It's the kinetochores, not the DNA, that initiate spindle assembly.
Abstract
During mitosis and meiosis in animal cells, chromosomes actively participate in spindle assembly by generating a gradient of Ran guanosine triphosphate (RanGTP). A high concentration of RanGTP promotes microtubule nucleation and stabilization in the vicinity of chromatin. However, the relative contributions of chromosome arms and centromeres/kinetochores in this process are not known. In this study, we address this issue using cells undergoing mitosis with unreplicated genomes (MUG). During MUG, chromatin is rapidly separated from the forming spindle, and both centrosomal and noncentrosomal spindle assembly pathways are active. MUG chromatin is coated with RCC1 and establishes a RanGTP gradient. However, a robust spindle forms around kinetochores/centromeres outside of the gradient peak. When stable kinetochore microtubule attachment is prevented by Nuf2 depletion in both MUG and normal mitosis, chromatin attracts astral microtubules but cannot induce spindle assembly. These results support a model in which kinetochores play the dominant role in the chromosome-mediated pathway of mitotic spindle assembly.
Introduction
Several microtubule-organizing mechanisms cooperate during mitotic spindle assembly in vertebrates (O'Connell and Khodjakov, 2007; Kaláb and Heald, 2008). These include direct “search and capture” of kinetochores by dynamic microtubules emanating from centrosomes (Kirschner and Mitchison, 1986) as well as nucleation and stabilization of microtubules in the vicinity of chromosomes (Carazo-Salas et al., 1999; Kaláb et al., 1999). Complete removal of centrosomes via laser irradiation or conventional micromanipulation does not prevent the formation of a functional bipolar spindle (Khodjakov et al., 2000; Hinchcliffe et al., 2001). Furthermore, recent computer models demonstrate that unbiased search and capture is not efficient enough to assemble a bipolar spindle in the typical time for human cells of 10–15 min (Wollman et al., 2005). Thus, microtubule nucleation and organization near the chromosomes appears to be a principal driver of spindle assembly even in centrosomal cells. This mechanism is thought to rely on the activity of RanGTP, which induces liberation of microtubule nucleation factors sequestered in the complex with importins. The Ran guanine nucleotide exchange factor (RanGEF) RCC1 converts RanGDP to RanGTP in the vicinity of chromatin, whereas Ran GTPase-activating protein (RanGAP) promotes GTP hydrolysis in the cytoplasm. Together, these activities establish a gradient of RanGTP centered on the chromatin (Kaláb et al., 2002, 2006), and it is generally assumed that microtubule nucleation and stabilization are promoted evenly in the volume of cytoplasm immediately surrounding the chromosomes (Bastiaens et al., 2006; Clarke and Zhang, 2008). Indeed, long strings of chromatin-coated beads in Xenopus laevis extracts exhibit microtubule nucleation and polymerization along the entire surface (Gaetz et al., 2006). In contrast, microtubules have been observed growing primarily at or near the primary constriction and rarely along the chromosome arms in cultured cells (Khodjakov et al., 2003; Maiato et al., 2004; Tulu et al., 2006). Thus, the centromere/kinetochore acts as a “hot spot” of microtubule nucleation within the gradient of RanGTP. This pathway is stimulated by the chromosomal passenger complex (CPC) at the centromere, which is probably involved in Aurora B–mediated microtubule stabilization (Sampath et al., 2004). RanGTP is required for kinetochore-mediated microtubule organization (Tulu et al., 2006; Torosantucci et al., 2008), although it is not clear whether its distribution in a gradient affects the ability of the centromere/kinetochore to serve as an efficient microtubule organizer. A recent study demonstrated that spindles can assemble without a RanGTP gradient in Xenopus egg extracts. However, assembly proceeds only when the CPC, which is normally concentrated within the centromere, is active (Maresca et al., 2009).
To better understand the relative contributions of the centrosome, chromatin, and centromeres/kinetochores toward spindle assembly, we turned to human cells undergoing mitosis with unreplicated genomes (MUG) as a model (Brinkley et al., 1988). In these cells, kinetochores with a minimal amount of centromeric DNA become spatially separated from the bulk of chromatin, allowing us to discern the relationships between, kinetochores, chromatin, and the RanGTP gradient.
Results and discussion
The centrosome-independent spindle assembly pathway is active during MUG
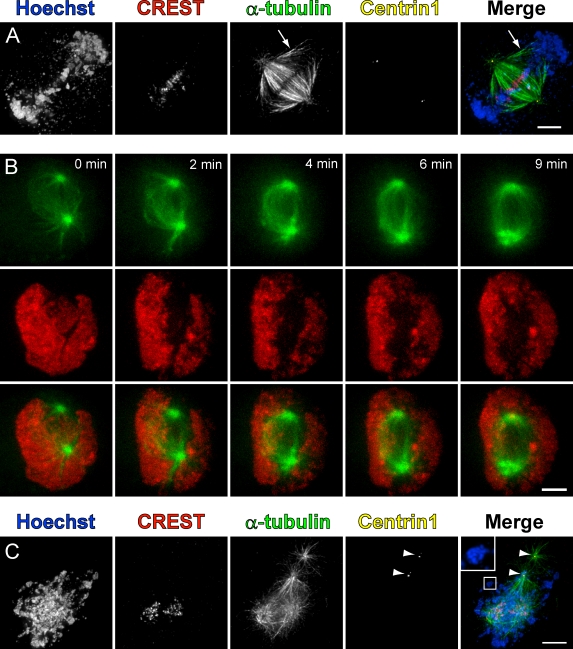
A unique feature of MUG is that the majority of unreplicated kinetochores achieve alignment at the metaphase plate, whereas the bulk of chromatin becomes spatially separated from the kinetochores and pushed to the periphery of the cell (Fig. 1 A). Exclusion of the chromatin from the spindle occurred rapidly upon nuclear envelope breakdown (NEB; Fig. 1 B). A few microtubules were consistently seen extending from the spindle poles toward the peripheral chromatin in fully assembled MUG spindles (Fig. 1 A, arrows). However, centrosomes and the main body of the spindle, which contains the majority of the microtubules, were always centered at kinetochores. This suggests that kinetochores play a larger role in the process of spindle assembly compared with chromatin.
Figure 1.
Organization and generation of spindle microtubules during MUG. (A) Centrin1-GFP–expressing HeLa cell in MUG metaphase fixed and stained for DNA (Hoechst), kinetochores (CREST), and microtubules (α-tubulin). Note the microtubules from the poles connecting to chromatin at the periphery (arrows). Images are maximum intensity projections of deconvolved z slices. (B) Time-lapse images of a cell in MUG expressing H2B-mRFP and α-tubulin–GFP. 0 min is immediately before NEB. (C) Centrin1-GFP–expressing HeLa cell in MUG after nocodazole washout. Microtubules nucleate from the two centriole-containing centrosomes (arrowheads) as well as a noncentrosomal source. A region containing peripheral chromatin is enlarged in the inset. Bars, 5 µm.
To establish whether or not cells retain a centrosome-independent spindle assembly pathway during MUG, hydroxyurea (HU)-arrested centrin1-GFP–expressing HeLa cells were treated with 5 µM nocodazole for 30 min followed by washout to allow microtubule regrowth. Cells were subsequently fixed and stained for DNA, kinetochores, and microtubules. Significant numbers of noncentrosomal microtubules were observed in addition to those emanating from the centrosomes (Fig. 1 C). In agreement with previous studies in live (Khodjakov et al., 2003; Tulu et al., 2006) and fixed cells (Witt et al., 1980), kinetochores were consistently found at or near the foci of noncentrosomal microtubules after nocodazole washout. It should be noted that, shortly after the removal of nocodazole, spatial separation between kinetochores and the chromatin was not pronounced. Thus, we cannot rule out the possibility that factors emitted by the chromatin are required for microtubule nucleation in the vicinity of the kinetochores. However, microtubules were rarely seen forming in association with the chromatin that was not intertwined with kinetochores (Fig. 1 C, inset), suggesting that kinetochores are needed for efficient microtubule nucleation during MUG.
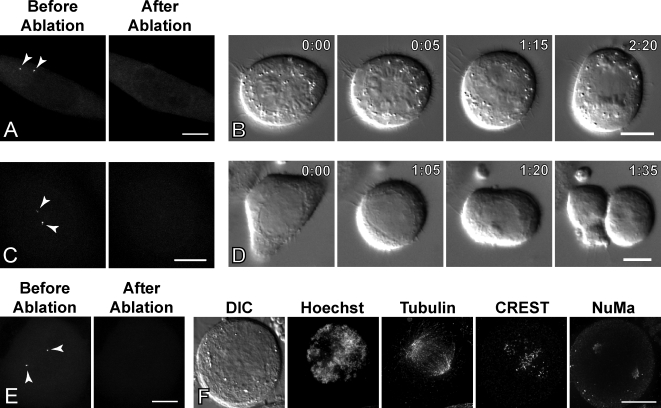
Thus far, our data demonstrate that microtubules can be nucleated at noncentrosomal sites during MUG. However, MUG cells lack the integration of kinetochores and chromatin into a normal, unified mitotic chromosome. This raises the question of whether a functional spindle can assemble purely via centrosome-independent pathways in this system. It has been shown that laser ablation of centrosomes does not prevent formation of a functional bipolar spindle in several types of vertebrate somatic cells (Khodjakov et al., 2000; La Terra et al., 2005). We confirmed this observation by ablating both centrosomes in untreated centrin1-GFP–expressing HeLa cells during G2 (Fig. 2 A). In all cases (n = 5), cells entered mitosis within 6 h after ablation and formed transient monopolar spindles, which subsequently bipolarized (Fig. 2 B, 0:05 and 1:15). The chromosomes achieved proper congression, and cells then exited mitosis after a moderate delay (3.0 ± 2.7 h).
Figure 2.
Bipolar spindle assembly in MUG does not require centrosomes. (A) Centrosomes were ablated in an untreated HeLa cell during G2 by a laser microbeam. (B) DIC images of the same cell throughout mitosis showing eventual chromosome congression and formation of a bipolar spindle (1:15) followed by mitotic exit (2:20). (C) The same approach was used to destroy centrosomes in an HU-arrested cell. (D) After NEB (0:00), a MUG spindle forms and appears as a clear zone in DIC (1:05). The checkpoint is satisfied, and the cell exits (1:20). A cleavage furrow bisects the cell during telophase, demonstrating that a spindle structure is assembled in MUG without centrosomes (1:35). (E) Another cell entered MUG without centrosomes and was fixed ∼30 min after NEB. (F) Distribution of chromatin (Hoechst), tubulin, kinetochores (CREST), and NuMA of the cell in E. Time is shown in hours:minutes. (A, C, and E) Arrowheads point to centrosomes that were ablated using laser microsurgery. Bars: (A, C, and E) 5 µm; (B, D, and F) 10 µm.
To test whether centrosome-independent spindle formation occurs during MUG, we ablated both centrosomes in HU-arrested HeLa cells (Fig. 2 C). The GFP intensity of daughter centrioles increases before MUG entry and is similar to the accumulation of centrin at the daughter centriole in normally cycling cells as they progress from S through G2. This feature was used as a prognostic factor to identify cells that were about to enter MUG. Cells that entered MUG in the absence of centrosomes also assembled spindles. Six cells, which were followed through the entire duration of MUG by time-lapse differential interference contrast (DIC) microscopy, exited slightly faster than controls under the same conditions (2.5 ± 1.5 h; Fig. 2 D). Timely exit from MUG indicates that all kinetochores become attached to microtubules and the mitotic checkpoint is satisfied. We have previously shown that this checkpoint is active during MUG (O'Connell et al., 2008). To determine the organization of microtubules during MUG without centrosomes, several additional cells were fixed and processed for immunofluorescence microscopy ∼30 min after entry into MUG. Tubulin staining in all fixed cells revealed that there were significant numbers of microtubules, accounting for the clear regions observed in DIC (Fig. 2 and Fig. S1). In two of the fixed cells, the spindle was clearly bipolar with well-focused poles. Similar to control spindles, polar regions in acentrosomal spindles during MUG contained the pole-organizing protein NuMA (nuclear–mitotic apparatus protein; Fig. 2 F). In another cell, NuMA localization was poorly defined, and a bipolar organization of microtubules was lacking (Fig. S1 F). We conclude that (a) the normal chromosome/centromere/kinetochore architecture is not an absolute prerequisite for the centrosome-independent spindle assembly and (b) kinetochores are the predominant contributors to this process.
The peak of the RanGTP gradient does not correlate with the position of the spindle during MUG
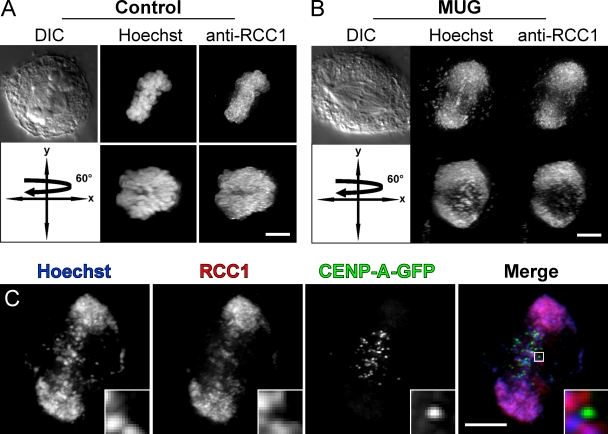
Our observations that centrosome-independent mechanisms for spindle formation are strong during MUG prompted us to examine the distribution of RanGTP relative to the spindle, chromosomes, and kinetochores. First, we compared the distribution of the RanGEF RCC1 between normal HeLa mitosis and MUG. In control cells, RCC1 coated the entire mass of chromosomes, and its distribution during metaphase resembled a disk positioned at the equator (Fig. 3 A). During MUG, RCC1 was predominantly bound to peripheral chromatin, often forming a toroid around the axis of the spindle (Fig. 3 B). Although there were some small pieces of chromatin within the central spindle that also possessed RCC1, kinetochores were consistently devoid of this protein (Fig. 3 C).
Figure 3.
RCC1 binds to chromatin but not kinetochores during MUG. (A) In a normal mitotic cell, the RanGEF RCC1 is evenly distributed on chromosomes. A view rotated by 60° around the y axis shows a uniform coating through the entire volume at which the mitotic spindle assembles. (B) During MUG, RCC1 is also bound to chromatin but is pushed to the side by the spindle that assembles around kinetochores. This is most evident as a hole created by spindle microtubules passing through the chromatin visible in the rotated view. (C) Although antibodies against RCC1 recognize small pieces of equatorial chromatin, centromeres/kinetochores labeled with CENP-A–GFP lack RCC1 staining (insets). Bars, 5 µm.
RCC1 is the principal source of RanGTP. Therefore, the concentration of RanGTP should be highest in the vicinity of the chromatin that becomes distributed to the periphery of the spindle during MUG. To directly test this prediction, we visualized the gradient of RanGTP via a fluorescence resonance energy transfer (FRET) biosensor, Rango-2 (Kaláb and Soderholm, 2009). Rango-2 was transiently transfected into wild-type HeLa cells before treatment with HU. Increased FRET between YPet and CyPet signals cargo liberation induced by the binding of RanGTP to importins and correlates with high local concentration of free RanGTP produced by RCC1.
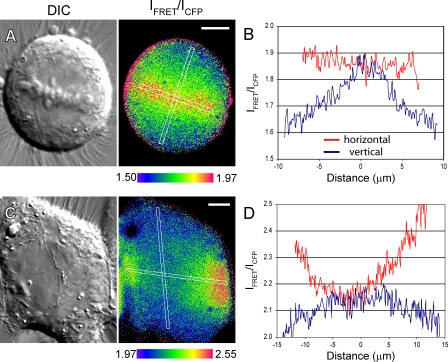
During normal mitosis in HeLa cells, the maximum concentration of RanGTP in the cell corresponds to the position of both the chromatin and centromeres/kinetochores. As a result, FRET was high and relatively uniform when plotted across the spindle equator (Fig. 4, A and B; and Fig. S2 A). When measured along the spindle axis, RanGTP levels gradually decreased toward the spindle poles, forming a gradient, as previously reported (Kaláb et al., 2006).
Figure 4.
The RanGTP gradient peak does not correlate with the spindle during MUG. (A) DIC and FRET/CyPet Rango-2 intensities of a normal mitotic HeLa cell. A uniform gradient of RanGTP-liberated cargo emanates from chromosomes along the entire width of the spindle. (B and D) Linear intensity plots of the cells in A and C averaged over a width of 7 pixels for FRET/CyPet ratios in the regions outlined in white in the vertical and horizontal spindle axis, with the center of each line scan designated as 0 µm. (C) In cells undergoing MUG, the gradient appears discontinuous. When analyzed perpendicular to the equator (red line in D), maximum FRET occurs at the peripheral chromatin and decreases in the volume occupied by the spindle. Bars, 5 µm.
Rango-2 also displayed a higher level of FRET around chromatin during MUG (Fig. 4, C and D; and Fig. S2 B), confirming that RanGTP is generated and induces release of cargo from importins in this system. As predicted by RCC1 immunostaining, the concentration of RanGTP in MUG was maximal near peripheral chromatin, which was identifiable in DIC by its rough granular appearance (Fig. S2 C). Consistent with MUG spindle morphology and in contrast to normal mitosis, cells undergoing MUG showed a dramatic drop in RanGTP levels perpendicular to the spindle axis at the equator (Fig. 4, compare B with D, red line). Even though some small chromatin fragments congressed at the metaphase plate, no detectable hot spots of RanGTP were visible there. Unfortunately, in our conditions, Rango-2 exhibited rapid photobleaching, a property precluding 3D reconstruction of the RanGTP gradient during MUG. Extrapolating from the distribution of chromatin and RCC1, the gradient likely had a toroidal shape as well. Together with time-lapse imaging showing the rapid exclusion of chromatin after NEB (Fig. 1 B), these data demonstrate that MUG spindles preferentially assemble at kinetochores outside the area of maximum RanGTP concentration.
RanGAP catalyzes the hydrolysis of GTP bound to Ran and is critical for maintenance of the directionality of nuclear transport. During mitosis, RanGAP is recruited to kinetochores (Joseph et al., 2002). Although there is no direct evidence that kinetochore-associated RanGAP1 is active, it does raise the possibility that the concentration of RanGTP within the microenvironment around kinetochores is lower as the result of locally increased RanGAP1 activity. RanGAP1 is also present on kinetochores of HeLa cells during MUG, although the staining is of low intensity, as it is in control cells (Fig. S3, A and B). Quantification of fluorescence intensities demonstrates that there is not a statistically significantly difference (P > 0.05) in RanGAP1 levels on kinetochores between mitosis and MUG (Fig. S3). The preferential incorporation of kinetochores into spindles and the marginalization of chromatin during MUG demonstrate that kinetochores remain active microtubule organizers even when they reside outside the peak of the RanGTP gradient. The morphology of spindles during MUG makes it an attractive system for future studies relating to RanGTP microenvironments.
The RanGTP gradient guides microtubule growth toward chromatin
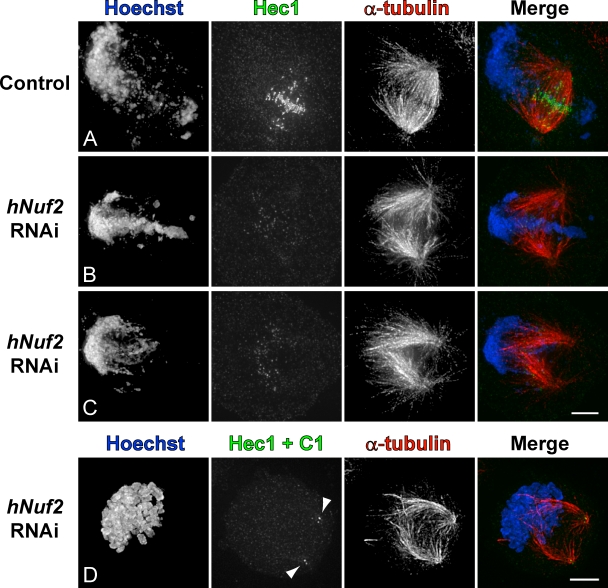
In Xenopus extracts, the RanGTP gradient serves a dual role. It promotes microtubule nucleation (Carazo-Salas et al., 1999; Kaláb et al., 1999) and guides microtubule growth toward the chromatin (Dogterom et al., 1996; Carazo-Salas and Karsenti, 2003; Athale et al., 2008). Less is known about the specific functions of the gradient in somatic cells, although it is clear that the gradient facilitates mitotic spindle assembly (Kaláb et al., 2006). It has been suggested that guiding the growth of centrosome-generated microtubules toward the chromosomes would significantly accelerate search and capture (Wollman et al., 2005). Formally, the RanGTP gradient can provide such guidance. However, the effects of RanGTP on the growth of astral microtubules in cells have not been directly investigated because kinetochores inevitably reside in close spatial proximity to the source of RanGTP (i.e., chromatin). As a result, the distribution of astral microtubules is obscured by the kinetochore fibers (K fibers). The exclusion of chromatin from the spindle during MUG provides a unique advantage for visualizing potential biases in astral microtubule growth (Fig. S3 C). This advantage can be extended by elimination of K fibers. To this end, we depleted Nuf2, a component of the microtubule-binding Ndc80 complex which is required for stable attachment of microtubules to the kinetochore (DeLuca et al., 2002). siRNA depletion reduced Nuf2 levels to ∼20–25%, as determined by quantifying the amount of Hec1, a protein in complex with Nuf2, on kinetochores.
We find that in the absence of K fibers, most astral microtubules grow toward the chromatin during MUG. In cells with a significant amount of chromatin intervening between the centrosomes, this can give the appearance of a spindle (Fig. 5 B). In some cells, the chromatin clustering toward one side is even more pronounced. In these cases, the space between separated centrosomes, where a spindle would be, is almost completely devoid of microtubules (Fig. 5 C), and kinetochores/centromeres are scattered throughout the area occupied by microtubules (Fig. S3 E). Such a microtubule and chromatin distribution in Nuf2-depleted cells is not unique to MUG. Normal mitotic cells depleted of Nuf2 with chromosomes biased toward one side also lack a spindle intervening between centrosomes (Fig. 5 D). It is important to note that this morphology was never observed in control depletions and is unlikely to represent an early stage of mitosis or MUG. Therefore, the RanGTP gradient guides microtubules toward the chromatin during mitosis in somatic cells.
Figure 5.
Chromatin in Nuf2-depleted cells attracts astral microtubules. (A–C) HeLa cells treated with HU were either mock transfected (A) or transfected with siRNA specific for human Nuf2 (hNuf2; B and C). After MUG entry, cells were fixed and stained for chromatin, Hec1, or α-tubulin. (D) Normal mitotic HeLa cell expressing centrin1-GFP (arrowheads) depleted of Nuf2. Bars, 5 µm.
Our three experimental conditions, MUG, acentrosomal MUG, and Nuf2-depleted MUG, allowed us to test various combinations of centrosomes, kinetochores, and chromatin for their ability to form a mitotic spindle. Although chromatin and kinetochores usually work together toward their integration into the spindle, when separated, the kinetochores are a dominant factor, as indicated by the formation of a functional spindle in cells with a mispositioned chromatin/RanGTP gradient during MUG. It is possible that the kinetochore-mediated mechanism is enhanced by the microtubule-nucleating activity of the CPC (Sampath et al., 2004; Maresca et al., 2009), which is enriched at the equator with kinetochores during MUG (O'Connell et al., 2008). However, we found that inhibition of Aurora B, a component of this complex involved in regulating microtubule stability, did not prevent the preferential formation of spindles around kinetochores during MUG (Fig. S3 F). This suggests that another CPC-mediated pathway is responsible.
Intriguingly, unpaired kinetochores detached from the bulk of chromatin can form a spindle even in the absence of centrosomes, albeit with relatively low efficiency. However, our results do not suggest that chromosome arms are irrelevant or the “corpse at a funeral” (Mazia, 1961). The gradient of RanGTP formed by the arms attracts astral microtubules, thus facilitating capture of kinetochores. Although a RanGTP gradient can bias most astral microtubules to grow toward the chromatin, this condition cannot result in the formation of a stable spindle in somatic cells. This is consistent with previous observations that, in the absence of kinetochores, two adjacent centrosomes cannot form a stable spindle structure and ultimately move away from one another (Faruki et al., 2002). However, when kinetochores and centrosomes work together, spindle assembly proceeds even without the RanGTP-mediated activity of the chromatin (Nishitani et al., 1991; Kaláb et al., 2006).
RanGTP-mediated microtubule nucleation and stabilization (Carazo-Salas et al., 1999; Wilde et al., 2001) are undeniably sufficient for the formation of a functional spindle around chromatin-coated beads in Xenopus egg extracts (Heald et al., 1996). However, until now, it has not been possible to determine whether a spindle assembles around “naked” chromatin in somatic cells; kinetochores have always been an integral component of the overall chromosomal structure. Similarly, the fate of kinetochores outside the peak of the RanGTP gradient could not be assessed. We demonstrate that chromatin in MUG is fully competent to liberate microtubule nucleation and stabilization factors through a gradient of RanGTP. However, the role of chromatin appears to be limited to a biasing of search and capture of centrosomal microtubules. Microtubule organization at the kinetochores plays a primary role in chromosome-mediated spindle assembly even when kinetochores are not supported by the maximum RanGTP concentration.
Materials and methods
Cell culture and microscopy
Wild-type and centrin1-GFP (Piel et al., 2000)–, CENP-A (centromere protein A)–GFP (O'Connell et al., 2008)-, and H2B–monomeric RFP (mRFP)/α-tubulin–GFP (Steigemann et al., 2009)-expressing HeLa cells were maintained in DME at 37°C in an atmosphere of 5% CO2. For live cell imaging, coverslips were mounted in Rose chambers containing CO2-independent medium (Invitrogen) and kept at 37°C with a heated enclosure. MUG was observed ∼40 h after mitotic shake off and treatment with 2 mM HU (Sigma-Aldrich; O'Connell et al., 2008). For depolymerization of microtubules, 5 µM nocodazole was added to the culture medium for a minimum of 30 min. Microtubule regrowth was initiated with two 7.5-ml changes of drug-free DME. HU-treated cells were incubated with 100 nM Hesperadin (a gift from T. Kapoor, The Rockefeller University, New York, NY) for >12 h before fixation.
5D time-lapse imaging of α-tubulin–GFP and H2B-mRFP was performed on a spinning disk confocal system (Revolution; Andor Technology) coupled to a microscope (TE2000-E; Nikon). Z series (0.5-µm steps) were obtained with a 100× NA 1.49 Apochromat total internal reflection fluorescence oil immersion objective lens, and maximum intensity projections were made with ImageJ software (National Institutes of Health). For fixed cells, z series (0.2-µm steps) were collected on a DeltaVision system (Applied Precision) with a 100× NA 1.35 UPlan-Apochromat oil immersion objective (Olympus) and subsequently deconvolved using SoftWoRx 2.5 software (Applied Precision). Quantification of RanGAP signal was performed by calculating the integrated pixel intensities in 9 × 9– and 13 × 13–pixel squares centered at a kinetochore. Background and final kinetochore intensity were then calculated as previously described (O'Connell et al., 2008).
RNAi
HeLa cells were shaken off and plated on coverslips and allowed to attach for 1 h before transfection of a siRNA oligonucleotide specific for human Nuf2 (DeLuca et al., 2002) using Oligofectamine (Invitrogen) according to the manufacturer's instructions. At this point, 2 mM HU was also added. An equally effective strategy to deplete Nuf2 in MUG was to transfect the oligonucleotide 6 h before shake off and HU addition. As a control, transfections were performed with Oligofectamine alone.
Immunofluorescence
To costain kinetochores, tubulin, and DNA, cells were washed once with warm PEM (100 mM Pipes, pH 6.9, 2.5 mM EGTA, and 5 mM MgCl2) and extracted with 0.5% Triton X-100 in prewarmed PEM for 1–2 min. This was followed by fixation for 10 min in 4% paraformaldehyde diluted in PEM. The same procedure was used to stain for kinetochores and RanGAP1. For immunofluorescence of cells after laser ablation, the fixation and extraction steps were combined. For RCC1 staining, cells were fixed and permeabilized for 20 min on ice in a 1:1 mixture of methanol and acetone chilled to −20°C. For Hec1 and tubulin staining, cells were extracted in warm PHEM (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 2 mM MgCl2, pH 7.0) for 2 min followed by fixation in PHEM with 4% paraformaldehyde and 0.1% glutaraldehyde for 10 min. Primary antibodies and dilutions used in this study were 1:200 monoclonal anti–α-tubulin (clone DM 1A; Sigma-Aldrich), 1:4,000 rat monoclonal anti–α-tubulin (clone YL1/2; Novus Biologicals), 1:10,000 human CREST (calcinosis, Raynaud's phenomenon, esophageal dysmotility, sclerodactyly, telangiectasia) antiserum (a gift from B. Brinkley, Baylor College of Medicine, Houston, TX), 1:50 polyclonal anti-RCC1 (Millipore), 1:350 monoclonal anti-RanGAP1 (Invitrogen), and 1:1,000 monoclonal anti-Hec1 (a gift from C. Walczak, Indiana University, Bloomington, IN).
Laser ablation
Centrosomes were ablated by targeted destruction of centrioles containing foci of centrin1-GFP. Normally cycling control cells in G2 were selected by the presence of replicated centrioles with bright daughters. A similar criterion was used to select HU-arrested cells that had an increased probability of entry into MUG. Each centriole was destroyed by a pulsed laser (0.56 ns and 532 nm; Teem Photonics) at a frequency of 250 Hz focused to a diffraction-limited spot. Laser power entering the back aperture of the objective lens was ∼0.5 µJ. The GFP signal was monitored after ablation to ensure that centrioles did not reappear before mitosis or MUG, confirming destruction of the centrosome.
FRET
The distribution of the RanGTP gradient in live cells was visualized with Rango-2, an improved version of the FRET biosensor Rango (Kaláb et al., 2006). In Rango-2, the EYFP and cerulean fluorophores were replaced with YPet and CyPet (Nguyen and Daugherty, 2005). Increased FRET results when RanGTP induces the release of Rango-2 from importin-β. The plasmid encoding Rango-2 was transfected into wild-type HeLa cells using FuGENE (Roche) according to the manufacturer's instructions. For FRET detection during MUG, cells were transfected with the same protocol. Approximately 6 h after the addition of DNA–lipid complexes, mitotic cells were collected by shake off and plated with 2 mM HU. Within 40 h, cells began to enter MUG, and YPet, CyPet, and FRET images were obtained with a microscope (TE200; Nikon) with a charge-coupled device camera (Orca II; Hamamatsu Photonics) and a 60× NA 1.4 Plan-Apochromat oil immersion lens. The excitation and emission filters of the CFP–YFP FRET filter set (Chroma Technology Corp.) were mounted in a motorized filter. After background subtraction, 32-bit ratio images were calculated using ImageJ software.
Online supplemental material
Fig. S1 shows the nonpolarized spindle morphology in some cells during MUG without centrosomes. Fig. S2 shows CyPet/YPet Rango-2 intensities during normal mitosis and MUG. Fig. S3 shows immunofluorescence images of cells during MUG in various experimental conditions. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200903076/DC1.
Acknowledgments
The Rango-2 sensor was created in the laboratories of K. Weis and R. Heald at the University of California, Berkeley, Berkeley, CA.
This work was supported by a grant from the National Institutes of Health (GMS 59363) to A. Khodjakov, a Kirschstein National Research Service Award postdoctoral fellowship (GM077911) to C.B. O'Connell, and National Institutes of Health Program Z01 (BC 011084) to P. Kaláb.
Footnotes
Abbreviations used in this paper:
- CPC
- chromosomal passenger complex
- DIC
- differential interference contrast
- FRET
- fluorescence resonance energy transfer
- HU
- hydroxyurea
- K fiber
- kinetochore fiber
- mRFP
- monomeric RFP
- MUG
- mitosis with unreplicated genomes
- NEB
- nuclear envelope breakdown
- RanGAP
- Ran GTPase-activating protein
- RanGEF
- Ran guanine nucleotide exchange factor
References
- Athale C.A., Dinarina A., Mora-Coral M., Pugieux C., Nedelec F., Karsenti E. 2008. Regulation of microtubule dynamics by reaction cascades around chromosomes.Science. 322:1243–1247 doi:10.1126/science.1161820 [DOI] [PubMed] [Google Scholar]
- Bastiaens P., Caudron M., Niethammer P., Karsenti E. 2006. Gradients in the self-organization of the mitotic spindle.Trends Cell Biol. 16:125–134 doi:10.1016/j.tcb.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Brinkley B.R., Zinkowski R.P., Mollon W.L., Davis F.M., Pisegna M.A., Pershouse M., Rao P.N. 1988. Movement and segregation of kinetochores experimentally detached from mammalian chromosomes.Nature. 336:251–254 doi:10.1038/336251a0 [DOI] [PubMed] [Google Scholar]
- Carazo-Salas R.E., Karsenti E. 2003. Long-range communication between chromatin and microtubules in Xenopus egg extracts.Curr. Biol. 13:1728–1733 doi:10.1016/j.cub.2003.09.006 [DOI] [PubMed] [Google Scholar]
- Carazo-Salas R.E., Guarguaglini G., Gruss O.J., Segref A., Karsenti E., Mattaj I.W. 1999. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation.Nature. 400:178–181 doi:10.1038/22133 [DOI] [PubMed] [Google Scholar]
- Clarke P.R., Zhang C. 2008. Spatial and temporal coordination of mitosis by Ran GTPase.Nat. Rev. Mol. Cell Biol. 9:464–477 doi:10.1038/nrm2410 [DOI] [PubMed] [Google Scholar]
- DeLuca J.G., Moree B., Hickey J.M., Kilmartin J.V., Salmon E.D. 2002. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells.J. Cell Biol. 159:549–555 doi:10.1083/jcb.200208159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogterom M., Félix M.A., Guet C.C., Leibler S. 1996. Influence of M-phase chromatin on the anisotropy of microtubule asters.J. Cell Biol. 133:125–140 doi:10.1083/jcb.133.1.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruki S., Cole R.W., Rieder C.L. 2002. Separating centrosomes interact in the absence of associated chromosomes during mitosis in cultured vertebrate cells.Cell Motil. Cytoskeleton. 52:107–121 doi:10.1002/cm.10036 [DOI] [PubMed] [Google Scholar]
- Gaetz J., Gueroui Z., Libchaber A., Kapoor T.M. 2006. Examining how the spatial organization of chromatin signals influences metaphase spindle assembly.Nat. Cell Biol. 8:924–932 doi:10.1038/ncb1455 [DOI] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Blank T., Sandaltzopoulos R., Becker P., Hyman A., Karsenti E. 1996. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts.Nature. 382:420–425 doi:10.1038/382420a0 [DOI] [PubMed] [Google Scholar]
- Hinchcliffe E.H., Miller F.J., Cham M., Khodjakov A., Sluder G. 2001. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase.Science. 291:1547–1550 doi:10.1126/science.1056866 [DOI] [PubMed] [Google Scholar]
- Joseph J., Tan S.H., Karpova T.S., McNally J.G., Dasso M. 2002. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles.J. Cell Biol. 156:595–602 doi:10.1083/jcb.200110109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaláb P., Heald R. 2008. The RanGTP gradient - a GPS for the mitotic spindle.J. Cell Sci. 121:1577–1586 doi:10.1242/jcs.005959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaláb P., Soderholm J. 2009. The design of Förster (fluorescence) resonance energy transfer (FRET)-based molecular sensors for Ran GTPase.Methods. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaláb P., Pu R.T., Dasso M. 1999. The ran GTPase regulates mitotic spindle assembly.Curr. Biol. 9:481–484 doi:10.1016/S0960-9822(99)80213-9 [DOI] [PubMed] [Google Scholar]
- Kaláb P., Weis K., Heald R. 2002. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts.Science. 295:2452–2456 doi:10.1126/science.1068798 [DOI] [PubMed] [Google Scholar]
- Kaláb P., Pralle A., Isacoff E.Y., Heald R., Weis K. 2006. Analysis of a RanGTP-regulated gradient in mitotic somatic cells.Nature. 440:697–701 doi:10.1038/nature04589 [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Cole R.W., Oakley B.R., Rieder C.L. 2000. Centrosome-independent mitotic spindle formation in vertebrates.Curr. Biol. 10:59–67 doi:10.1016/S0960-9822(99)00276-6 [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Copenagle L., Gordon M.B., Compton D.A., Kapoor T.M. 2003. Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis.J. Cell Biol. 160:671–683 doi:10.1083/jcb.200208143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M., Mitchison T. 1986. Beyond self-assembly: from microtubules to morphogenesis.Cell. 45:329–342 doi:10.1016/0092-8674(86)90318-1 [DOI] [PubMed] [Google Scholar]
- La Terra S., English C.N., Hergert P., McEwen B.F., Sluder G., Khodjakov A. 2005. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation.J. Cell Biol. 168:713–722 doi:10.1083/jcb.200411126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Rieder C.L., Khodjakov A. 2004. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis.J. Cell Biol. 167:831–840 doi:10.1083/jcb.200407090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca T.J., Groen A.C., Gatlin J.C., Ohi R., Mitchison T.J., Salmon E.D. 2009. Spindle assembly in the absence of a RanGTP gradient requires localized CPC activity.Curr. Biol. 19:1210–1215 doi:10.1016/j.cub.2009.05.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazia D. 1961. Mitosis and the physiology of cell division. The Cell: Biochemistry, Physiology, Morphology. Brachet J., Mirsky A.E., Academic Press, New York: 77–412 [Google Scholar]
- Nguyen A.W., Daugherty P.S. 2005. Evolutionary optimization of fluorescent proteins for intracellular FRET.Nat. Biotechnol. 23:355–360 doi:10.1038/nbt1066 [DOI] [PubMed] [Google Scholar]
- Nishitani H., Ohtsubo M., Yamashita K., Iida H., Pines J., Yasudo H., Shibata Y., Hunter T., Nishimoto T. 1991. Loss of RCC1, a nuclear DNA-binding protein, uncouples the completion of DNA replication from the activation of cdc2 protein kinase and mitosis.EMBO J. 10:1555–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell C.B., Khodjakov A.L. 2007. Cooperative mechanisms of mitotic spindle formation.J. Cell Sci. 120:1717–1722 doi:10.1242/jcs.03442 [DOI] [PubMed] [Google Scholar]
- O'Connell C.B., Loncarek J., Hergert P., Kourtidis A., Conklin D.S., Khodjakov A. 2008. The spindle assembly checkpoint is satisfied in the absence of interkinetochore tension during mitosis with unreplicated genomes.J. Cell Biol. 183:29–36 doi:10.1083/jcb.200801038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M., Meyer P., Khodjakov A., Rieder C.L., Bornens M. 2000. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells.J. Cell Biol. 149:317–330 doi:10.1083/jcb.149.2.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath S.C., Ohi R., Leismann O., Salic A., Pozniakovski A., Funabiki H. 2004. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly.Cell. 118:187–202 doi:10.1016/j.cell.2004.06.026 [DOI] [PubMed] [Google Scholar]
- Steigemann P., Wurzenberger C., Schmitz M.H., Held M., Guizetti J., Maar S., Gerlich D.W. 2009. Aurora B-mediated abscission checkpoint protects against tetraploidization.Cell. 136:473–484 doi:10.1016/j.cell.2008.12.020 [DOI] [PubMed] [Google Scholar]
- Torosantucci L., De Luca M., Guarguaglini G., Lavia P., Degrassi F. 2008. Localized RanGTP accumulation promotes microtubule nucleation at kinetochores in somatic mammalian cells.Mol. Biol. Cell. 19:1873–1882 doi:10.1091/mbc.E07-10-1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulu U.S., Fagerstrom C., Ferenz N.P., Wadsworth P. 2006. Molecular requirements for kinetochore-associated microtubule formation in mammalian cells.Curr. Biol. 16:536–541 doi:10.1016/j.cub.2006.01.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde A., Lizarraga S.B., Zhang L., Wiese C., Gliksman N.R., Walczak C.E., Zheng Y. 2001. Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities.Nat. Cell Biol. 3:221–227 doi:10.1038/35060000 [DOI] [PubMed] [Google Scholar]
- Witt P.L., Ris H., Borisy G.G. 1980. Origin of kinetochore microtubules in Chinese hamster ovary cells.Chromosoma. 81:483–505 doi:10.1007/BF00368158 [DOI] [PubMed] [Google Scholar]
- Wollman R., Cytrynbaum E.N., Jones J.T., Meyer T., Scholey J.M., Mogilner A. 2005. Efficient chromosome capture requires a bias in the ‘search-and-capture’ process during mitotic-spindle assembly.Curr. Biol. 15:828–832 doi:10.1016/j.cub.2005.03.019 [DOI] [PubMed] [Google Scholar]