Deletion of S phase disrupts mitotic timing in maternally regulated cycles, but it doesn't alter the cell cycle once zygotic transcription has begun.
Abstract
We examined the contribution of S phase in timing cell cycle progression during Drosophila embryogenesis using an approach that deletes S phase rather than arresting its progress. Injection of Drosophila Geminin, an inhibitor of replication licensing, prevented subsequent replication so that the following mitosis occurred with uninemic chromosomes, which failed to align. The effect of S phase deletion on interphase length changed with development. During the maternally regulated syncytial blastoderm cycles, deleting S phase shortened interphase, and deletion of the last of blastoderm S phase (cycle 14) induced an extra synchronous division and temporarily deferred mid-blastula transition (MBT) events. In contrast, deleting S phase after the MBT in cycle 15 did not dramatically affect mitotic timing, which appears to retain its dependence on developmentally programmed zygotic transcription. We conclude that normal S phase and replication checkpoint activities are important timers of the undisturbed cell cycle before, but not after, the MBT.
Introduction
The first 13 nuclear divisions in the Drosophila embryo have a streamlined cell cycle in which nuclei synchronously oscillate between S phase and mitosis independent of zygotic transcription (Foe et al., 1993). Maternal RNAs and proteins deposited during oogenesis guide cell cycle progression. Cycles 1–9 occur deep in a common cytoplasm and are of equal duration. At the close of cycle 9, nuclei reach the embryo surface forming a blastoderm, and undergo 4 additional mitotic cycles where interphase (and S phase) gets progressively longer. At cycle 14 bulk zygotic transcription begins, many maternal products are degraded, nuclei are cellularized, and S phase significantly lengthens (Edgar and Schubiger, 1986; Edgar et al., 1986). These events in cycle 14 are collectively known as the mid-blastula transition (MBT).
The MBT is accompanied by changes in cell cycle regulation (O'Farrell, 2001). In cycle 14, maternal Cdc25 phosphatase is degraded and cells arrest in the first G2 due to inhibitory phosphorylation of cyclin-dependent kinase 1 (Cdk1) (Edgar and O'Farrell, 1989; Edgar et al., 1994b). Patterned zygotic transcription of Cdc25string during G2 in cycle 14 relieves Cdk1 inhibition and directs mitosis in a matching pattern (Edgar and O'Farrell, 1990). In contrast, Cdc25 is abundant during the maternally driven cycles and there is little inhibitory phosphorylation of Cdk1 (Edgar et al., 1994b; Stumpff et al., 2004). It has been suggested that accumulation of cyclin to some critical threshold times Cdk1 activation (Murray and Kirschner, 1989; Edgar et al., 1994b; Stiffler et al., 1999; Ji et al., 2004; Crest et al., 2007). However, the bulk levels of mitotic cyclin exhibit little oscillation during the Drosophila maternally driven cycles (Edgar et al., 1994b), and recent experiments ruled out mitotic cyclin accumulation as the direct timer of mitotic entry (McCleland and O'Farrell, 2008; McCleland et al., 2009).
The fast maternally driven cycles have altered checkpoint responsiveness. Arresting DNA replication during the maternal cycles extends interphase, but does not block nuclei from entering mitoses (Raff and Glover, 1988; Sibon et al., 1997). Flies mutant for genes in the DNA damage checkpoint, mei41, grapes (grp), or mus304, are viable, but eggs laid from homozygous mutant mothers fail to normally prolong interphase during cycles 10–13 and ultimately enter a catastrophic mitosis in cycle 13 (Fogarty et al., 1997; Sibon et al., 1997, 1999). Models attribute the normal interphase extension to prolongation of S phase and consequent activation of the checkpoint genes. Although the results indeed showed that Grapes is required to delay mitosis, the idea that the duration of DNA replication is the master timer of the early cycles is inferred from the common involvement of S phase in checkpoint activation. Thus, the contribution of a normal S phase in timing the early embryonic cycles has not been directly demonstrated.
In contrast to inhibition of DNA replication, which generally activates the S phase checkpoint, some modes of preventing S phase do not arrest the cell cycle. Cells mutant in genes required for the initial licensing of origins, such as the yeast gene CDC6 and the Drosophila gene double-parked (dup, a homologue of yeast CDT1) fail to replicate their DNA, yet they enter mitosis with unreplicated sister chromatids (Kelly et al., 1993; Piatti et al., 1995; Whittaker et al., 2000). Addition of the Dup inhibitor, Geminin, to Xenopus egg extracts or its overexpression in flies prevents the loading of the MCM helicase complex, and blocks the licensing step of DNA replication (McGarry and Kirschner, 1998; Quinn et al., 2001). We reasoned that injection of recombinant Geminin into syncytial Drosophila embryos would similarly block DNA replication licensing, delete S phase (Fig. 1 B), and thereby allow us to directly assess an S phase contribution in timing the cell cycle.
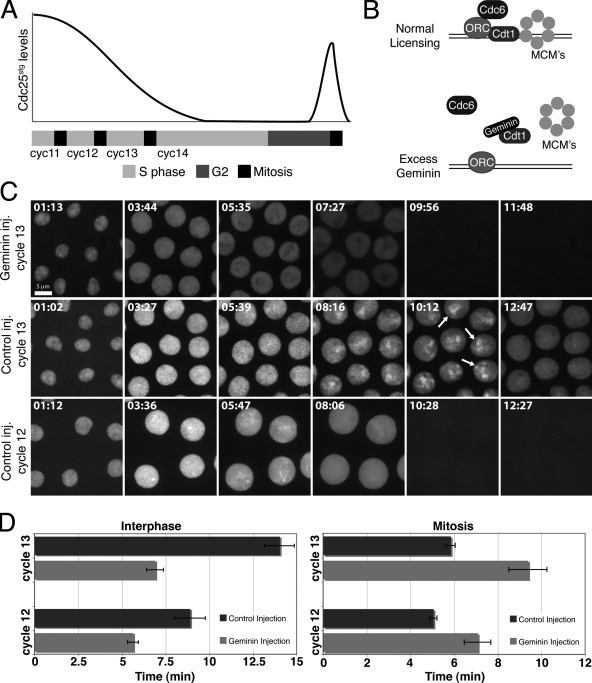
Figure 1.
Injection of Geminin during the Drosophila blastoderm divisions deletes S phase and advances mitosis. (A) Schematic illustrating progression through the blastoderm divisions and the MBT. The bar beneath the graph represents time. Cdc25stg protein declines during the blastoderm cycles. Zygotic transcription of Cdc25stg during G2 of cycle 14 promotes mitosis. (B) Schematic illustrating DNA replication initiation and experimental expectation from Geminin injection. (C) Nuclei of YFP-PCNA–expressing embryos in the cycle after injection of Geminin or control buffer. YFP-PCNA exhibits distinct nuclear foci in control injections. Arrows highlight foci in the apical region of nuclei at the end of S phase. Note that YFP-PCNA foci are absent throughout S phase after Geminin injection. Frames 9:56 and 11:48 of the Geminin-injected embryo correspond to premature mitotic entry and dispersal of nuclear YFP-PCNA in the cytoplasm. (D) YFP-PCNA–expressing embryos were injected in cycle 11 or cycle 12 with Geminin and interphase length and mitosis were measured in the subsequent cycle (cycle 12 or cycle 13, respectively). Interphase was defined by nuclear localization of PCNA and mitosis was defined by its dispersal. Average cell cycle times are shown for cycle 12 (n = 7 embryos) and cycle 13 (n = 10 embryos). Error bars represent SD.
Injection of Geminin prevented S phase in the ensuing cycle and advanced mitotic entry during the syncytial blastoderm divisions to give interphase durations similar to those in grp mutant embryos. After S phase deletion, nuclei entered mitosis with uninemic chromosomes, failed to achieve a metaphase alignment, and subsequently exited mitosis after a delay. We conclude that normal S phase delays mitotic entry during the maternal blastoderm divisions. Preventing S phase at the MBT in cycle 14 caused an extra syncytial division, uncovering a role for S phase 14 in the termination of the maternal mitotic program. Interestingly, deleting S phase in cycle 15 did not significantly alter the timing of mitosis. We suggest that the shift in regulation of Cdk1 activation to dependency on transcription of Cdc25 at the MBT supersedes the inputs from DNA replication.
Results and discussion
Geminin injection deletes S phase and advances blastoderm mitoses
If Geminin injection blocks the licensing of origins, it should affect the S phase occurring subsequent to licensing. Recruitment of MCM proteins to anaphase chromosomes during the early embryonic cycles marks the time of licensing (Su and O'Farrell, 1997). Geminin injection during interphase 13 did not affect the progression of nuclei through mitosis 13, but did block the incorporation of dUTP in cycle 14, demonstrating the expected second cycle effect (Fig. S1).
Embryos expressing YFP-PCNA were used to follow S phase in real time. PCNA, a sliding clamp for DNA polymerase, localizes to the nucleus during interphase and forms foci at active sites of DNA replication (Hingorani and O'Donnell, 2000). Although the syncytial S phases of Drosophila are incredibly rapid, distinct foci of YFP-PCNA were evident throughout S phase and were especially prominent during the closing minutes of cycle 13 in the apical region of nuclei (Fig. 1 C and Video 1).
Consistent with a second cycle block to S phase, Geminin injection did not immediately disturb YFP-PCNA foci and a successful mitosis followed (Video 2 and unpublished data). However, YFP-PCNA foci were not evident in the subsequent interphase (Fig. 1 C and Video 2). Likewise, when Geminin was injected at the beginning of cycle 14, defects appeared only 2.5 h later when cells began to collect in a prolonged mitosis 15 with unreplicated chromatids. The successful execution of cellularization, gastrulation, mitosis 14, and germband extension in these embryos implies that gene expression and general cell biological functions are not disturbed by Geminin injection, attesting to the specificity of its action.
We used real-time records of YFP-PCNA to determine how S phase deletion affects interphase duration. After Geminin injection, interphase was shortened by 37% in cycle 12 and by 51% in cycle 13 (Fig. 1 D). We note that a brief interphase remains in the absence of S phase, suggesting that inhibitors such as phosphatases or low mitotic activators can limit progress to mitosis in a brief post-mitotic period. However, the advancement of mitosis shows that the levels of mitotic cyclin and other mitotic activators are sufficient for mitosis midway through interphase. Thus, these factors do not normally limit final entry into mitosis during the blastoderm cycles, consistent with previous analysis suggesting that cyclin accumulation is not the direct timer of mitotic entry (McCleland and O'Farrell, 2008; McCleland et al., 2009). Importantly, the data suggest that DNA replication produces a negative signal that defines interphase length and directs its normal extension during the blastoderm divisions.
Geminin injection promotes premature chromatin condensation and mitosis with unreplicated chromatids
Embryos coexpressing Cid-GFP and Histone-RFP were injected with Geminin to examine whether deletion of S phase affected chromatin dynamics. In the affected interphase, nuclei decondensed normally after telophase, but less than 3.5 min into interphase chromatin began to recondense (Fig. 2 B and Video 4). Within 1–2 min the chromatin was a compact mass in the center of the nucleus. Shortly thereafter, nuclear envelope breakdown and initial chromatid movements occurred. The early compaction contrasts to wild-type embryos, wherein chromatin begins to condense 8 min into cycle 12 or 13.5 min into cycle 13, just before mitotic entry (Fig. 2 A, Video 3, and unpublished data). Hence, deleting S phase in the early mitotic cycles promotes premature DNA condensation and advances mitotic entry.
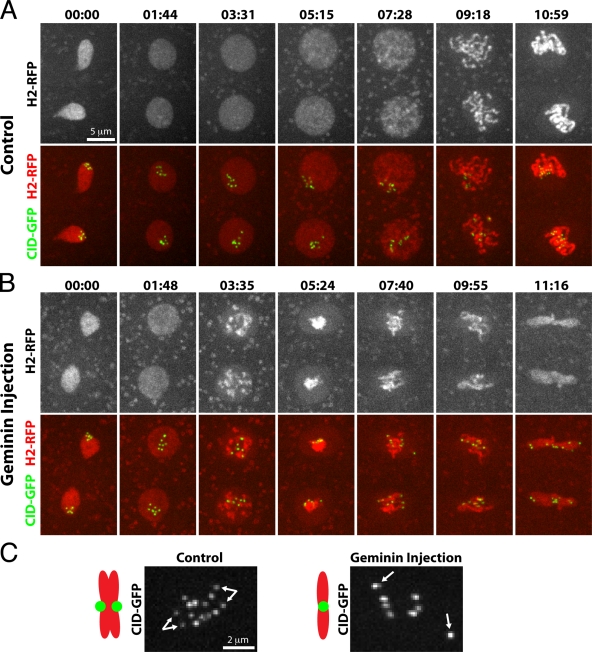
Figure 2.
Geminin injection promotes premature chromatin condensation and mitosis with unreplicated chromatids. Frames showing nuclei from embryos expressing Histone-RFP and Cid-GFP in the cycle after injection with control buffer (A) or Geminin (B). In the Geminin-injected embryo, chromatin begins to condense ∼3 min into interphase, kinetochores are unpaired in mitosis, and chromosomes fail to congress to the metaphase plate. Time is presented as min:sec. (C) Higher magnification view of Cid-GFP during mitosis in control and Geminin injections. Bifurcated arrows in the control nucleus point to paired kinetochores at metaphase. Single arrows in Geminin injection highlight unpaired kinetochores.
Control metaphases exhibit eight stably aligned binemic chromosomes whose paired centromeres are detected as two dots of Cid-GFP (Fig. 2 and Video 3). In contrast, after S phase deletion, mitotic chromosomes appeared uninemic, oscillated unstably, and possessed a single Cid-GFP dot (Fig. 2 and Video 4). Thus, in a cycle after Geminin injection, nuclei enter mitosis with single sister chromatids and fail to bi-orient. Real-time imaging of other mitotic proteins, GFP-Polo and Aurora B-GFP, supports the conclusion that S phase deletion advances mitosis with uninemic chromosomes (Fig. S2 and Videos 5 and 6).
Despite the lack of metaphase alignment, nuclei of Geminin-injected embryos exited mitosis after a brief mitotic delay. The total length of mitosis was nearly doubled in cycle 13 (Fig. 1 D), as chromosomes oscillated for ∼8 min before anaphase onset (Fig. 2 B and Video 4). We conclude that unreplicated chromosomes delay mitotic progression.
S phase deletion does not advance mitosis in grp mutant embryos
It has been argued that the exponential increase in DNA titrates replication components and slows S phase, which, via checkpoint activation, delays mitosis to create the gradual lengthening of blastoderm cycles (Sibon et al., 1997; Crest et al., 2007). If the activity of a checkpoint gene, such as grp, relies on DNA replication, the deletion of S phase by Geminin injection should prevent its activation. In this case, a grp mutation will have no additional phenotype in a cycle lacking S phase. Furthermore, if all of the effects of DNA replication on mitosis are mediated by grp, the mitotic phenotype of grp and deletion of S phase ought to be the same. To probe this we compared grp mutant embryos to those in which S phase was deleted.
Histone-GFP–expressing embryos, which were wild type, mutant in grp, or injected with Geminin, were filmed at high resolution. In contrast to the very early condensation of chromatin after S phase deletion, chromatin in grp mutant embryos remain dispersed until the close of interphase (Fig. 3 A). Furthermore, Geminin injection into grp mutant embryos results in chromatin condensation behavior indistinguishable from Geminin injection into wild-type embryos. Thus, DNA replication promotes chromatin dispersal, and dispersal does not require checkpoint function. In cycle 14, the dispersed interphase appearance of chromatin is not dependent on S phase (see below and Fig. 4), suggesting a change in the dependence on S phase for chromatin dispersal.
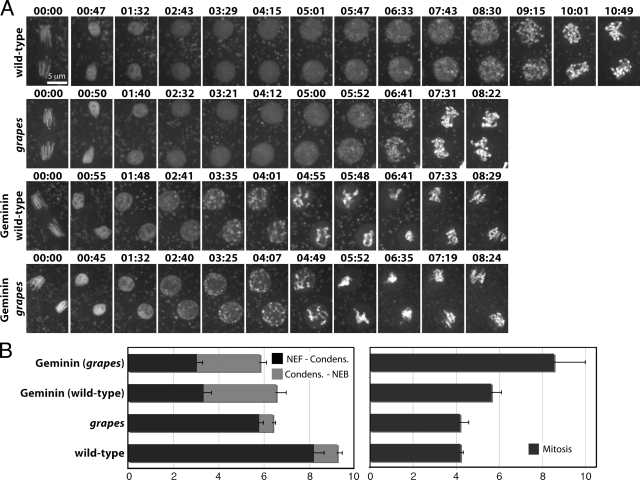
Figure 3.
Direct cell cycle comparison between embryos injected with Geminin and grp mutant embryos. (A) Either wild-type or grp mutant embryos expressing Histone-GFP were injected with control buffer or Geminin. Time is presented in min:sec. (B) Quantification of interphase chromatin dynamics and mitotic duration from embryos in A. Nuclear envelope formation (NEF) to the beginning of chromatin condensation was measured and the time from chromatin condensation to the onset of mitosis was measured (onset of mitosis was defined by disappearance of the Histone-GFP halo around the chromatin and the first chromatid movements). Average cell cycle times are shown for wild-type (n = 8 embryos), grp (n = 18 embryos), Geminin injected (n = 10 embryos), and Geminin injected into grp (n = 11 embryos). Error bars represent SD.
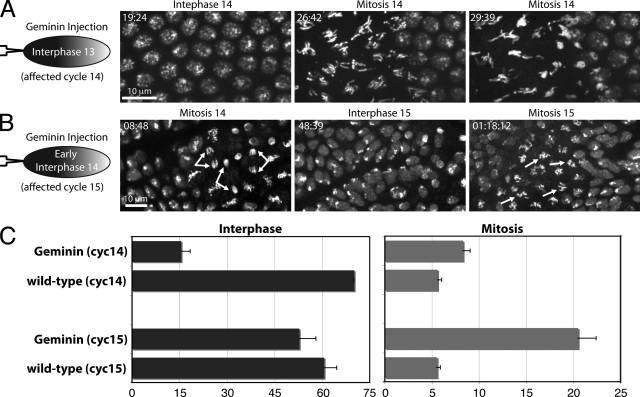
Figure 4.
Geminin injection induces an extra syncytial division, but does not affect interphase length after the MBT. (A) Histone-GFP–expressing embryos were injected at one pole (left) during cycle 13 with Geminin. Note that nuclei synchronously and prematurely enter mitosis 14 in the injected region. (B) Histone-GFP–expressing embryos were injected with Geminin during interphase 14 before cellularization. Bifurcated arrows highlight normal anaphases during mitosis 14 and arrows show unreplicated chromosomes in mitosis 15 that fail to reach the metaphase plate. (C) Quantification of interphase and mitotic duration from embryos as in A and B. Average interphase and mitotic times are shown for cycle 14 (n = 13 embryos) and cycle 15 in domain 5 (n = 33 embryos). Error bars represent SD.
Despite differences in chromatin condensation, Geminin-injected embryos and grp mutant embryos exhibited similar mitotic entry times (Fig. 3 B). Furthermore, grp mutants injected with Geminin exhibited similar interphase duration as wild-type embryos injected with Geminin (Fig. 3). This result suggests that Grapes has no activity after Geminin injection, and that the shortening of interphase in the cycle after Geminin injection might be attributable to the absence of the checkpoint pathway when S phase is deleted. These findings support the previous inference that S phase delays entry into mitosis until its completion by activating Grapes-dependent checkpoint function.
Although Geminin injection into grp mutants barely altered interphase length, it still prolonged mitosis, presumably because it induced spindle checkpoint function in response to uninemic chromatids. However, the mitotic delay caused by Geminin injection into grp embryos was almost twice that caused by Geminin injection into wild-type embryos (Fig. 3 B). Perhaps the additional mitotic extension in grp mutants reflects a role of Grapes in mitosis or the spindle checkpoint (Su et al., 1999).
DNA replication at the MBT is required for prompt termination of the syncytial divisions
The switch from maternal to zygotic control at the MBT is accompanied by changes in cell cycle regulation (O'Farrell, 2001). The literature has emphasized that a G2 phase is added to the cell cycle, and that zygotic transcription times subsequent mitoses. However, the first MBT change in the cell cycle is prolongation of S phase. We directly tested whether this S phase has a role in the MBT.
Geminin was injected into embryos expressing Histone-GFP during cycle 13 (Fig. 4 A). After progressing normally through mitosis 13, nuclei at the injected pole synchronously entered mitosis ∼16.5 min after the onset of cycle 14 (Fig. 4, A and C; and Video 7), whereas mitosis 14 normally occurs in a spatial program beginning 70 min after mitosis 13. These data indicate that S phase 14 is required to terminate the syncytial mitotic cycles.
To assess the influence of S phase 14 deletion on MBT events, we monitored cellularization. Geminin was injected during cycle 13 into embryos expressing Histone-RFP and Sqh-GFP. Only one extra synchronous division was observed, and this was followed by cellularization, only slightly delayed compared with the uninjected end of the embryo (Video 8). Surprisingly, a subsequent division exhibited a normal mitosis 14 domain pattern (unpublished data). Apparently, S phase 14 forestalls mitosis, but other MBT events, which introduce zygotic programming of gastrulation and the cell cycle, continue in its absence (Edgar and Datar, 1996).
S phase is not required for the zygotic programming of a post-MBT mitosis
Geminin injection in cycle 14 had no effect until the next cycle, when cells entered a normally patterned mitosis 15 with unreplicated chromatids (Fig. 4, B and C; Fig. S3 and Video 9). To probe for subtle timing defects, we tracked individual cells during cycle 15. The fifth group of cells to initiate mitosis 14, domain 5, does so relatively synchronously (Foe et al., 1993), but the program is more complex at entry into mitosis 15 (Fig. S3 and Video 10). Cells proximal to the cephalic furrow divide earliest at ∼60.5 min after mitosis 14. Geminin-injected embryos showed a slight advancement of mitosis in these cells to 52.75 min. Notably, deletion of S phase 15 did not normalize interphase duration in domain 5, as cells anterior to the first-dividing cells still showed a later mitosis (Fig. S3 and Video 9). Thus, deletion of S phase may have slightly shortened cycle 15, but it did not override inputs that spatially pattern mitosis.
Other observations also support an interpretation that zygotic programming of post-MBT mitoses is independent of S phase. First, although deletion of S phase 14 caused rapid entry into an extra mitosis, this was followed by MBT events and a mitosis that closely followed the normal temporal program of mitosis 14. Second, previous analyses of dup mutant embryos, which lack S phase 16, show a normally patterned mitosis 16 (Garner et al., 2001). Post-MBT divisions are normally promoted by Cdc25string expression (Edgar and O'Farrell, 1990), which is guided by the patterning regulators (Edgar et al., 1994a). This zygotic programming of divisions appears relatively resistant to S phase deletion.
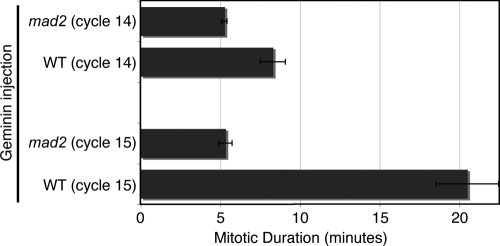
As in the earlier cycles, deletion of S phase resulted in a prolonged mitosis; however, the duration of the prolonged mitoses increased from ∼8.3 min in mitosis 14 to ∼20.5 min in mitosis 15. We do not know the basis of this difference; however, the mitotic delay in both cycles depended on the spindle checkpoint protein Mad2 (Fig. 5), as has similarly been observed in dup mutants in cycle 16 (Garner et al., 2001).
Figure 5.
The mitotic delay in response to unreplicated sister chromatids is dependent on the spindle checkpoint. Embryos that were wild type or mad2 mutant and expressing Histone-GFP were injected with Geminin protein. Mitotic duration was recorded during cycle 14 and cycle 15 in which mitoses occurred with unreplicated sister chromatids. Mitosis 15 duration was measured in multiple mitotic domains and identical results were found. Average mitotic duration is shown for cycle 14 (n = 6 embryos) and cycle 15 (n = 12 embryos). Error bars represent SD.
S phase as a mitotic timer
We developed a means to examine the contribution of a normal S phase in timing mitotic entry during early Drosophila embryogenesis. Previous work suggests that an S phase checkpoint is activated during a normal S phase (Grallert and Boye, 2008), and that S phase extension is key to the gradual prolongation of the maternally driven blastoderm cycles (Fogarty et al., 1997; Sibon et al., 1997, 1999); however, before the results shown here, the role of an undisturbed S phase in timing mitotic entry had not previously been demonstrated. Our results show that a normal S phase is an important timer of interphase length during the syncytial blastoderm cycles.
The role of S phase as a mitotic timer during the maternal divisions does not persist in the zygotically controlled divisions after the MBT. We suggest that this change reflects complete transition to a different mitotic timer. Before the MBT, Cdc25 and other mitotic activators are abundant throughout the cell cycle, there is no evident gap between S phase and mitosis, and nuclei rely on S phase to forestall mitotic entry. After the MBT, many mitotic activators are degraded upon mitotic exit, and restoration of one of these, Cdc25string, requires new zygotic transcription (Lehner and O'Farrell, 1989; Edgar and O'Farrell, 1990). Mitosis awaits Cdc25string expression, which usually occurs after completion of S phase, thus introducing a G2 and absolving DNA replication of responsibility for timing mitosis. Notably, this proposal offers an explanation for why the checkpoint genes, which couple mitosis to the completion of S phase, are essential during the early embryonic divisions but subsequently dispensable.
How then does S phase contribute during the beginning of cycle 14 at the MBT? The prolongation of interphase is an important part of the MBT, allowing cellularization and providing enough time for complete transcription of genes with long transcription units (Shermoen and O'Farrell, 1991; Foe et al., 1993). The prolongation of interphase and production of G2 phase requires down-regulation of mitotic activators, particularly the activating phosphatases encoded by string and twine. For example, the inhibitors of String action, Tribbles and Früshstart, which promote Cdc25string destruction and inhibit cyclin:Cdk1, respectively, are transcribed during the long S phase of cycle 14 (Grosshans and Wieschaus, 2000; Seher and Leptin, 2000). Although it has been attractive to view the MBT as a sharp switch from maternal to zygotic control, we suggest that the retooling of cell cycle regulation requires considerable time within interphase 14 and that the prolonged S phase 14 enforces Cdk1 inactivation while MBT transition events progress.
Materials and methods
Fly stocks
Drosophila melanogaster strains were grown on standard cornmeal-yeast medium. Flies expressing Histone H2AvD-GFP, GFP-Polo (Fly Trap #CC01326), Sqh-GFP, Cid-GFP, or Histone H2AvD-RFP were used for live-embryo analysis (Clarkson and Saint, 1999; Royou et al., 2004; Buszczak et al., 2007; Schuh et al., 2007). For checkpoint deficient experiments, the following stocks were generated: w; grp06034; H2AvD-GFP and w; H2AvD-GFP/Cyo; mad2p (Buffin et al., 2007).
The PCNA ORF was amplified using primers 5′-CACCATGTTCGAGGCACGCCTGGGT-3′ and 5′-TGTCTCGTTGTCCTCGATCTTG-3′ and cloned into the gateway system pENTR-D-TOPO vector (Invitrogen) according to the manufacturer's instructions. The resulting clone was used to recombine the PCNA ORF into pPVW (UASp promoter with N-terminal venus fusion from the Drosophila Gateway collection; T. Murphy Laboratory, Carnegie Institute of Washington, Baltimore, MD). P{UASp-YFP-PCNA} lines were obtained by standard germ-line transformation (BestGene, Inc.). Resulting lines were crossed to Da-Gal4 flies (Daughterless promoter) for YFP-PCNA expression.
Similarly, P{UASp-Aurora B-GFP} lines were constructed. The Aurora B ORF was amplified using primers 5′-CACCATGACGCTTTCCCGCGCGAAG-3′ and 5′-ATTTCTGGCCGTGTTCTCC-3′ and ultimately recombined into pPWG (UASp promoter with C-terminal GFP fusion). Resulting lines were crossed to Tub-Gal4 flies (Lee and Luo, 1999) for Aurora B-GFP expression.
Embryo manipulation
Drosophila embryos were collected on agar plates containing grape juice, aged for the appropriate developmental time, dechorionated for 2 min in 50% bleach, and then extensively washed in water. For injections, embryos were aligned on agar plates and transferred to coverslips. Embryos were desiccated for ∼8–10 min and then overlaid with Halocarbon oil (Sigma-Aldrich). Alexa 546–conjugated dUTP (Invitrogen) was injected at a needle concentration of 100 µM.
Geminin protein purification
The Drosophila Geminin ORF was amplified using primers 5′-AAACATATGTCTTCGAGCGCTGCCAGG-3′ and 5′-AAACTCGAGCTAGGCGTTGACCTTGTCCTC-3′ and cloned into Pet28a as an NdeI–Xho fragment. 6XHis-Geminin was expressed in BL-21 DE3 pLysS bacteria (Agilent Technologies) and purified on nickel agarose beads according to the manufacturer's instructions (QIAGEN). Geminin protein was dialyzed into 40 mM Hepes, pH 7.4, and 150 mM KCl and concentrated to ∼20 mg/ml before injection.
Image acquisition and processing
Embryos were imaged on an inverted microscope (DM 1RB; Leica) equipped with a spinning-disk confocal unit (CSU10; Yokagowa), 100x Plan Fluotar 1.3 NA and 40x Plan Fluotar 0.7 NA objectives (Lecia), a camera (Orca AG; Hamamatsu Photonics), and Volocity 4 acquisition software (PerkinElmer). Image stacks were collected using 1.5-µm steps over an 8–12-µm range using a controlled stage (MS-2000; Applied Scientific Instrumentation). All images and videos were processed in Volocity 4 software and presented as extended focused images. Image capture rates are indicated per video. Time is displayed in hours:minutes:seconds.
At least three independent experiments were performed for each experiment shown. In each experiment an X-Y stage facilitated the filming of multiple embryos (usually greater than five embryos).
Fixed analysis and immunofluorescence
Embryos injected with Geminin were incubated for the appropriate time, fixed in a mixture of 37% formaldehyde and heptane for 10 min, and immediately hand devitellinized. Embryos were washed into phosphate-buffered saline containing 2 µg/ml Hoescht 33258 (Invitrogen) and 2 µg/ml Oregon green–conjugated wheat germ agglutinin (Invitrogen).
Online supplemental material
Fig. S1 illustrates the absence of DNA replication after Geminin injection. Fig. S2 highlights the dynamics of mitotic proteins during the blastoderm division after the deletion of S phase. Fig. S3 provides a wide-field perspective of the post-MBT divisions after deletion of S phase 15. Videos 1–6 show real-time videos of blastoderm embryos progressing through interphase and mitosis after deletion of S phase. Videos 7–8 highlight the effects of S phase deletion on MBT events. Videos 9–10 highlight developmental and cell cycle behavior upon deletion of S phase 15. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200906191/DC1.
Acknowledgments
We thank members of the O'Farrell laboratory for helpful comments.
This research was supported by National Institutes of Health training grant GM078710 to M.L. McCleland and National Institutes of Health grant GM037193 to P.H. O'Farrell.
Footnotes
Abbreviation used in this paper:
- MBT
- mid-blastula transition
References
- Buffin E., Emre D., Karess R.E. 2007. Flies without a spindle checkpoint.Nat. Cell Biol. 9:565–572 doi:10.1038/ncb1570 [DOI] [PubMed] [Google Scholar]
- Buszczak M., Paterno S., Lighthouse D., Bachman J., Planck J., Owen S., Skora A.D., Nystul T.G., Ohlstein B., Allen A., et al. 2007. The carnegie protein trap library: a versatile tool for Drosophila developmental studies.Genetics. 175:1505–1531 doi:10.1534/genetics.106.065961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson M., Saint R. 1999. A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior.DNA Cell Biol. 18:457–462 doi:10.1089/104454999315178 [DOI] [PubMed] [Google Scholar]
- Crest J., Oxnard N., Ji J.Y., Schubiger G. 2007. Onset of the DNA replication checkpoint in the early Drosophila embryo.Genetics. 175:567–584 doi:10.1534/genetics.106.065219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A., Datar S.A. 1996. Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila's early cell cycle program.Genes Dev. 10:1966–1977 doi:10.1101/gad.10.15.1966 [DOI] [PubMed] [Google Scholar]
- Edgar B.A., O'Farrell P.H. 1989. Genetic control of cell division patterns in the Drosophila embryo.Cell. 57:177–187 doi:10.1016/0092-8674(89)90183-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A., O'Farrell P.H. 1990. The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string.Cell. 62:469–480 doi:10.1016/0092-8674(90)90012-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A., Schubiger G. 1986. Parameters controlling transcriptional activation during early Drosophila development.Cell. 44:871–877 doi:10.1016/0092-8674(86)90009-7 [DOI] [PubMed] [Google Scholar]
- Edgar B.A., Kiehle C.P., Schubiger G. 1986. Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development.Cell. 44:365–372 doi:10.1016/0092-8674(86)90771-3 [DOI] [PubMed] [Google Scholar]
- Edgar B.A., Lehman D.A., O'Farrell P.H. 1994a. Transcriptional regulation of string (cdc25): a link between developmental programming and the cell cycle.Development. 120:3131–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A., Sprenger F., Duronio R.J., Leopold P., O'Farrell P.H. 1994b. Distinct molecular mechanism regulate cell cycle timing at successive stages of Drosophila embryogenesis.Genes Dev. 8:440–452 doi:10.1101/gad.8.4.440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe V.E., Odell G.M., Edgar B.A. 1993. Mitosis and morphogenesis in the Drosophila embryo: point and counterpoint. The Development of Drosophila melanogaster. Bate M., Martinez-Arias A., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 149–300 [Google Scholar]
- Fogarty P., Campbell S.D., Abu-Shumays R., Phalle B.S., Yu K.R., Uy G.L., Goldberg M.L., Sullivan W. 1997. The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity.Curr. Biol. 7:418–426 doi:10.1016/S0960-9822(06)00189-8 [DOI] [PubMed] [Google Scholar]
- Garner M., van Kreeveld S., Su T.T. 2001. mei-41 and bub1 block mitosis at two distinct steps in response to incomplete DNA replication in Drosophila embryos.Curr. Biol. 11:1595–1599 doi:10.1016/S0960-9822(01)00483-3 [DOI] [PubMed] [Google Scholar]
- Grallert B., Boye E. 2008. The multiple facets of the intra-S checkpoint.Cell Cycle. 7:2315–2320 [DOI] [PubMed] [Google Scholar]
- Grosshans J., Wieschaus E. 2000. A genetic link between morphogenesis and cell division during formation of the ventral furrow in Drosophila.Cell. 101:523–531 doi:10.1016/S0092-8674(00)80862-4 [DOI] [PubMed] [Google Scholar]
- Hingorani M.M., O'Donnell M. 2000. Sliding clamps: a (tail)ored fit.Curr. Biol. 10:R25–R29 doi:10.1016/S0960-9822(99)00252-3 [DOI] [PubMed] [Google Scholar]
- Ji J.Y., Squirrell J.M., Schubiger G. 2004. Both cyclin B levels and DNA-replication checkpoint control the early embryonic mitoses in Drosophila.Development. 131:401–411 doi:10.1242/dev.00944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T.J., Martin G.S., Forsburg S.L., Stephen R.J., Russo A., Nurse P. 1993. The fission yeast cdc18+ gene product couples S phase to START and mitosis.Cell. 74:371–382 doi:10.1016/0092-8674(93)90427-R [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L. 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis.Neuron. 22:451–461 doi:10.1016/S0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- Lehner C.F., O'Farrell P.H. 1989. Expression and function of Drosophila cyclin A during embryonic cell cycle progression.Cell. 56:957–968 doi:10.1016/0092-8674(89)90629-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland M.L., O'Farrell P.H. 2008. RNAi of mitotic cyclins in Drosophila uncouples the nuclear and centrosome cycle.Curr. Biol. 18:245–254 doi:10.1016/j.cub.2008.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland M.L., Farrell J.A., O'Farrell P.H. 2009. Influence of cyclin type and dose on mitotic entry and progression in the early Drosophila embryo.J. Cell Biol. 184:639–646 doi:10.1083/jcb.200810012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry T.J., Kirschner M.W. 1998. Geminin, an inhibitor of DNA replication, is degraded during mitosis.Cell. 93:1043–1053 doi:10.1016/S0092-8674(00)81209-X [DOI] [PubMed] [Google Scholar]
- Murray A.W., Kirschner M.W. 1989. Cyclin synthesis drives the early embryonic cell cycle.Nature. 339:275–280 doi:10.1038/339275a0 [DOI] [PubMed] [Google Scholar]
- O'Farrell P.H. 2001. Triggering the all-or-nothing switch into mitosis.Trends Cell Biol. 11:512–519 doi:10.1016/S0962-8924(01)02142-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S., Lengauer C., Nasmyth K. 1995. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae.EMBO J. 14:3788–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn L.M., Herr A., McGarry T.J., Richardson H. 2001. The Drosophila Geminin homolog: roles for Geminin in limiting DNA replication, in anaphase and in neurogenesis.Genes Dev. 15:2741–2754 doi:10.1101/gad.916201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff J.W., Glover D.M. 1988. Nuclear and cytoplasmic mitotic cycles continue in Drosophila embryos in which DNA synthesis is inhibited with aphidicolin.J. Cell Biol. 107:2009–2019 doi:10.1083/jcb.107.6.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royou A., Field C., Sisson J.C., Sullivan W., Karess R. 2004. Reassessing the role and dynamics of nonmuscle myosin II during furrow formation in early Drosophila embryos.Mol. Biol. Cell. 15:838–850 doi:10.1091/mbc.E03-06-0440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M., Lehner C.F., Heidmann S. 2007. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase.Curr. Biol. 17:237–243 doi:10.1016/j.cub.2006.11.051 [DOI] [PubMed] [Google Scholar]
- Seher T.C., Leptin M. 2000. Tribbles, a cell-cycle brake that coordinates proliferation and morphogenesis during Drosophila gastrulation.Curr. Biol. 10:623–629 doi:10.1016/S0960-9822(00)00502-9 [DOI] [PubMed] [Google Scholar]
- Shermoen A.W., O'Farrell P.H. 1991. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts.Cell. 67:303–310 doi:10.1016/0092-8674(91)90182-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibon O.C., Stevenson V.A., Theurkauf W.E. 1997. DNA-replication checkpoint control at the Drosophila midblastula transition.Nature. 388:93–97 doi:10.1038/40439 [DOI] [PubMed] [Google Scholar]
- Sibon O.C., Laurençon A., Hawley R., Theurkauf W.E. 1999. The Drosophila ATM homologue Mei-41 has an essential checkpoint function at the midblastula transition.Curr. Biol. 9:302–312 doi:10.1016/S0960-9822(99)80138-9 [DOI] [PubMed] [Google Scholar]
- Stiffler L.A., Ji J.Y., Trautmann S., Trusty C., Schubiger G. 1999. Cyclin A and B functions in the early Drosophila embryo.Development. 126:5505–5513 [DOI] [PubMed] [Google Scholar]
- Stumpff J., Duncan T., Homola E., Campbell S.D., Su T.T. 2004. Drosophila Wee1 kinase regulates Cdk1 and mitotic entry during embryogenesis.Curr. Biol. 14:2143–2148 doi:10.1016/j.cub.2004.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T.T., O'Farrell P.H. 1997. Chromosome association of minichromosome maintenance proteins in Drosophila mitotic cycles.J. Cell Biol. 139:13–21 doi:10.1083/jcb.139.1.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T.T., Campbell S.D., O'Farrell P.H. 1999. Drosophila grapes/CHK1 mutants are defective in cyclin proteolysis and coordination of mitotic events.Curr. Biol. 9:919–922 doi:10.1016/S0960-9822(99)80399-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker A.J., Royzman I., Orr-Weaver T.L. 2000. Drosophila double parked: a conserved, essential replication protein that colocalizes with the origin recognition complex and links DNA replication with mitosis and the down-regulation of S phase transcripts.Genes Dev. 14:1765–1776 [PMC free article] [PubMed] [Google Scholar]