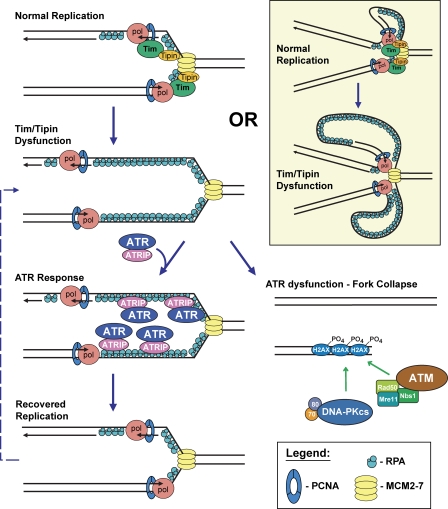
Increased amounts of single-stranded DNA accumulate at replication forks when Tim–Tipin isn't around, activating an ATR-mediated DNA damage response necessary for DNA replication to proceed.
Abstract
The Tim (Timeless)–Tipin complex has been proposed to maintain genome stability by facilitating ATR-mediated Chk1 activation. However, as a replisome component, Tim–Tipin has also been suggested to couple DNA unwinding to synthesis, an activity expected to suppress single-stranded DNA (ssDNA) accumulation and limit ATR–Chk1 pathway engagement. We now demonstrate that Tim–Tipin depletion is sufficient to increase ssDNA accumulation at replication forks and stimulate ATR activity during otherwise unperturbed DNA replication. Notably, suppression of the ATR–Chk1 pathway in Tim–Tipin-deficient cells completely abrogates nucleotide incorporation in S phase, indicating that the ATR-dependent response to Tim–Tipin depletion is indispensible for continued DNA synthesis. Replication failure in ATR/Tim-deficient cells is strongly associated with synergistic increases in H2AX phosphorylation and DNA double-strand breaks, suggesting that ATR pathway activation preserves fork stability in instances of Tim–Tipin dysfunction. Together, these experiments indicate that the Tim–Tipin complex stabilizes replication forks both by preventing the accumulation of ssDNA upstream of ATR–Chk1 function and by facilitating phosphorylation of Chk1 by ATR.
Introduction
As part of an interdependent complex, Tim (Timeless) and Tipin associate with replisome components (MCM subunits, Pol δ/ϵ, and Claspin) and perform important functions in both DNA replication and genome maintenance (Gotter, 2003; Unsal-Kaçmaz et al., 2005; Chou and Elledge, 2006; Errico et al., 2007; Gotter et al., 2007; Yoshizawa-Sugata and Masai, 2007; Urtishak et al., 2009). Replisome-associated functions for Tim–Tipin have been proposed to include the coupling of DNA unwinding with DNA synthesis. In support of this function, DNA polymerase inhibition in yeast strains harboring mutations in Tim–Tipin orthologues (Tof1–Csm3 in Saccharomyces cerevisiae; Swi1–Swi3 in Schizosaccharomyces pombe) causes replisome components to separate from newly synthesized daughter strands and leads to the accumulation of single-stranded DNA (ssDNA; Katou et al., 2003; Noguchi et al., 2004; Sommariva et al., 2005). Consistent with a similar function in vertebrates, depletion of Tim–Tipin in Xenopus laevis extracts causes a twofold increase in chromatin-associated replication protein A (RPA) after DNA polymerase inhibition (Errico et al., 2007). Together, these data imply that the Tim–Tipin complex may possess similar helicase–polymerase coupling functions in mammals.
Accumulation of ssDNA at replication forks and resected double-strand breaks (DSBs) activates the ATR–Chk1 checkpoint pathway (Costanzo et al., 2003; Zou and Elledge, 2003; Byun et al., 2005). At the replication fork, this pathway prevents replication fork collapse, a process which leads to chromatid breaks (Lopes et al., 2001; Tercero and Diffley, 2001; Casper et al., 2002; Brown and Baltimore, 2003; Zachos et al., 2003; Paulsen and Cimprich, 2007; Chanoux et al., 2009). If the Tim–Tipin complex is indeed required for helicase and polymerase coupling, Tim–Tipin reduction would be expected to enhance ATR pathway activation as a result of ssDNA accumulation. However, paradoxically, recent studies have demonstrated that Tim–Tipin is required to facilitate ATR-mediated phosphorylation of Chk1 in response to DNA damage or polymerase inhibition (Unsal-Kaçmaz et al., 2005, 2007; Chou and Elledge, 2006; Errico et al., 2007; Gotter et al., 2007; Yoshizawa-Sugata and Masai, 2007). Similar dependencies on Tim–Tipin orthologues in yeast have also been reported (Foss, 2001; Noguchi et al., 2003, 2004). Nevertheless, the requirement for these complexes in yeast and mammalian checkpoint signaling appears to be only partial, likely because of the participation of overlapping parallel pathways (Foss, 2001; Noguchi et al., 2003).
Deficiency in Tim–Tipin and yeast orthologues results in replication fork instability, increased sister chromatid exchange, and chromatid breaks during otherwise unperturbed DNA replication (Katou et al., 2003; Noguchi et al., 2003, 2004; Urtishak et al., 2009). Because the ATR–Chk1 pathway prevents replication fork collapse, it is conceivable that the effect of Tim–Tipin reduction on genome stability in S phase may be solely a function of its participation in mediating Chk1 activation by ATR. Alternatively, Tim–Tipin could possess dual functions, both suppressing ssDNA generation at the replication fork as a replisome component and acting as a localized adapter to facilitate the transmission of signaling from ATR to Chk1. In this regard, the elevated levels of chromatid breaks, translocations, and sister chromatid exchange observed in Tim–Tipin-reduced cells (Urtishak et al., 2009) may result from the combined effects of increased ssDNA accumulation and a decreased ability to activate Chk1 to maintain fork stability.
We reasoned that if Tim–Tipin plays a role in replication fork stability solely through facilitating Chk1 phosphorylation, Tim–Tipin reduction combined with ATR deletion should produce a level of genomic instability that is no greater than Tim–Tipin reduction or ATR deletion alone. Alternatively, if Tim–Tipin suppresses the formation of ssDNA, creating a reliance on ATR–Chk1 for fork stability, reduction of Tim–Tipin in combination with complete elimination of ATR-mediated Chk1 activation should synergistically increase genomic instability. Through this genetic approach, we demonstrate that Tim–Tipin dysfunction during otherwise unperturbed DNA replication leads to ssDNA accumulation at replication forks and dramatically increases dependence on the ATR pathway to maintain genome stability and permit the continuation of DNA synthesis.
Results and discussion
Tim–Tipin reduction leads to ssDNA generation at DNA replication forks, causing ATR pathway activation
Because polymerase stalling and DNA damage are each sufficient to cause ssDNA accumulation at the replication fork even in the presence of Tim–Tipin (Byun et al., 2005), we reasoned that the effects of Tim–Tipin reduction on ssDNA generation and ATR activation may be more readily observable during otherwise unperturbed DNA replication. To determine Tim–Tipin's role in suppressing ssDNA accumulation in S phase, Tim-reduced cells were prelabeled with IdU, which afforded the detection of parental ssDNA at replication forks under nondenaturing conditions (Raderschall et al., 1999). Just before cell harvest, active sites of DNA replication were marked by EdU labeling (Buck et al., 2008; Salic and Mitchison, 2008).
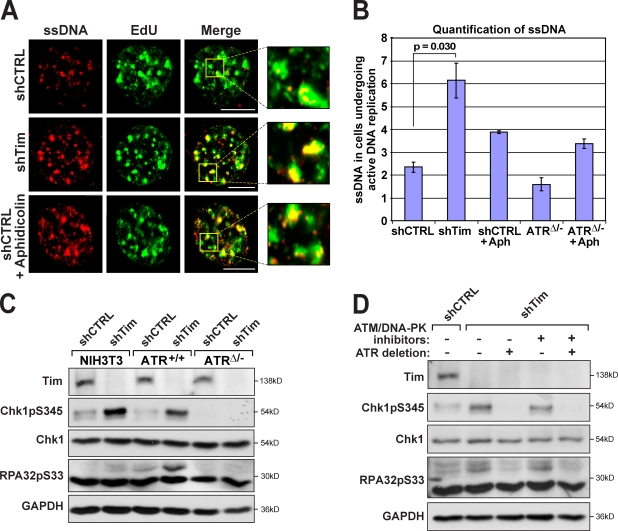
Quantification of ssDNA in cells undergoing DNA replication (EdU positive) indicated that Tim reduction alone was sufficient to cause a 2.6-fold increase in DNA replication–associated ssDNA over that observed in control cells (Fig. 1, A and B; and Fig. S1, A–C). Notably, ssDNA generation occurred specifically at sites of DNA replication, as 70–80% of IdU foci were coincident with regions of EdU incorporation (Fig. 1 A, inset). Because EdU was incorporated just before cell fixation, overlap between ssDNA and EdU-positive regions was unlikely to be caused by DSB resection and, instead, was consistent with a function for Tim–Tipin in suppressing helicase–polymerase uncoupling at active DNA replication forks. Importantly, the increase in replication-associated ssDNA achieved by Tim reduction exceeded that observed in control cells treated with the DNA polymerase inhibitor aphidicolin and was not phenocopied by ATR deletion, either in the absence or presence of aphidicolin (Fig. 1 B). These data indicate that Tim–Tipin functions in suppressing ssDNA generation at DNA replication forks, and this function is distinct from its role in ATR–Chk1 signaling.
Figure 1.
Tim–Tipin reduction leads to ssDNA generation and ATR pathway activation. (A) ssDNA detection at sites of active DNA replication. IdU-labeled cells were subjected to shRNA-mediated Tim (shTim) reduction or control shRNA expression (shCTRL), fixed, and visualized for ssDNA by immunodetection of IdU (red) without prior DNA denaturation. Areas of active DNA replication were marked by pulse labeling with EdU (green) 20 min before collection. EdU detection does not require denaturation. The effect of 5 µM aphidicolin treatment of control cells (30 min) is also shown. Insets show higher magnifications of IdU–EdU overlapping regions (yellow). (B) Quantification of ssDNA levels. Fold increase in ssDNA in EdU-positive cells over ssDNA in EdU-negative cells (three to four independent experiments). The effects of ATR deletion (48 h) on ssDNA accumulation with or without 5 µM aphidicolin (Aph) treatment were also quantified. Error bars represent SEM. (C) Phosphorylation of ATR substrates upon Tim suppression. Phospho-S345 Chk1 (Chk1pS345) and phospho-S33 RPA32 (RPA32pS33; Fanning et al., 2006; Olson et al., 2006) were detected by Western blotting. Tim and ATR suppression was performed as described in Materials and methods. S-phase entry was similar for all conditions, as determined by BrdU incorporation/PI staining (not depicted). Chk1 and GAPDH were detected as loading controls. (D) Phosphorylation of Chk1 and RPA32 upon Tim suppression is dependent on ATR and refractory to ATM/DNA-PK inhibition. Phospho-S345 Chk1 and phospho-S33 RPA32 were detected after a 6-h treatment with ATM and DNA-PK inhibitors, where indicated. Bars, 10 µm.
Although Tim–Tipin reduction partially inhibited aphidicolin-induced phosphorylation of Chk1 by ATR, it did not suppress aphidicolin-induced phosphorylation of RPA (Fig. S2 B). These results demonstrate that Tim–Tipin specifically participates in signaling from ATR to Chk1 but does not facilitate ATR-mediated phosphorylation of more proximal substrates like RPA. To determine whether the high level of ssDNA generated by Tim–Tipin reduction stimulates ATR activity during otherwise unperturbed DNA replication, both RPA and Chk1 phosphorylation were assessed. Consistent with the observed increased ssDNA generation (Fig. 1, A and B), Tim–Tipin reduction caused a significant increase in ATR-mediated phosphorylation of RPA after entry into S phase (Fig. 1, C and D; and Fig. S2 B). Remarkably, even ATR-mediated phosphorylation of Chk1 was increased by Tim–Tipin reduction, demonstrating that the level of ATR activation produced by Tim–Tipin reduction was sufficiently elevated to overcome the partial deficiency in mediating signaling to Chk1 (Fig. 1, C and D; and Fig. S2 B). Together, these findings indicate that Tim–Tipin dysfunction leads to the generation of ssDNA at replication forks, which then promotes activation of ATR.
ATR is required for continued DNA synthesis in instances of Tim–Tipin failure
Reduced expression of Tim–Tipin has previously been shown to slow the overall rate of DNA synthesis (Chou and Elledge, 2006; Gotter et al., 2007; Unsal-Kaçmaz et al., 2007; Yoshizawa-Sugata and Masai, 2007). ATR activation in Tim–Tipin-reduced cells might contribute to this inhibition by initiating a cell cycle checkpoint that prevents subsequent origin firing. Alternatively, ATR–Chk1 pathway activation may sustain the progression of replication forks that have uncoupled as a result of Tim–Tipin dysfunction, which is consistent with ATR's role in preventing the collapse of forks that have stalled because of polymerase inhibition.
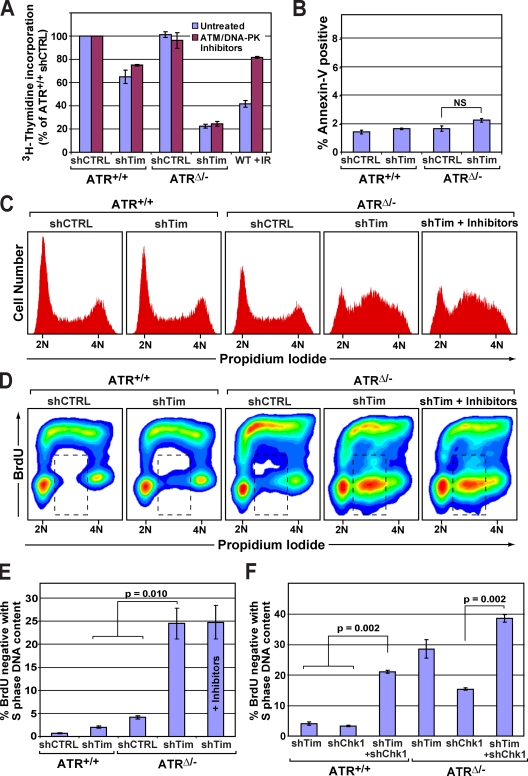
To discriminate between these models, ATR was deleted in Tim–Tipin-reduced cells, and DNA synthesis rates were measured. Consistent with previous studies (Chou and Elledge, 2006; Gotter et al., 2007; Unsal-Kaçmaz et al., 2007; Yoshizawa-Sugata and Masai, 2007), the overall rate of nucleotide incorporation was modestly decreased after Tim–Tipin reduction alone (65% of controls; Fig. 2 A). However, upon ATR deletion, Tim–Tipin-reduced cells exhibited a dramatically reduced rate of DNA synthesis, three- to fivefold slower than that observed after Tim–Tipin reduction or ATR deletion alone (Fig. 2 A).
Figure 2.
The ATR–Chk1 pathway is required for continued DNA synthesis after Tim–Tipin reduction. (A) [3H]thymidine incorporation (60-min pulse) in ATR- and Tim–Tipin-depleted cells. ATM and DNA-PK inhibitors were added 5 h before [3H]thymidine pulse, where indicated. (B) Frequency of apoptosis in ATR- and Tim–Tipin-depleted cells, as determined by Annexin-V and 7-AAD staining. No significant difference was observed between ATR- and Tim–Tipin-depleted cells. (C) Cell cycle analysis in ATR/Tim-deficient and control cells by PI staining. ATM/DNA-PK inhibitor treatments were performed as described in A. ATM/DNA-PK inhibition before S-phase accumulation (18-h treatment) also failed to recover DNA synthesis in ATR/Tim-deficient cells (not depicted). (D) Flow cytometric detection of DNA synthesis in ATR/Tim-deficient and control (CTRL) cells. Cells were labeled with BrdU (20 min), fixed, and costained for BrdU incorporation and DNA content (PI). ATM/DNA-PK inhibitor treatments were performed as described in A. (E and F) Quantification of BrdU-negative cells with S-phase DNA content (between 2N and 4N; dashed boxes in D). The percentage of total cells is shown. (A, B, E, and F) Error bars represent SEM.
It has previously been shown that ATM and DNA-PK are activated after replication fork collapse in ATR-deleted cells (Chanoux et al., 2009). Therefore, it is conceivable that ATM and DNA-PK activation might suppress DNA synthesis in ATR/Tim-reduced cells either by engaging the intra–S phase checkpoint or by inducing apoptosis. However, strongly arguing against these possibilities, treatment of ATR/Tim-reduced cells with ATM and DNA-PK inhibitors failed to lead to any recovery of DNA synthesis despite the ability of these compounds to prevent H2AX phosphorylation and suppress the intra–S phase checkpoint response to ionizing radiation (IR; Fig. 2 A and Fig. S2 C). Furthermore, the frequency of apoptosis was generally low (≤2%) and not significantly elevated in ATR/Tim-deficient cells (Fig. 2 B). Therefore, inhibited DNA replication in ATR/Tim-deficient cells does not appear to be the effect of checkpoint-mediated cell cycle inhibition or apoptosis.
We next investigated whether general inhibition of DNA synthesis in ATR/Tim-deficient cells was caused by reduced rates of DNA synthesis within S phase or failed progression into S phase (G1/G2 buildup). ATR/Tim-deficient and control cells were pulse labeled with BrdU and then fixed and detected for BrdU incorporation and DNA content. As determined by propidium iodide (PI) staining, the frequency of cells with S-phase DNA content (between 2N and 4N) was significantly increased after dual depletion of ATR and Tim–Tipin in comparison with controls (Fig. 2 C and Fig. S1 D). This S-phase enrichment demonstrated that ATR/Tim-depleted cells had the capacity to progress into S phase but were unable to complete it. Remarkably, ATR/Tim-depleted cells demonstrated a complete abrogation of DNA synthesis at mid to late S phase, as indicated by the inability to efficiently incorporate BrdU (Fig. 2, D and E; and Fig. S1 E). The frequency of BrdU-negative cells with S-phase DNA content increased more than fivefold in ATR/Tim-depleted cells over wild-type controls, and this increase was synergistically greater than the additive effects of Tim–Tipin reduction or ATR-deletion alone (Fig. 2, D and E). Notably, treatment of ATR/Tim-depleted cells with ATM and DNA-PK inhibitors failed to rescue arrested DNA synthesis (Fig. 2, C and D), again arguing against the involvement of ATM/DNA-PK–mediated checkpoint mechanisms. These results indicate that combined deficiency in Tim–Tipin and ATR leads to a complete inhibition of DNA synthesis in S phase.
Because Chk1 phosphorylation increased after Tim–Tipin reduction (Fig. 1, C and D; and Fig. S2 B), it was conceivable that Chk1 activation also participates in maintaining DNA synthesis in the absence of Tim–Tipin. Indeed, coreduction of Chk1 and Tim–Tipin abrogated DNA synthesis, largely phenocopying the effect of ATR/Tim deficiency (Fig. 2 F and Fig. S2 D). Similar effects were observed upon inhibition of Chk1 kinase activity (1 µM Gö6976) for as little as 3–6 h (unpublished data), indicating that ATR–Chk1 pathway engagement is required soon after the effects of Tim–Tipin reduction arise.
Recent studies suggest that Chk1 possesses functions that are independent of ATR activation (Rodríguez-Bravo et al., 2006; Yang et al., 2008). Consistent with such roles, ATR deletion in combination with near-complete elimination of Chk1 (Fig. S2 D) led to an elevated frequency of cells with abrogated DNA synthesis (Fig. 2 F). However, this frequency was substantially lower than that observed in ATR/Tim-deficient cells (Fig. 2 F), demonstrating that near-complete Chk1 suppression in ATR-deleted cells does not phenocopy dual deficiency of ATR and Tim–Tipin. Moreover, reduction of Tim–Tipin in ATR/Chk1-depleted cells elevated the frequency of cells with arrested DNA replication well beyond that observed in ATR/Chk1-depleted controls (2.5-fold greater; Fig. 2 F). These results once again indicate a function for Tim–Tipin that is nonepistatic with its participation in ATR–Chk1 signaling and, collectively, demonstrate that activation of the ATR–Chk1 pathway is indispensible for continued DNA replication in instances of Tim–Tipin dysfunction.
Upon Tim–Tipin dysfunction, ATR prevents catastrophic loss of replication fork stability
ATR and ATR orthologues in yeast have previously been shown to stabilize replication forks that have stalled as the result of DNA polymerase inhibition (Tercero and Diffley, 2001; Casper et al., 2002; Brown and Baltimore, 2003; Chanoux et al., 2009). Because polymerase inhibition activates ATR through the generation of ssDNA (Costanzo et al., 2003; Zou and Elledge, 2003; Byun et al., 2005), we reasoned that helicase–polymerase uncoupling that results from Tim–Tipin reduction may similarly lead to an increased reliance on ATR for replication fork stability. Therefore, the complete inhibition of DNA synthesis observed in ATR/Tim-deficient cells may be the product of exacerbated replication fork collapse, degenerating to a point in which reinitiation of replication is no longer possible. If this model is valid, elimination of ATR in Tim–Tipin-reduced cells should cause a synergistic increase in DSBs during otherwise unperturbed DNA replication, and this increase will be associated with DNA replication failure.
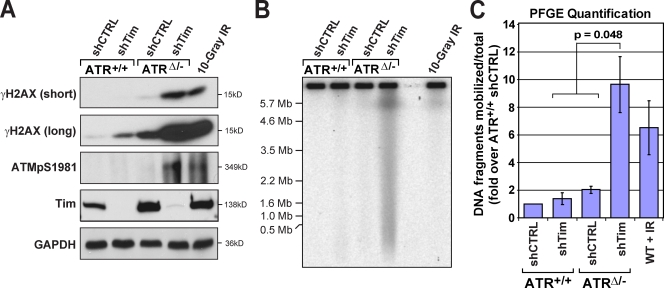
To determine whether combined suppression of ATR and Tim–Tipin is sufficient to cause increased DSB formation during DNA replication, H2AX phosphorylation (γ-H2AX) and the generation of subchromosomal DNA fragments were quantified. Consistent with previous studies (Chou and Elledge, 2006; Chanoux et al., 2009; Urtishak et al., 2009), a small but detectable increase in γ-H2AX was observed after reduction of Tim–Tipin or ATR alone (Fig. 3 A). However, γ-H2AX was dramatically elevated upon deletion of ATR in Tim–Tipin-reduced cells (Fig. 3 A). This increase took place in the absence of exogenous DNA replication inhibitors, indicating that combined suppression of ATR and Tim–Tipin was sufficient to cause increased DSB formation. Similarly, the absence of ATR in Tim–Tipin-reduced cells led to a greater-than-additive increase in the production of subchromosomal DNA fragments (Fig. 3, B and C), as determined by native pulsed field gel electrophoresis (PFGE). The number of DSBs generated after combined suppression of ATR and Tim–Tipin was relatively high. For example, both γ-H2AX and DNA fragment mobilization were generally elevated more by ATR/Tim deficiency than 10 Gy IR (Fig. 3, B and C), a dose estimated to produce hundreds of breaks per mouse diploid genome. Together, these data indicate that ATR strongly suppresses DSB formation upon Tim–Tipin reduction.
Figure 3.
ATR prevents replication fork collapse after Tim–Tipin dysfunction. (A) H2AX and ATM phosphorylation in ATR/Tim-deficient and control (CTRL) cells. Asynchronous ATR/Tim-reduced and control cell lysates were detected for phospho-S139 H2AX (γ-H2AX) and phospho-S1981 ATM (ATMpS1981) by Western blotting. Long and short exposures of γ-H2AX are shown. IR indicates control cells 10 min after treatment with 10 Gy. GAPDH was detected as a loading control. (B) PFGE detection of subchromosomal DNA fragments in asynchronous ATR/Tim-deficient and control cells. A representative SYBR green I–stained agarose gel is shown. (C) Quantification of mobilized DNA fragments shown in B. Mean DNA fragmentation, fold increases over control shRNA–infected ATR+/+ cells, is shown. Error bars represent SEM. WT, wild type.
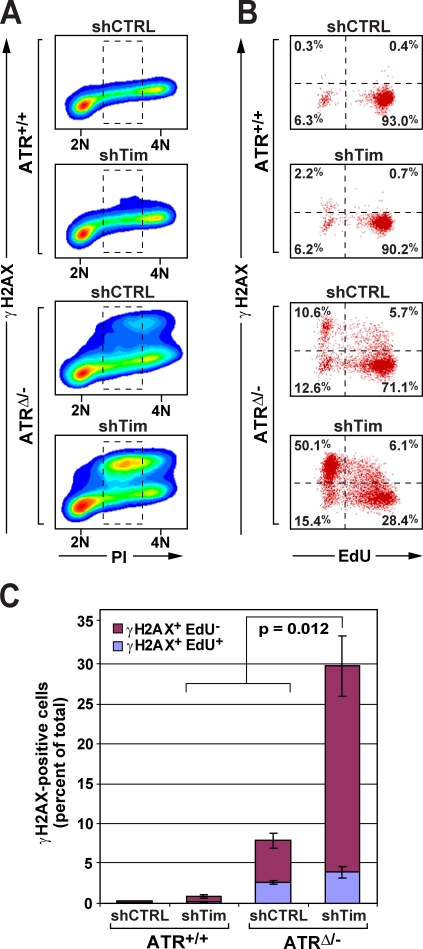
We next examined whether DSB formation correlated with replication failure in ATR/Tim-deficient cells. To do so, ATR/Tim-deficient and control cells were pulse labeled with EdU to measure DNA replication rates just before fixation, and γ-H2AX, DNA content (PI), and nucleotide incorporation (EdU) were then quantified by flow cytometry. H2AX phosphorylation in ATR/Tim-deficient cells was predominantly observed in cells exhibiting S-phase DNA content (Fig. 4 A), confirming that DSBs in ATR/Tim-deficient cells were generated during S phase. Moreover, although cells with undetectable levels of γ-H2AX were largely proficient in DNA synthesis (EdU positive), the vast majority of γ-H2AX–positive ATR/Tim-deficient cells did not incorporate EdU (Fig. 4, B and C). Similar results were observed in ATR/Tipin-deficient cells (Fig. S1, F and G). These data indicate that ATR's indispensible role in promoting continued DNA synthesis upon Tim–Tipin dysfunction may involve the suppression of replication fork collapse.
Figure 4.
DSB generation is associated with replication failure in ATR/Tim-deficient cells. (A) H2AX phosphorylation in ATR/Tim-deficient and control (CTRL) cells. γ-H2AX was immunodetected in asynchronous ATR/Tim-deficient and control cells, costained for DNA content (PI), and visualized by flow cytometry. (B) Quantification of cells undergoing DNA synthesis in γ-H2AX–positive and –negative populations. Cells treated as in A were pulse labeled with EdU (20 min), fixed, and detected for γ-H2AX, EdU incorporation, and DNA content (PI) by flow cytometry. Cells exhibiting S-phase DNA content (dashed boxes in A) were quantified for γ-H2AX staining and EdU incorporation by dot plot. Inset numbers represent quadrant means. (C) DNA replication in γ-H2AX–positive cells. Percentages of total γ-H2AX–positive cells in A that incorporated or failed to incorporate EdU. Error bars represent SEM.
The Tim–Tipin complex plays critical roles in replisome stability, acting both upstream and downstream of ATR
The Tim–Tipin complex has previously been shown to facilitate Chk1 phosphorylation by ATR after exposure to exogenous DNA damage or DNA replication inhibitors (see Introduction; Fig. S2 B). However, in regards to maintaining DNA synthesis and genome stability, our results demonstrate that Tim–Tipin reduction is synthetic (nonepistatic) with ATR–Chk1 deficiency (Figs. 2–4). Therefore, Tim–Tipin must possess functions in S phase that extend beyond simple facilitation of Chk1 phosphorylation. Indeed, our results demonstrate that the Tim–Tipin complex prevents the accumulation of ssDNA at replication forks during otherwise unperturbed DNA replication and that this function does not appear to involve ATR–Chk1 signaling (Fig. 1). These results support a role for the Tim–Tipin complex in maintaining helicase–polymerase coupling and show that deficiency in this function results in ssDNA accumulation and an increased reliance on the ATR–Chk1 pathway to prevent replication fork collapse (Fig. 5).
Figure 5.
Tim–Tipin dysfunction creates an indispensible reliance on the ATR–Chk1 pathway for continued DNA synthesis. Model depicting the proposed relationship between the Tim–Tipin complex and ATR during DNA replication. Tim–Tipin deficiency leads to helicase and polymerase uncoupling, which generates ssDNA and elicits an ATR-dependent response to prevent replication fork collapse. Tim deficiency may cause unregulated DNA unwinding activity while helicase–polymerase physical interactions are maintained, leading to ssDNA accumulation (tan inset). PCNA, proliferating cell nuclear antigen.
Consistent with this model, the genomic instability that results from Tim deficiency (Urtishak et al., 2009) may be the effect of increased ssDNA accumulation in combination with a partial reduction in ATR–Chk1 signaling. According to this model, the level of Chk1 phosphorylation achieved in Tim–Tipin-reduced cells is insufficient to counter all of the effects of ssDNA accumulation, leading to some fork collapse. In this context, deletion of ATR or near-complete suppression of Chk1 in Tim-reduced cells (Fig. 2, E and F; and Fig. S2 D) eliminates residual Chk1 activation, which exacerbates replication fork collapse and ultimately leads to an inability to continue DNA synthesis (Fig. 5). It is possible that the chromosome breaks observed in Tim–Tipin-deficient cells (Urtishak et al., 2009) occur at regions of the genome that may be particularly reliant on this complex to suppress uncoupling events such as replication fork barriers and common fragile sites. Collectively, our findings indicate that the Tim–Tipin complex plays dual roles in replication fork stability by both preventing the accumulation of ssDNA upstream of ATR–Chk1 function and facilitating Chk1 activation downstream of ATR.
ATR has previously been shown to maintain genome stability during S phase even in the absence of exogenous DNA-damaging agents (Brown and Baltimore, 2000, 2003; Chanoux et al., 2009). The requirement for ATR under such circumstances may partly involve polymerase stalling at bases damaged by normal DNA metabolism. Alternatively, the increased reliance of Tim–Tipin-reduced cells on the ATR–Chk1 pathway to maintain genome stability suggests that instances of replisome dysfunction that produce helicase–polymerase uncoupling may contribute to a requirement for ATR in the absence of exogenous DNA damage. Therefore, it is possible that during unperturbed DNA replication, the ATR–Chk1 pathway primarily acts as a homeostatic safeguard, sustaining replication fork integrity upon inevitable stochastic deficiencies in replisome integrity. Thus, by suppressing helicase–polymerase uncoupling, Tim, Tipin, and functionally similar replisome components perform a critical genome stabilizing activity that is largely upstream of that governed by the ATR–Chk1 pathway.
Materials and methods
Lentiviral short hairpin RNA (shRNA) infections, ATR deletion, and cell synchronization
shRNA-expressing lentiviral vectors targeting a control sequence (Addgene plasmid 1864) or Tim (TRCN0000097989; Thermo Fisher Scientific) were used as previously described (Urtishak et al., 2009). shRNA targeting Tipin (TRCN0000123979) was obtained from Thermo Fisher Scientific. shRNA targeting Chk1 (5′-GAAGTTGTTGTAGTGAAAA-3′) was cloned into the H1UG1 vector (provided by F. Xiao-Feng Qin, The University of Texas M.D. Anderson Cancer Center, Houston, TX). Lentiviruses were produced by transfecting lentiviral packaging vectors (pMDLg/pRRE, CMV-VSVG, and RSV-Rev) into 293T cells through calcium phosphate precipitation as previously described (Dull et al., 1998). Lentivirus was then delivered to the cells at a multiplicity of infection of 5–10. ATR expression was eliminated via a Cre/lox-conditional deletion in a 3T3-protocol immortalized cell line derived from ATRflox/−Cre-ERT2+ murine embryonic fibroblasts (Brown and Baltimore, 2003; Ruzankina et al., 2007; Chanoux et al., 2009). ATR+/+Cre-ERT2+ 3T3 cells were generated as a control and treated similarly with 4-hydroxytamoxifen (4-OHT; EMD) treatment for all experiments. Deletion was achieved by treating cells with 0.5 µM 4-OHT at the time of lentiviral infection and incubating for a minimum of 24 h. For experiments in Figs. 2–4, asynchronous ATR+/+Cre-ERT2+ and ATRflox/−Cre-ERT2+ 3T3 cells were infected with shRNA-expressing lentiviruses at the beginning of 4-OHT treatment and collected 48 h later. The level of ATR protein depletion and functional deficiency achieved 48 and 72 h after Cre activation was assessed by Western blotting (Fig. S2, E and F). Cell synchronization for Fig. 1 (B and C) was achieved by infecting cells at ∼80% confluence, followed by a 48-h incubation to induce contact inhibition. To stimulate cell cycle reentry, cells were then replated at low density and then collected at early S phase (between 12 and 15 h depending on the cell line). Similar levels of S-phase entry were controlled by BrdU incorporation/PI staining and flow cytometric analysis.
Immunocytochemical detection of IdU and EdU
For ssDNA detection in NIH3T3 cells, lentiviral infections were performed as described in the previous section. After 24 h, cells were plated onto round coverslips in medium containing 10 µM IdU (Sigma-Aldrich). For ssDNA detection in ATRflox/−Cre-ERT2+ 3T3 cells, cells were incubated in medium containing 10 µM IdU at the time of 4-OHT treatment and then plated onto round coverslips after 24 h. 48 h after ATR deletion or 72 h after lentiviral infection, the cells were pulsed with 10 µM EdU (Invitrogen) for 20 min and then immediately pre-extracted in low salt buffer (10 mM Hepes, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, and 10% glycerol, with 0.1% Triton X-100 added immediately before use) for 5 min at 4°C and fixed with 3% paraformaldehyde/2% sucrose in PBS for 10 min at RT. As a positive control, cells were treated with 5 µM aphidicolin for 30 min immediately after the EdU pulse. Coverslips were permeabilized in 0.5% Triton X-100 in PBS for 5 min at 4°C, stained with anti-BrdU antibodies (BD) without denaturation or nuclease treatment (Raderschall et al., 1999), followed by Rhodamine red-X secondary antibody (Jackson ImmunoResearch Laboratories, Inc.). Nondenaturing “click” chemistry detection of EdU was performed according to the manufacturer's instructions using an Alexa Fluor 488–azide (Invitrogen; Buck et al., 2008; Salic and Mitchison, 2008). Coverslips were then mounted with Vectashield mounting medium containing DAPI (Vector Laboratories). Cells were visualized at RT using a fluorescent microscope (Eclipse 80i; Nikon) using a 60× NA 1.40 objective lens. Images were acquired using a camera (Retiga-SRV; QImaging) and Image-Pro 6.2 software (Media Cybernetics). Image-Pro SharpStack Image Deconvolution software (Media Cybernetics) was used for generating figure images. Mean IdU fluorescence per nuclei was determined by ImageJ (National Institutes of Health) in cells undergoing active DNA replication (EdU-positive cells) and expressed as fold increase over mean fluorescence in cells not undergoing active DNA replication (EdU-negative cells). Fluorescence in 100–250 cells was quantified per condition. Overlap of ssDNA and EdU-incorporating foci indicated sufficient proximity for linear distances to be microscopically indistinguishable, possibly because of the formation of intermingled tertiary structures.
Western blots
Cells were lysed in SDS sample buffer, and protein concentration was determined by bicinchoninic acid assay (Thermo Fisher Scientific). Whole cell lysates were separated by SDS-PAGE and transferred to 0.45-µm polyvinylidene fluoride membranes. Blots were detected for Chk1 (Santa Cruz Biotechnology, Inc.), phospho-S345 Chk1 (Cell Signaling Technology), phospho-S33 RPA32 (Bethyl Laboratories, Inc.), glyceraldehyde 3-phosphate dehydrogenase (GAPDH; United States Biological), γ-H2AX (Millipore), phospho-S1981 ATM (Rockland Immunochemicals), and ATR (Santa Cruz Biotechnology, Inc.). Timeless antibody was provided by P. Minoo (University of Southern California, Los Angeles, CA). Tipin antibody was produced by Josman LLC using full-length recombinant murine Tipin. IR-treated cells (10 Gy γ-irradiation) were collected 10 min after IR treatment (Fig. 3 A).
[3H]thymidine labeling
Cells were grown in the presence of 5 nCi/ml methyl-[14C]thymidine (PerkinElmer) for 48 h, pulsed for 60 min with 4 µCi/ml methyl-[3H]thymidine (PerkinElmer), and collected by scraping, and DNA was isolated using a DNeasy Blood and Tissue kit (QIAGEN). As a control, cells were exposed to 5 Gy IR before the [3H]thymidine pulse. Radioactive thymidine incorporation into DNA was quantified using a liquid scintillation counter (LS-6500; Beckman Coulter). Results were normalized for percent S-phase content and expressed relative to ATR+/+ + control shRNA expression values for each treatment.
PFGE
Cells were collected by trypsinization, washed in PBS, and embedded into agarose plugs in L buffer (100 mM EDTA, 20 mM NaCl, and 10 mM Tris, pH 7.5) with 4 × 105 cells per plug. Plugs were incubated in lysis buffer (L buffer with 1% SDS and 1 mg/ml proteinase K) at 50°C for 24 h. Fresh lysis buffer was added, and the plugs were incubated for an additional 24 h at 50°C. Plugs were washed one time with 10 mM EDTA and 10 mM Tris-Cl, pH 8.0 (TE*), and incubated in TE* for 24 h at RT and then in 1× TAE (40 mM Tris-acetate, 20 mM sodium acetate, and 1 mM EDTA, pH 8.0) for a minimum of 24 h at 4°C. Electrophoresis was performed using a pulsed field electrophoresis system (CHEF-DR II; Bio-Rad Laboratories) in 0.8% PFGE-certified agarose in 1× TAE at 2.8 V/cm with a 400–1,800-s switch time for 60 h at 4°C. DNA was stained using SYBR green I (Invitrogen). Imaging was performed on a scanner (Storm 860; Molecular Dynamics), and quantification was performed using ImageQuant (Molecular Dynamics). S. cerevisiae and S. pombe molecular weight standards (Bio-Rad Laboratories) were used for fragment size estimation. Results represent the DNA mobilized over total (mobilized + well) and are expressed as fold increase over ATR+/+ + control shRNA. IR-treated cells were collected immediately after exposure to 10 Gy γ-irradiation.
Flow cytometric quantification of DNA content, BrdU, γ-H2AX, and EdU
For analysis of DNA content, cells were collected and fixed in 70% EtOH and then stained with PI solution (50 µg/ml PI, 0.1% Triton X-100, 50 µg/ml RNase, and 5 mM EDTA in PBS). For quantification of BrdU incorporation, cells were incubated in 10 µM BrdU (Roche) for 20 min just before collection, fixed in 70% EtOH, acid denatured (3N HCl containing 0.5% Tween 20), and neutralized (0.1 M sodium borate, pH 8.5). After staining with anti-BrdU primary and FITC-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.), cells were stained with PI as described in this section. To determine the percentage of γ-H2AX–positive cells, cells were incubated in 10 µM EdU for 20 min and then collected and fixed in 70% EtOH. Cells were permeabilized (1% BSA and 0.25% Triton X-100 in PBS) and then stained with FITC-conjugated γ-H2AX antibody (Millipore; Zhu and Weiss, 2007). EdU detection was performed according to the manufacturer's instructions with Alexa Fluor 647–azide (Invitrogen; Buck et al., 2008; Salic and Mitchison, 2008); cells were then stained with PI. For each procedure, cells were analyzed by flow cytometry using a FACScalibur (BD). All imaging and quantifications were performed using CellQuest (BD) or FlowJo (Tree Star, Inc.) software.
Apoptosis assays
1 × 105 cells were stained with allophycocyanin-conjugated Annexin-V and 7-AAD in Annexin-V–binding buffer (BD) for 15 min. Apoptosis rates were determined by flow cytometry and gating for Annexin-V–positive/7-AAD–negative events using CellQuest software.
Chemical inhibitors
Aphidicolin (EMD) was used at a final concentration of 5 µM for the indicated times. The ATM inhibitor KU-55933 (EMD) and DNA-PK inhibitor NU7026 (EMD) were each used at a final concentration of 10 µM for the indicated times. The Chk1 inhibitor Gö6976 (EMD) was used at a final concentration of 1 µM for 3 and 6 h.
Replicates and statistical tests
For all data represented in the figures, three to four independent experiments were performed. Error bars represent SEM, and p-values were calculated by the Student's t test.
Online supplemental material
Fig. S1 shows that Tipin reduction causes increased ssDNA generation and an increased reliance on ATR. Fig. S2 shows additional control experiments. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200905006/DC1.
Acknowledgments
We are grateful to the following individuals for their kind assistance: T. Mitchison and A. Salic (EdU protocol), F.B. Johnson and R. Greenberg (PFGE apparatus and fluorescent microscope use, respectively), and P. Minoo (anti-Tim antibody).
This study was funded by the National Institute on Aging (grant R01AG027376) and the Abramson Family Cancer Research Institute.
Footnotes
Abbreviations used in this paper:
- 4-OHT
- 4-hydroxytamoxifen
- DSB
- double-strand break
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- IR
- ionizing radiation
- PFGE
- pulsed field gel electrophoresis
- PI
- propidium iodide
- RPA
- replication protein A
- shRNA
- short hairpin RNA
- ssDNA
- single-stranded DNA
References
- Brown E.J., Baltimore D. 2000. ATR disruption leads to chromosomal fragmentation and early embryonic lethality.Genes Dev. 14:397–402 [PMC free article] [PubMed] [Google Scholar]
- Brown E.J., Baltimore D. 2003. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance.Genes Dev. 17:615–628 doi:10.1101/gad.1067403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck S.B., Bradford J., Gee K.R., Agnew B.J., Clarke S.T., Salic A. 2008. Detection of S-phase cell cycle progression using 5-ethynyl-2′-deoxyuridine incorporation with click chemistry, an alternative to using 5-bromo-2′-deoxyuridine antibodies.Biotechniques. 44:927–929 doi:10.2144/000112812 [DOI] [PubMed] [Google Scholar]
- Byun T.S., Pacek M., Yee M.C., Walter J.C., Cimprich K.A. 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint.Genes Dev. 19:1040–1052 doi:10.1101/gad.1301205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper A.M., Nghiem P., Arlt M.F., Glover T.W. 2002. ATR regulates fragile site stability.Cell. 111:779–789 doi:10.1016/S0092-8674(02)01113-3 [DOI] [PubMed] [Google Scholar]
- Chanoux R.A., Yin B., Urtishak K.A., Asare A., Bassing C.H., Brown E.J. 2009. ATR and H2AX cooperate in maintaining genome stability under replication stress.J. Biol. Chem. 284:5994–6003 doi:10.1074/jbc.M806739200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou D.M., Elledge S.J. 2006. Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function.Proc. Natl. Acad. Sci. USA. 103:18143–18147 doi:10.1073/pnas.0609251103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V., Shechter D., Lupardus P.J., Cimprich K.A., Gottesman M., Gautier J. 2003. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication.Mol. Cell. 11:203–213 doi:10.1016/S1097-2765(02)00799-2 [DOI] [PubMed] [Google Scholar]
- Dull T., Zufferey R., Kelly M., Mandel R.J., Nguyen M., Trono D., Naldini L. 1998. A third-generation lentivirus vector with a conditional packaging system.J. Virol. 72:8463–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico A., Costanzo V., Hunt T. 2007. Tipin is required for stalled replication forks to resume DNA replication after removal of aphidicolin in Xenopus egg extracts.Proc. Natl. Acad. Sci. USA. 104:14929–14934 doi:10.1073/pnas.0706347104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning E., Klimovich V., Nager A.R. 2006. A dynamic model for replication protein A (RPA) function in DNA processing pathways.Nucleic Acids Res. 34:4126–4137 doi:10.1093/nar/gkl550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss E.J. 2001. Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae.Genetics. 157:567–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotter A.L. 2003. Tipin, a novel timeless-interacting protein, is developmentally co-expressed with timeless and disrupts its self-association.J. Mol. Biol. 331:167–176 doi:10.1016/S0022-2836(03)00633-8 [DOI] [PubMed] [Google Scholar]
- Gotter A.L., Suppa C., Emanuel B.S. 2007. Mammalian TIMELESS and Tipin are evolutionarily conserved replication fork-associated factors.J. Mol. Biol. 366:36–52 doi:10.1016/j.jmb.2006.10.097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou Y., Kanoh Y., Bando M., Noguchi H., Tanaka H., Ashikari T., Sugimoto K., Shirahige K. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex.Nature. 424:1078–1083 doi:10.1038/nature01900 [DOI] [PubMed] [Google Scholar]
- Lopes M., Cotta-Ramusino C., Pellicioli A., Liberi G., Plevani P., Muzi-Falconi M., Newlon C.S., Foiani M. 2001. The DNA replication checkpoint response stabilizes stalled replication forks.Nature. 412:557–561 doi:10.1038/35087613 [DOI] [PubMed] [Google Scholar]
- Noguchi E., Noguchi C., Du L.L., Russell P. 2003. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1.Mol. Cell. Biol. 23:7861–7874 doi:10.1128/MCB.23.21.7861-7874.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E., Noguchi C., McDonald W.H., Yates J.R., III, Russell P. 2004. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast.Mol. Cell. Biol. 24:8342–8355 doi:10.1128/MCB.24.19.8342-8355.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E., Nievera C.J., Klimovich V., Fanning E., Wu X. 2006. RPA2 is a direct downstream target for ATR to regulate the S-phase checkpoint.J. Biol. Chem. 281:39517–39533 doi:10.1074/jbc.M605121200 [DOI] [PubMed] [Google Scholar]
- Paulsen R.D., Cimprich K.A. 2007. The ATR pathway: fine-tuning the fork.DNA Repair (Amst.). 6:953–966 doi:10.1016/j.dnarep.2007.02.015 [DOI] [PubMed] [Google Scholar]
- Raderschall E., Golub E.I., Haaf T. 1999. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage.Proc. Natl. Acad. Sci. USA. 96:1921–1926 doi:10.1073/pnas.96.5.1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Bravo V., Guaita-Esteruelas S., Florensa R., Bachs O., Agell N. 2006. Chk1- and claspin-dependent but ATR/ATM- and Rad17-independent DNA replication checkpoint response in HeLa cells.Cancer Res. 66:8672–8679 doi:10.1158/0008-5472.CAN-05-4443 [DOI] [PubMed] [Google Scholar]
- Ruzankina Y., Pinzon-Guzman C., Asare A., Ong T., Pontano L., Cotsarelis G., Zediak V.P., Velez M., Bhandoola A., Brown E.J. 2007. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss.Cell Stem Cell. 1:113–126 doi:10.1016/j.stem.2007.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A., Mitchison T.J. 2008. A chemical method for fast and sensitive detection of DNA synthesis in vivo.Proc. Natl. Acad. Sci. USA. 105:2415–2420 doi:10.1073/pnas.0712168105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommariva E., Pellny T.K., Karahan N., Kumar S., Huberman J.A., Dalgaard J.Z. 2005. Schizosaccharomyces pombe Swi1, Swi3, and Hsk1 are components of a novel S-phase response pathway to alkylation damage.Mol. Cell. Biol. 25:2770–2784 doi:10.1128/MCB.25.7.2770-2784.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero J.A., Diffley J.F.X. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint.Nature. 412:553–557 doi:10.1038/35087607 [DOI] [PubMed] [Google Scholar]
- Unsal-Kaçmaz K., Mullen T.E., Kaufmann W.K., Sancar A. 2005. Coupling of human circadian and cell cycles by the timeless protein.Mol. Cell. Biol. 25:3109–3116 doi:10.1128/MCB.25.8.3109-3116.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsal-Kaçmaz K., Chastain P.D., Qu P.P., Minoo P., Cordeiro-Stone M., Sancar A., Kaufmann W.K. 2007. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement.Mol. Cell. Biol. 27:3131–3142 doi:10.1128/MCB.02190-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urtishak K.A., Smith K.D., Chanoux R.A., Greenberg R.A., Johnson F.B., Brown E.J. 2009. Timeless maintains genomic stability and suppresses sister chromatid exchange during unperturbed DNA replication.J. Biol. Chem. 284:8777–8785 doi:10.1074/jbc.M806103200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.H., Shiotani B., Classon M., Zou L. 2008. Chk1 and Claspin potentiate PCNA ubiquitination.Genes Dev. 22:1147–1152 doi:10.1101/gad.1632808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa-Sugata N., Masai H. 2007. Human Tim/Timeless-interacting protein, Tipin, is required for efficient progression of S phase and DNA replication checkpoint.J. Biol. Chem. 282:2729–2740 doi:10.1074/jbc.M605596200 [DOI] [PubMed] [Google Scholar]
- Zachos G., Rainey M.D., Gillespie D.A.F. 2003. Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects.EMBO J. 22:713–723 doi:10.1093/emboj/cdg060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., Weiss R.S. 2007. Increased common fragile site expression, cell proliferation defects, and apoptosis following conditional inactivation of mouse Hus1 in primary cultured cells.Mol. Biol. Cell. 18:1044–1055 doi:10.1091/mbc.E06-10-0957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Elledge S.J. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes.Science. 300:1542–1548 doi:10.1126/science.1083430 [DOI] [PubMed] [Google Scholar]