixative (2% formaldehyde for 10 min at 22°C) before its interaction with the dimer-coated nanospheres (Fig. 2, b and c). Note that on occasion, a nanosphere coated with myosin VI lands on a retraction fiber at the rear end of the cell (green trace at the bottom of Fig. 2 b) and moves toward the nucleus.
Figure 2.
Myosin VI–coated nanosphere movement on extracted keratocyte actin networks does not result in or require actin filament reorganization. (a, left) Platinum replica micrograph of an extracted keratocyte prepared after movement of myosin VI artificial dimer-coated nanospheres on its surface. (a, middle) A higher resolution image showing the size of nanospheres relative to the F-actin network size. (a, right) A schematic diagram depicting the relative sizes of a nanosphere (highlighted by the green circle in the center and right panels), the myosin VI dimers, and the keratocyte actin network. The right panel shows two dimers bound to actin filaments, coupled through the nanosphere (green line), and bound to the nanosphere at the yellow circles. (b) Trajectories of movement of myosin VI dimer-coated nanospheres on a keratocyte extracted and fixed with 2% formaldehyde for 10 min. (c) Sample trajectories from panel b without exception show movement toward the cell center. Bars, 1 µm.
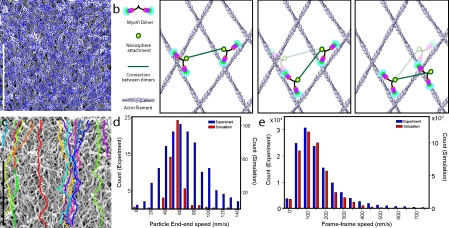
To understand how multiple motors coordinate to bring about linear directed motion of nanospheres, we used our detailed knowledge of myosin VI artificial dimer function, obtained from single molecule and kinetic studies, to simulate their movement on the keratocyte actin mesh. To capture the interlaced nature of the keratocyte lamellipodium, we digitized platinum replica electron micrographs of extracted keratocytes to obtain the actin tracks encountered by the myosin motors (Fig. 3 a). We created a simplified model for movement of nanospheres brought about by two myosin VI dimers (Fig. 3 b) using the following steps: (1) each myosin VI dimer steps stochastically at a rate limited by the release of ADP from its rear head (5 s−1; De La Cruz et al., 2001); (2) each myosin VI dimer takes 36 ± 4-nm steps (Rock et al., 2001); (3) the next binding site of the lead head of the stepping dimer is constrained such that the distance between the points where myosin VI attaches to the nanospheres (yellow circles in Fig. 3 b) is conserved; (4) the nanosphere constrains myosin movement such that more than one step by a dimer, before the movement of its counterpart, is not productive; and (5) the rear head of a dimer always releases before the lead head.
Figure 3.
Movement of myosin VI artificial dimer-coated nanospheres on extracted keratocyte actin network assessed from experimentation and simulation. (a) A digitized interlaced actin network used in the simulation (blue) overlaid on the platinum replica micrograph from which it was obtained. (b) A schematic drawing of the simplified model used for the simulation. (b, left) Two dimers bound to actin filaments, coupled through the nanosphere (green line), and bound to the nanosphere at the yellow circle. (b, center) The dimer on the left steps first to its next binding site. (b, right) The dimer on the right steps to the next binding site. Binding sites are selected to maintain the distance between the nanosphere attachment points (length of the green line). (c) Sample trajectories of simulated movement of nanospheres on the extracted keratocyte lamellipodium. (d) Comparison of end–end speeds from the experiment (blue) and simulation (red). (e) Comparison of frame–frame speeds between the experiment (blue) and simulation (red). Bars, 1 µm.
Our simplified simulation shows that coordinated movement of two myosin dimers on the keratocyte mesh is indeed possible and results in linear directed motion toward the cell center (Fig. 3 c). This model also reproduces the quality of the tracks seen by myosin VI dimer-coated nanospheres seen in Fig. 1 (e–g), and the end–end speed of the nanospheres (ratio of distance between the ends of the trajectory to the time for covering that distance; Fig. 3 d). The end–end speed includes the linearity of the movement path, kinetics of the individual myosins, their step sizes, and their coordination, therefore suggesting that our simplified model captures the essential features of the myosin VI–nanosphere–network interaction.
In addition to the end–end speed, our model accurately captures the frame–frame speed (the ratio of the distance moved between two successive image acquisition frames to the time between frames acquired during imaging on an epifluorescent microscope) distribution observed in the simulation (Fig. 3 e). Occasional release of the lead head before release of the trailing head of a dimer results in nonproductive steps that slow down the nanosphere. However, occasional multiple steps by a single dimer increase the mean speed of the nanosphere. Effects such as these, which are not modeled in our simulation, likely explain the differences in the width of the end–end speed distributions (Fig. 3 d). Despite the simplifying assumptions, our model verifies that the underlying coordinated movement of the two myosin dimer molecules enables the nanospheres to traverse the entire actin network.
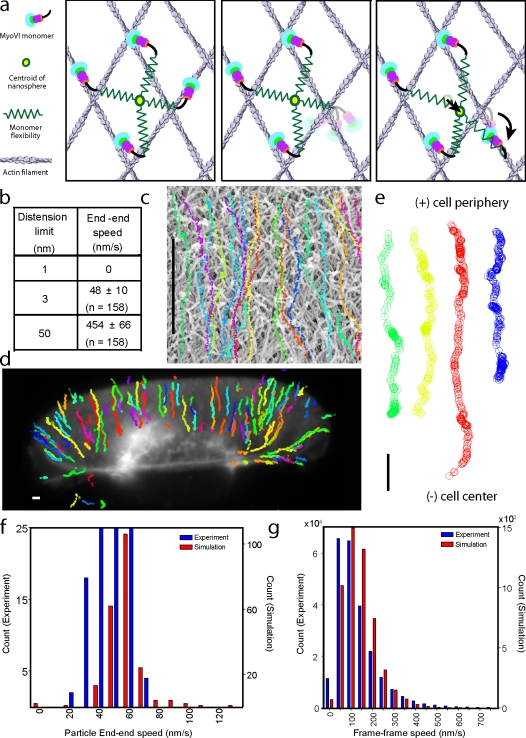
A similar simulation was used to predict the movement of a nanosphere with four bound myosin VI monomers across an interlaced actin network (Fig. 4 a). Each monomer head releases stochastically from the actin network at a rate of 5 s−1. After its release, it undergoes ATP hydrolysis and a search to find the next actin-binding site at random, although the structure of the head and the polarity of the actin filaments lead to heads binding forward in the pointed-end direction (Fig. 4 a, middle). Upon binding to the actin filament, it strokes and attempts to move the nanosphere forward (Fig. 4 a, right). We modeled the flexibility in the myosin molecule, between the nanosphere and the actin attachments, as an elastic spring that allows finite linear distension. The flexibility in the myosin molecule refers to the effective spring constant of all compliant elements in it. The compliant elements are modeled as linear Hookean springs (fixed spring constant) for small distensions with a transition to a very high spring constant for deformations beyond this finite linear distension. This model predicts two extremes, with very little distension (∼1 nm) resulting in an unproductive stroke of the myosin and hence no movement of the nanosphere (Fig. 4 b). The second is for very large distensions of the springs (∼50 nm) comparable to the size of the nanosphere. Under these conditions, the elastic energy of each myosin VI stroke is stored in the system of springs. The full stroke from each of the four motors contributes to movement of the nanosphere, though with a cumulative dwell time that is considerably less than four times the dwell time of each motor. The net effect is a very large simulated speed (∼450 nm/s), which exceeds the speed of a single myosin VI monomer (150 nm/s). We estimate a finite flexibility of ∼3 nm for the myosin VI monomer (Sivaramakrishnan et al., 2008b). This estimation results in a speed of ∼50 nm/s for the monomer-coated nanosphere. The simulated trajectories of the monomer-coated nanosphere, similar to the dimer, are linear and directed toward the nucleus (Fig. 4 c).
Figure 4.
Movement of myosin VI monomer-coated nanospheres on extracted keratocyte actin networks assessed from the experiment and simulation. (a) A schematic of the simplified model used. (a, left) Four monomers are coupled through the nanospheres, all bound to the actin filaments (the myosin VI flexibility is depicted as a spring and the centroid of the nanosphere is shown as a yellow circle). (a, middle) The monomer on the right releases from actin upon binding to ATP and searches for the next binding site. (a, right) The monomer binds at random to an actin filament, and then strokes (curved arrow). The stroke causes a distension of all monomers (depicted as distension of connecting springs) along with movement of the centroid (depicted by movement of the yellow circle). The extent of movement is determined by the balance of force between the four connected monomers. (b) Simulated end–end speed for limited (1 nm), moderate (3 nm), and very large (50 nm) distensions permitted. (c) Simulated trajectories of monomer-coated nanosphere motion for moderate distensions. (d) Experimental trajectories of myosin VI monomer-coated nanospheres moving along an extracted keratocyte. (e) Sample trajectories from panel d aligned such that the cell periphery is at the top and the cell center is at the bottom. All movement, without exception, is toward the cell center. (f) Comparison of end–end speeds from the experiment (blue) and simulation (red). (g) Comparison of frame–frame speeds between the experiment (blue) and simulation (red). Bars, 1 µm.
These model predictions were verified by experiments using the myosin VI monomer (Fig. 4, d and e). The distribution of speeds from the simulation overlaps well with that observed in the experiment (Fig. 4, f and g). Our simulation does not account for changes in kinetics of monomers when restrained by the finite distensibility of bound heads, which likely reduce the nanosphere speed and lead to broadening of the distribution, as observed in the experiment (Fig. 4 f). The end–end speeds of dimer (65 ± 30 nm/s; n = 156)- and monomer (47 ± 10 nm/s; n = 157)-coated nanospheres are similar. Furthermore, the movement trajectories for dimer and monomer are qualitatively similar (Fig. 1, e–g compared with Fig. 3, d and e). Thus, we infer that dimerization, which gates the two heads of an unconventional myosin and renders a single dimer processive in a single-molecule assay, is not essential for processive movement so long as multiple motor heads simultaneously interact with the actin network.
To gain insight into the effect of motor density on movement of vesicular cargo, we examined the interaction of nanospheres with ∼100 dimers or ∼200 monomers on the actin network. Simulation and experimentation both confirm that nanospheres with large numbers of dimers or monomers are not stalled, but instead creep along at speeds of ∼2-3 nm/s. Thus, although there appears to be no limit to motor density for vesicle movement, large numbers of motors do impede efficiency of cargo transport.
In summary, we used myosin VI to directly test the widely proposed hypothesis that the keratocyte lamellipodium is uniformly polarized, with barbed ends pointed toward the leading edge of the cell. Our study demonstrates that the extracted keratocyte lamellipodium is a model polarized interlaced actin network with a large area (∼10 × 30 µm) to study the coupled function of unconventional myosins. We find that multiple myosin VI molecules, either artificial dimers or monomers, can cooperate to move endosome-sized nanospheres over the entire length of this model F-actin mesh. Using simplified simulations, we show that the known stroke size, duty ratio, and kinetics of myosin VI, combined with a digitized model of the keratocyte lamellipodium, is sufficient to explain the observed movement of nanospheres coupled to myosin molecules. In the context of multiple myosin VI motors interacting with multiple actin filaments in vivo, we find that dimerization of the myosin VI is not essential for processive movement of cargo vesicles under the low load conditions of the assay. The effect of external load on the movement of the dimeric myosin VI–bound nanospheres compared with the monomeric myosin VI–bound nanospheres will be an important future study. This model system will enable the characterization of the function of unconventional myosins, both in their purified form, as in this study, as well as isolated from cells, bound to their native cargo.
Materials and methods
Keratocyte preparation
Keratocytes were isolated from the scales of the Central American cichlid Hypsophrys nicaraguensis (Cooper and Schliwa, 1986; Kolega, 1986). Keratocytes were washed gently with 85% PBS solution before treatment with extraction buffer (50 mM imidazole, pH 6.8, 50 mM KCl, 0.5 mM MgCl2, 0.1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 4% polyethylene glycol mol wt 40,000, and 250 nM tetramethylrhodamine [TMR]-phalloidin [Sigma-Aldrich]) for 3–4 min. Extracted cells were incubated with 85% PBS with 250 nM TMR-phalloidin in the dark for 30–45 min before use in motility assays.
Platinum replica electron microscopy
Platinum replicas of the extracted cells were prepared as previously described (Svitkina et al., 1995). In brief, cells were fixed before or after movement of nanospheres with 2.5% glutaraldehyde in 100 mM sodium cacodylate, pH 7.3, followed by sequential treatment with 0.1% aqueous tannic acid and 0.2% uranyl acetate. Cells were prepared for critical point drying by graded washes into 100% ethanol. Critical point drying was performed in a Samdri critical point dryer (Tousimis Research Corporation). Dried specimens were shadowed with platinum and carbon before transfer to formvar-coated grids for viewing on a transmission electron microscope (JEOL 1230; JEOL Ltd.). Images were captured by a cooled charge-coupled device camera (Gatan 967; Gatan).
Myosin VI: constructs and protein preparation
Artificial myosin VI dimer was prepared from porcine myosin VI cDNA truncated at Arg 992 followed by a leucine zipper (GCN4) to induce artificial dimerization (Trybus et al., 1997; Altman et al., 2004). Myosin VI monomer was prepared from porcine myosin VI cDNA truncated at Arg 981. Both sequences have C-terminal monomeric YFP to facilitate protein quantification and a biotin carboxy carrier protein (bccp) sequence, at which a biotin molecule is attached in Sf9 cells (Tyska et al., 1999). Protein was purified using FLAG resin (Sweeney et al., 1998; Bryant et al., 2007). Biotinylation was verified by a Neutravidin gel shift assay (Fig. S1 b). Protein concentration was determined using a Nanodrop at 514 nm (Thermo Fisher Scientific).
Nanosphere preparation
200-nm-diameter, yellow-green, polystyrene nanospheres (Polysciences, Inc.) were coated with NeutrAvidin (Invitrogen) by passive adsorption. Nanospheres were washed with 50 mM Tris, pH 8.0, and 1 mg/ml BSA (Sigma-Aldrich) using a tabletop centrifuge (17,000 g, 1 min). After washing, the nanospheres were bath sonicated (Branson Ultrasonics Sonifier; Emerson Industrial Automation). Monodispersity was verified using an epifluorescence microscope (100× magnification; Nikon). Concentration was determined using a Nanodrop at 400 nm (Fig. S1 c).
Coating nanospheres with myosin VI
A known number of myosin VI monomer/dimer molecules were mixed with nanospheres and incubated at 22°C for 20–30 min. We used nanospheres coated with either 20 monomers or 10 dimers to compare movement with the same approximately four heads interacting with the keratocyte (Fig. S1 d). To obtain nanospheres coated with a single dimer, nanosphere/myosin VI dimer concentrations were adjusted so there were 10 nanospheres for each dimer.
Myosin VI: keratocyte interaction
Myosin VI–coated nanospheres were mixed into motility buffer (25 mM imidazole HCl, pH 7.4, 25 mM KCl, 4.5 µM calmodulin, 1 mM EGTA, 10 mM DTT, 4 mM MgCl2, an oxygen-scavenging system to retard photo damage [25 µg/ml glucose oxidase, 45 µg/ml catalase, 31 mM β-mercaptoethanol, and 0.5% glucose], an ATP regeneration system [0.1 mg/ml creatine phosphokinase and 1 mM creatine phosphate], 2 mM ATP, and 1 mg/ml BSA) and transferred to extracted keratocytes. Nanospheres were imaged at 10 Hz on an upright epifluorescence microsphere (Nikon) with a Coolsnap HQ camera (Photometrics).
Image analysis of moving myosin VI nanospheres
A custom Matlab particle-tracking program (The MathWorks, Inc.) was used to identify and track the movement of individual nanospheres. Nanosphere centers were found by least squares fit to a 2D elliptical Gaussian function with a background (Churchman et al., 2005). To compensate for stage drift, fiducial (stuck to the coverslip surface outside the keratocyte) nanospheres were tracked and their movement was subtracted.
Simulation of nanosphere movement
Platinum replica images were digitized to obtain filament tracks using a custom Matlab program. Rotary shadowing with platinum/carbon results in progressively smaller quantities of platinum/carbon coating away from the apical surface of the keratocyte lamellipodium. This is reflected in actin filament darkness in the transmission electron microscope. Myosin VI molecules have a limited reach (∼20 nm), hence we preferentially selected filaments, in the images, in the first 2–3 layers of the lamellipodium (∼30 nm; Fig. 3 a). Nanosphere movement was simulated as described in the text (see the following section for details).
Nanosphere simulation algorithm
Dimer.
Movement of each dimer is considered stochastic, with a single exponential dwell time distribution having a mean of 0.2 s (derived from kinetic measurements; De La Cruz et al., 2001). Based on the geometry of the nanosphere (see Fig. S1), ∼2 out of 10 myosin VI dimers coupled to a nanosphere interact with the actin network. Two independent stochastic dwell time sequences for the two dimers were created, and the order of movement was determined based on the dwell times of successive events. Distance between the dimers at the start of the simulation varies randomly from 10 to 100 nm depending on the location of actin filaments at the top of the image.
Movement of a dimer consisted of the following steps. (1) The trailing head (determined by relative “y” location) releases after a specified dwell time and samples a new binding site 32 ± 4 nm below the leading head, assuming uniform polarity from the cell periphery to the cell center. (2) A new binding site is selected such that the distance between the nanospheres attachment point (the attachment point is the midpoint between the two heads of a dimer) of the moving dimer and its coupled partner is maintained relative to its initial value at the start of the simulation. (3) Centroid of the nanosphere (halfway between the attachment points of the two myosin VI dimers) is updated. Release, stroke, and rebinding of the moving dimer are considered instantaneous, which is consistent with the high duty ratio of this motor (>0.8; De La Cruz et al., 2001).
Upon completing its dwell time, the second dimer now moves as per the first three steps outlined above. We assume that in events when a dimer undergoes two or more steps while the other remains bound, only the first step is successful and this dimer has to wait for its partner to step. This is consistent with the finite flexibility of the coupling between the two dimers and the large step size of myosin VI (∼36 nm). Speed statistics from the simulation are based on movement of the centroid of the nanosphere, as observed under the microscope. We simulate the movement of ∼20 nanospheres, thrice, over five separate digitized micrographs of the platinum replica network.
Monomer.
Similar to the dimer simulation, monomer movement is considered stochastic, with a single exponential dwell time distribution with a mean of 0.2 s. Based on the geometry of the nanospheres (Fig. S1), ∼4 out of the 20 monomers coupled to the nanosphere are properly positioned to interact with the F-actin network. Four independent stochastic dwell time sequences were created, and the order of movements was determined based on the dwell times of successive events. The distance between the monomers at the start of the simulation varies randomly, with a minimum separation of 10 nm and a maximum separation of 100 nm for a group of four monomers. The separation distances are based on the area of the nanosphere that can access the keratocyte surface actin network.
Monomer movement consisted of the following steps. (1) The head detaches from the actin filament. Strain between the remaining myosins is now redistributed such that the forces between the remaining heads are balanced. The flexibility (effective spring constant keff) of each head is the same, hence the value of keff is not required for calculating the force balance. (2) The head rebinds below the nanosphere attachment point, assuming uniform F-actin polarity from the cell periphery to the cell center. (3) The head attempts to stroke through 30 ± 4 nm. The flexibility of all heads is identical, hence a force balance between all heads is used to calculate the net movement of each nanosphere attachment point and hence the movement of the bead centroid. (4) The strain in each head is monitored throughout the simulation. If the strain exceeds the permissible distension (variable, see Fig. 4 b), stiffness of that particular head–nanosphere attachment connection is set to infinity, and the strains in the system are calculated accordingly. This is consistent with the finite compliance in any head–nanosphere attachment being ironed out, leading to its nonlinear behavior. (5) After its stroke, the myosin head completes its subsequent dwell before releasing and continuing its stroke. (6) Unlike the two dimer case discussed above, where we assume steps between the two dimers are sequential, no explicit assumption of the order of strokes is made for the four monomers. The strain buildup in the system automatically adjusts the movement of successive strokes, either by the same myosin head stroking in succession or one head stroking after another. (7) To simplify our model, the only parameter we have introduced is the permissible distension in the heads. A stroking head that encounters a nonlinear distension of another head is assumed to complete a partial stroke and then release from the actin filament with a dwell time identical to what would occur in the absence of the load. This simplifying assumption, as stated in the text, causes differences between simulation and experimentation. However, in the absence of information on the kinetics of motors under these conditions, we have chosen no changes in kinetics as a first approximation.
After this monomer stroke, the next monomer releases, rebinds, and strokes in exactly the same sequence, and the movement is continued. The centroid of a nanosphere is the centroid of the attachment points of the four monomers. Speed statistics from the simulation are based on movement of the centroid of the nanospheres, as observed under the microscope. We simulate the movement of 35–40 nanospheres over five separate digitized micrographs of the platinum replica network.
Quantifying the number of myosin-binding sites per nanosphere
Nanospheres were prepared as described in Nanosphere preparation. Nanosphere preparations were mixed with increasing amounts of d-biotin (unlabeled) for 20 min at 20°C. Subsequently, they were incubated with a constant amount of biotin-4-fluorescein (B4F; Sigma-Aldrich). Nanospheres were pelleted by centrifugation (35,000 g, 15 min), and the fluorescence in the supernatant was quantified using a Fluorolog fluorimeter (Horiba Scientific). With increasing d-biotin in solution, fewer B4F molecules bind to the nanosphere, and hence B4F fluorescence in the supernatant increases with increasing d-biotin addition. When all of the binding sites on the nanosphere are saturated with d-biotin, B4F fluorescence is unaffected with further d-biotin addition. This can be used to compute the amount of d-biotin that can bind to the nanospheres and hence the number of biotin-binding sites per nanosphere. We estimated the number of biotin-binding sites per nanosphere to be 13,600. From this, assuming there is no cooperative binding of biotin to neutravidin, we estimated that only one in 1,000 monomers are bound to the same neutravidin molecule on the surface of the nanosphere.
Fluorescent images showing the actin filament organization in the extracted keratocyte lamellipodium were visualized using TMR phalloidin and taken on a microscope (Eclipse 80i) with a Plan-Apochromat VC 60×, 1.40 NA oil immersion objective (Nikon) at 20°C. The imaging medium was 25 mM imidazole HCl, pH 7.4, 25 mM KCl, 4.5 µM calmodulin, 1 mM EGTA, 10 mM DTT, 4 mM MgCl2, an oxygen-scavenging system to retard photo damage [25 µg/ml glucose oxidase, 45 µg/ml catalase, 31 mM β-mercaptoethanol, and 0.5% glucose], and an ATP regeneration system [0.1 mg/ml creatine phosphokinase and 1 mM creatine phosphate], 2 mM ATP, and 1 mg/ml BSA. Images were acquired using a CoolSnap HQ camera (Photometrics) with MetaMorph software (MDS Analytical Technologies). Image intensity levels were adjusted linearly, using the Matlab image processing toolbox in order to make the maximum display intensity equal to the maximum intensity in the image. Scale bars are included in all images and their lengths are indicated in the figure legends.
Transmission electron micrographs of platinum replica specimens of extracted keratocytes showing the organization of the actin filaments in the extracted keratocyte lamellipodium were taken with a transmission electron microscope (TEM1230; JEOL Ltd.). Different magnification settings were used and are indicated using the image scale bar. Images were acquired at 20°C. Samples were coated with platinum and carbon for contrast. Images were acquired on a Gatan 967 slow-scan, cooled charge-coupled device camera with Gatan software. Image intensity levels were adjusted linearly, using Photoshop software (Adobe), to make the make the maximum display intensity equal to the maximum intensity in the image.
Online supplemental material
Fig. S1 shows preparation of the myosin VI–coated nanospheres and estimation of the number of myosins interacting with the extracted keratocyte actin network. Video 1 shows a small sample of movement, seen in detail in Fig. 1 f, of myosin VI–coated nanospheres on an extracted keratocyte actin network. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200906133/DC1.
Acknowledgments
We thank G. Allen, M. Tsuchida, and J. Theriot for help with keratocyte isolation and A. Dunn, M. Elting, and R. Sommese for useful discussions.
S. Sivaramakrishnan was supported by an American Cancer Society postdoctoral fellowship and J.A. Spudich was supported by grant GM33289 (from the National Institutes of Health).
Footnotes
Abbreviation used in this paper:
- TMR
- tetramethylrhodamine
References
- Altman D., Sweeney H.L., Spudich J.A. 2004. The mechanism of myosin VI translocation and its load-induced anchoring.Cell. 116:737–749 doi:10.1016/S0092-8674(04)00211-9 [DOI] [PubMed] [Google Scholar]
- Altman D., Goswami D., Hasson T., Spudich J.A., Mayor S. 2007. Precise positioning of myosin VI on endocytic vesicles in vivo.PLoS Biol. 5:e210 doi:10.1371/journal.pbio.0050210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawley C.M., Rock R.S. 2009. Unconventional myosin traffic in cells reveals a selective actin cytoskeleton.Proc. Natl. Acad. Sci. USA. 106:9685–9690 doi:10.1073/pnas.0810451106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant Z., Altman D., Spudich J.A. 2007. The power stroke of myosin VI and the basis of reverse directionality.Proc. Natl. Acad. Sci. USA. 104:772–777 doi:10.1073/pnas.0610144104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss F., Kendrick-Jones J. 2008. How are the cellular functions of myosin VI regulated within the cell? Biochem. Biophys. Res. Commun. 369:165–175 doi:10.1016/j.bbrc.2007.11.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss F., Arden S.D., Lindsay M., Luzio J.P., Kendrick-Jones J. 2001. Myosin VI isoform localized to clathrin-coated vesicles with a role in clathrin-mediated endocytosis.EMBO J. 20:3676–3684 doi:10.1093/emboj/20.14.3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman L.S., Okten Z., Rock R.S., Dawson J.F., Spudich J.A. 2005. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time.Proc. Natl. Acad. Sci. USA. 102:1419–1423 doi:10.1073/pnas.0409487102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M.S., Schliwa M. 1986. Motility of cultured fish epidermal cells in the presence and absence of direct current electric fields.J. Cell Biol. 102:1384–1399 doi:10.1083/jcb.102.4.1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz E.M., Ostap E.M., Sweeney H.L. 2001. Kinetic mechanism and regulation of myosin VI.J. Biol. Chem. 276:32373–32381 doi:10.1074/jbc.M104136200 [DOI] [PubMed] [Google Scholar]
- Eichler T.W., Kögel T., Bukoreshtliev N.V., Gerdes H.H. 2006. The role of myosin Va in secretory granule trafficking and exocytosis.Biochem. Soc. Trans. 34:671–674 doi:10.1042/BST0340671 [DOI] [PubMed] [Google Scholar]
- Hasson T. 2003. Myosin VI: two distinct roles in endocytosis.J. Cell Sci. 116:3453–3461 doi:10.1242/jcs.00669 [DOI] [PubMed] [Google Scholar]
- Kim S.V., Flavell R.A. 2008. Myosin I: from yeast to human.Cell. Mol. Life Sci. 65:2128–2137 doi:10.1007/s00018-008-7435-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolega J. 1986. Effects of mechanical tension on protrusive activity and microfilament and intermediate filament organization in an epidermal epithelium moving in culture.J. Cell Biol. 102:1400–1411 doi:10.1083/jcb.102.4.1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krendel M., Osterweil E.K., Mooseker M.S. 2007. Myosin 1E interacts with synaptojanin-1 and dynamin and is involved in endocytosis.FEBS Lett. 581:644–650 doi:10.1016/j.febslet.2007.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laakso J.M., Lewis J.H., Shuman H., Ostap E.M. 2008. Myosin I can act as a molecular force sensor.Science. 321:133–136 doi:10.1126/science.1159419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister I., Schmitz S., Walker M., Trinick J., Buss F., Veigel C., Kendrick-Jones J. 2004. A monomeric myosin VI with a large working stroke.EMBO J. 23:1729–1738 doi:10.1038/sj.emboj.7600180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou X., Yano H., Lee F., Chao M.V., Farquhar M.G. 2001. GIPC and GAIP form a complex with TrkA: a putative link between G protein and receptor tyrosine kinase pathways.Mol. Biol. Cell. 12:615–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loubéry S., Coudrier E. 2008. Myosins in the secretory pathway: tethers or transporters? Cell. Mol. Life Sci. 65:2790–2800 doi:10.1007/s00018-008-8350-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménétrey J., Llinas P., Mukherjea M., Sweeney H.L., Houdusse A. 2007. The structural basis for the large powerstroke of myosin VI.Cell. 131:300–308 doi:10.1016/j.cell.2007.08.027 [DOI] [PubMed] [Google Scholar]
- Moore J.R., Krementsova E.B., Trybus K.M., Warshaw D.M. 2001. Myosin V exhibits a high duty cycle and large unitary displacement.J. Cell Biol. 155:625–635 doi:10.1083/jcb.200103128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooseker M.S. 1985. Organization, chemistry, and assembly of the cytoskeletal apparatus of the intestinal brush border.Annu. Rev. Cell Biol. 1:209–241 doi:10.1146/annurev.cb.01.110185.001233 [DOI] [PubMed] [Google Scholar]
- Mooseker M.S., Tilney L.G. 1975. Organization of an actin filament-membrane complex. Filament polarity and membrane attachment in the microvilli of intestinal epithelial cells.J. Cell Biol. 67:725–743 doi:10.1083/jcb.67.3.725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okten Z., Churchman L.S., Rock R.S., Spudich J.A. 2004. Myosin VI walks hand-over-hand along actin.Nat. Struct. Mol. Biol. 11:884–887 doi:10.1038/nsmb815 [DOI] [PubMed] [Google Scholar]
- Rock R.S., Rice S.E., Wells A.L., Purcell T.J., Spudich J.A., Sweeney H.L. 2001. Myosin VI is a processive motor with a large step size.Proc. Natl. Acad. Sci. USA. 98:13655–13659 doi:10.1073/pnas.191512398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y., Sheetz M.P. 2002. Force transduction by Triton cytoskeletons.J. Cell Biol. 156:609–615 doi:10.1083/jcb.200110068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub S., Meister J.J., Verkhovsky A.B. 2007. Analysis of actin filament network organization in lamellipodia by comparing experimental and simulated images.J. Cell Sci. 120:1491–1500 doi:10.1242/jcs.03379 [DOI] [PubMed] [Google Scholar]
- Sheetz M.P., Spudich J.A. 1983. Movement of myosin-coated fluorescent beads on actin cables in vitro.Nature. 303:31–35 doi:10.1038/303031a0 [DOI] [PubMed] [Google Scholar]
- Sivaramakrishnan S., DeGiulio J.V., Lorand L., Goldman R.D., Ridge K.M. 2008a. Micromechanical properties of keratin intermediate filament networks.Proc. Natl. Acad. Sci. USA. 105:889–894 doi:10.1073/pnas.0710728105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaramakrishnan S., Spink B.J., Sim A.Y., Doniach S., Spudich J.A. 2008b. Dynamic charge interactions create surprising rigidity in the ER/K alpha-helical protein motif.Proc. Natl. Acad. Sci. USA. 105:13356–13361 doi:10.1073/pnas.0806256105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small J.V., Herzog M., Anderson K. 1995. Actin filament organization in the fish keratocyte lamellipodium.J. Cell Biol. 129:1275–1286 doi:10.1083/jcb.129.5.1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spink B.J., Sivaramakrishnan S., Lipfert J., Doniach S., Spudich J.A. 2008. Long single alpha-helical tail domains bridge the gap between structure and function of myosin VI.Nat. Struct. Mol. Biol. 15:591–597 doi:10.1038/nsmb.1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina T.M., Borisy G.G. 1998. Correlative light and electron microscopy of the cytoskeleton of cultured cells.Methods Enzymol. 298:570–592 doi:10.1016/S0076-6879(98)98045-4 [DOI] [PubMed] [Google Scholar]
- Svitkina T.M., Verkhovsky A.B., Borisy G.G. 1995. Improved procedures for electron microscopic visualization of the cytoskeleton of cultured cells.J. Struct. Biol. 115:290–303 doi:10.1006/jsbi.1995.1054 [DOI] [PubMed] [Google Scholar]
- Sweeney H.L., Rosenfeld S.S., Brown F., Faust L., Smith J., Xing J., Stein L.A., Sellers J.R. 1998. Kinetic tuning of myosin via a flexible loop adjacent to the nucleotide binding pocket.J. Biol. Chem. 273:6262–6270 doi:10.1074/jbc.273.11.6262 [DOI] [PubMed] [Google Scholar]
- Sweeney H.L., Park H., Zong A.B., Yang Z., Selvin P.R., Rosenfeld S.S. 2007. How myosin VI coordinates its heads during processive movement.EMBO J. 26:2682–2692 doi:10.1038/sj.emboj.7601720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trybus K.M., Freyzon Y., Faust L.Z., Sweeney H.L. 1997. Spare the rod, spoil the regulation: necessity for a myosin rod.Proc. Natl. Acad. Sci. USA. 94:48–52 doi:10.1073/pnas.94.1.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska M.J., Dupuis D.E., Guilford W.H., Patlak J.B., Waller G.S., Trybus K.M., Warshaw D.M., Lowey S. 1999. Two heads of myosin are better than one for generating force and motion.Proc. Natl. Acad. Sci. USA. 96:4402–4407 doi:10.1073/pnas.96.8.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi A., Tsuboi T., Rutter G.A. 2005. Myosin Va transports dense core secretory vesicles in pancreatic MIN6 beta-cells.Mol. Biol. Cell. 16:2670–2680 doi:10.1091/mbc.E04-11-1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells A.L., Lin A.W., Chen L.Q., Safer D., Cain S.M., Hasson T., Carragher B.O., Milligan R.A., Sweeney H.L. 1999. Myosin VI is an actin-based motor that moves backwards.Nature. 401:505–508 doi:10.1038/46835 [DOI] [PubMed] [Google Scholar]
- Xu X.X., Yi T., Tang B., Lambeth J.D. 1998. Disabled-2 (Dab2) is an SH3 domain-binding partner of Grb2.Oncogene. 16:1561–1569 doi:10.1038/sj.onc.1201678 [DOI] [PubMed] [Google Scholar]