Abstract
Mammalian nucleostemin (NS) is a nucleolar guanosine triphosphate-binding protein implicated in cell cycle progression, stem cell proliferation, and ribosome assembly. Drosophila melanogaster contains a four-member nucleostemin family (NS1–4). NS1 is the closest orthologue to human NS; it shares 33% identity and 67% similarity with human NS. We show that NS1 has intrinsic GTPase and ATPase activity and that it is present within nucleoli of most larval and adult cells. Endogenous NS1 and lightly expressed green fluorescent protein (GFP)-NS1 enrich within the nucleolar granular regions as expected, whereas overexpressed GFP-NS1 localized throughout the nucleolus and nucleoplasm, and to several transcriptionally active interbands of polytene chromosomes. Severe overexpression correlated with the appearance of melanotic tumors and larval/pupal lethality. Depletion of 60% of NS1 transcripts also lead to larval and pupal lethality. NS1 protein depletion>95 correlated with the loss of imaginal island (precursor) cells in the larval midgut and to an apparent block in the nucleolar release of large ribosomal subunits in terminally differentiated larval midgut polyploid cells. Ultrastructural examination of larval Malpighian tubule cells depleted for NS1 showed a loss of cytoplasmic ribosomes and a concomitant appearance of cytoplasmic preautophagosomes and lysosomes. We interpret the appearance of these structures as indicators of cell stress response.
INTRODUCTION
Mammalian nucleostemin (NS) is a nucleolar guanosine triphosphate (GTP)-binding protein first characterized in embryonic and neuronal stem cells and in certain cancer cells where it probably plays regulatory roles in cell cycle progression and ribosome biogenesis (Tsai and McKay, 2002, 2005; reviewed by Ma and Pederson, 2008b). Steady-state concentrations of NS drop to undetectable levels just before rat cortical stem cell differentiation (Tsai and McKay, 2002), suggesting that reduced expression of NS regulates stem cell proliferation and differentiation by triggering exit from the cell cycle (Normile, 2002; Tsai and McKay, 2002; Beekman et al., 2006). Mammalian NS rapidly cycles between the nucleolus and the nucleoplasm. Retention of NS within the nucleolus is prolonged when it binds GTP (Misteli, 2005; Tsai and McKay, 2005; Meng et al., 2006, Meng et al., 2007). GTP bound to the central region of NS is thought to stabilize interactions between its amino-terminal basic domain and other nucleolar components, whereas NS redistributes to the nucleoplasm when GTP binding is impaired by mutation (Tsai and McKay, 2005). Coimmunoprecipitation, yeast two-hybrid, and bimolecular fluorescence complementation assays demonstrated an interaction between NS and nucleophosmin (B23) (Ma and Pederson, 2008a), a multifunctional chaperone that probably participates in the later stages of ribosome assembly within the granular component of nucleoli. Similar to NS, nucleophosmin probably plays a role in cell proliferation (Szebeni et al., 2003; Grisendi et al., 2006). Recent studies suggest that NS plays a direct role in ribosome assembly or transport; mammalian NS associates with ribosome assembly factors Pes1, DDX21, and EBP2 in large RNP complexes (Romanova et al., 2009), and the loss of nucleostemin in HeLa cells and Caenorhabditis elegans reduces rRNA levels, perhaps by blocking ribosome release from the nucleoli (Kudron and Reinke, 2008; Romanova et al., 2009). NS displays functional complexity within nucleoli; NS resides in rRNA-deficient subcompartments of nucleolar granular components, suggesting its role in nucleolar functions other than ribosomal subunit assembly and transport (Politz et al., 2005).
NS can interact with p53 (Tsai and McKay, 2002) and MDM2, the ubiquitin E3 ligase specific for p53 (Dai et al., 2008). When overexpressed, NS binds and inhibits MDM2 to stabilize p53, which then arrests cell cycle progression. Conversely, when NS is depleted, MDM2 binds ribosomal proteins L5 and L11, again resulting in p53 activation and cell cycle arrest (Dai et al., 2008). The emerging hypothesis, therefore, describes mammalian NS as a dual-function protein, interacting with nucleolar factors necessary for ribosome biogenesis and with nucleoplasmic factors necessary for cell cycle regulation.
NS is a member of the YawG/YlqF/MMR_HSR1 family of GTPases (Daigle et al., 2002; Leipe et al., 2002; Reynaud et al., 2005). Prokaryotic and eukaryotic members of the family are characterized by circular permutation of their GTP-binding motifs, the order being G5′, G4, G1, G2, and G3 (this is the MMR_HSR1 domain). Comparison of these signature motifs in select eukaryotic family members, including Drosophila NS1, shows a high degree of sequence conservation (Du et al., 2006; Meng et al., 2007). Although the precise functions of eukaryotic MMR_HSR1-containing GTPases remain uncertain, many in yeast (Nug1p and Nog2p/Nug2p in budding yeast; Grn1p in fission yeast) are nucleolar or nuclear proteins that probably participate in the assembly or export of the large ribosomal subunit (Saveanu et al., 2001; Johnson, 2003; Kallstrom et al., 2003; Hedges et al., 2005; Bassler et al., 2006; Du et al., 2006).
Kaplan et al., (2008) described the four-member nucleostemin family in Drosophila. NS1 and NS2 are nucleolar proteins required for viability. NS3 is cytoplasmic, but related to yeast Lsg1p/Kre35, a cytoplasmic GTPase required for the efficient export of the large ribosomal subunit. Loss of NS3 blocks the release of serotonin from a small subset of larval brain neurons that signal the release of insulin from closely apposed neurons. Failure in insulin release from these brain cells causes growth defects in peripheral tissues (Kaplan et al., 2008). Much less is known about NS4; it is apparently dispensable for viability. Of the four family members, NS1 is the closest in sequence homology to mammalian NS. Here, we describe our initial findings of NS1. Although similar in sequence to mammalian NS, we find it expressed in all tissues examined. As a first approach we used RNA interference (RNAi) to deplete NS1 in Drosophila. Depletion of the NS1 causes an apparent loss of larval midgut imaginal island (adult precursor) cells; a block in large ribosomal subunit biogenesis in terminally differentiated polyploid midgut cells of the larva; and a loss of cytoplasmic ribosomes in Malpighian tubule cells leading to autophagy, a stress response.
MATERIALS AND METHODS
Fly Stocks
All Drosophila stocks were maintained at 21–24°C. Stocks obtained from the Bloomington Drosophila Stock Center (Department of Biology, Indiana University, Bloomington, IN) included the homozygous w1118 line (Bloomington stock 3605) used for fly transformation and the homozygous daughterless-GAL4 line (Bloomington stock 8641) used as the principal driver line in most studies reported here. We also used the he third chromosome salivary gland GAL4 driver, P{GawB}AB1 (Bloomington stock 1824). Standard chromosome segregation analyses were performed using the second chromosome balancer stock w*/w*; Sp1/CyO originally from W. M. Saxton (Indiana University), and the third chromosome balancer stock w−/w−; ScmEt50 e/TM3 Sb originally from J. A. Simon (University of Minnesota, Minneapolis, MN).
Protein Expression and Purification
For nucleotide hydrolysis assays, the cDNA encoding full-length Drosophila melanogaster NS1 (AT23067; Drosophila Genomics Resource Center, Bloomington, IN) was amplified by polymerase chain reaction (PCR) with flanking NdeI and HindIII restriction sites and inserted into the NdeI and HindIII sites of the T7 promoter-based expression vector pJES307 (Tabor and Richardson, 1985). The protein was overexpressed in Escherichia coli Rosetta (DE3) cells (Novagen, Madison, WI) by growing the cells in Luria-Bertani media supplemented with 100 μg/ml ampicillin at 37°C to mid-log phase at which point isopropyl-β-d-thiogalactopyranoside was added to 1 mM. The cells were then cultured for 18 h at 16°C and subsequently harvested by centrifugation.
Purification of NS1 from E. coli cells was carried out at 4°C by using standard chromatographic techniques, including Q Sepharose ion exchange, hydroxyapatite, and gel filtration chromatography. The purified protein was concentrated to 1–5 mg/ml in a buffer containing 20 mM Tris-HCl, pH 7.5, 200 mM NaCl, and 5 mM dithiothreitol. The purity of the protein was >95% as judged by standard SDS-polyacrylamide gel electrophoresis (Laemmli, 1970).
Steady-State Nucleotide Hydrolysis Assays
Nucleotide hydrolysis activities of NS1 were determined by measuring the release of free phosphate using the malachite green/ammonium molybdate colorimetric assay (Lanzetta et al., 1979). Reactions were performed using 1 μM NS1 and varying nucleotide concentrations ranging from 10- to 5000-fold molar excess of protein. Hydrolysis reactions were carried out at 37°C in 20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 10 mM MgCl2, and 2 mM dithiothreitol. Initial rates were measured from reactions that proceeded for 30 min before being quenched with malachite green/ammonium molybdate reagent. After 30 min of incubation, color formation was measured at 660 nm by using a DU 640 spectrophotometer (Beckman Coulter, Fullerton, CA). Reactions were repeated a minimum of three times. Values for Km and kcat were determined by fitting the data to the Michaelis–Menton equation with nonlinear regression curve fitting using GraphPad Prism, version 4.0 (GraphPad Software, San Diego, CA).
Antibody Production and Immunoblot Analysis
For antibody production, the full-length Drosophila NS1 cDNA (AT23067) was ligated into pET30a (Novagen). NS1 with the 6XHis tag at its amino terminus was expressed in BL21 (DE3) E. coli cells and purified using a Ni+ affinity column as described by Novagen. A chicken polyclonal antibody was prepared against the purified NS1 by Aves Labs (Tigard, OR).
Standard SDS-polyacrylamide gels were used to resolve proteins from whole larval, adult, or Schneider S2 (S2) culture cell lysates. The proteins were blotted to nitrocellulose for 1 h using the semidry system (Bio-Rad Laboratories, Hercules, CA). Blots were blocked for 1 h in 3% nonfat dry milk that had been reconstituted in 0.9% NaCl (wt/vol), 100 mM Tris, pH 7.4, and 0.1% Tween 20 (TTBS). Blots were probed with the chicken polyclonal antibody described above at 1/1000. The secondary antibody was an affinity-purified, peroxidase-conjugated goat anti-chicken immunoglobulin G (Pierce Chemical, Rockford, IL) diluted 1/500 with TTBS. Peroxidase staining used 3, 3′-diaminobenzidine at 0.8 mg/ml, cobalt chloride at 0.4 mg/ml, and 0.1% H2O2 in 100 mM Tris, pH 7.5.
Plasmid Constructions and Fly Transformations
The Drosophila NS1 cDNA (AT23067) was amplified using 5′-CGACCTCGAGCTCAAGCTTATGGC as the forward primer and 5′-GTCGACGGTACCATAGTGTACTATCTACAG-3′ as the reverse primer. HindIII and KpnI sites (underlined) were included in the forward and reverse primers, respectively. The amplified sequence was ligated into pEGFP-C3 (Clontech, Mountain View, CA). The ATG start codon for NS1 is italicized in the forward primer. We checked continuity of the open reading frame between the green fluorescent protein (GFP) cDNA and the NS1 cDNA by transfecting HeLa cells with this plasmid; GFP-NS1 localized well to human nucleoli. DNA encoding GFP-tagged NS1 was then removed from pEGFP-C3 using NheI and KpnI, and ligated into pBluescript(−) at its SpeI and KpnI sites. DNA encoding GFP-NS1 was next removed from pBS(−) by using NotI and KpnI, and ligated into the same sites within pUAST, a Drosophila P-element–based transformation plasmid (Brand and Perrimon, 1993; Duffy, 2002) that contains the selectable mini-white+ gene with its own promoter, along with tandem yeast GAL4 UASs and the Drosophila Hsp70 promoter that can drive transgene expression when induced with GAL4 or heat shock, respectively. The final pUAST recombinant plasmid and a helper plasmid (pUChsΔ2) encoding transposase were coinjected into homozygous w1118 Drosophila embryos according to established techniques (Spradling, 1986; Kiehart et al., 2000). Seven independent insertion lines were recovered that expressed GFP-NS1. Insertions were mapped to the X, second, or third chromosomes by using standard segregation analyses. The GFP-NS1 A1 line was used most frequently in these studies; it maps to the third chromosome. Homozygous A1 flies were crossed to flies homozygous for daughterless (da)-GAL4 transgene (also on the third chromosome) to induce ectopic expression of GFP-NS1 in heterozygous progeny larvae.
To express mRFP-fibrillarin, mRFP-ribosomal protein (Rp)S6, or mRFP-RpL26 in transgenic flies, we used the topoisomerase-mediated cloning method (Invitrogen, Carlsbad, CA) to ligate the PCR-amplified, full-length cDNAs encoding these various Drosophila proteins (fibrillarin, GM13963; RpS6, UT01917; and RpL26, RE17611, all three from the Drosophila Genomics Resource Center) into pENTR/D-TOPO (Invitrogen). We next recombined the respective cDNAs into the destination plasmid, pTRW, as described by Invitrogen. pTRW (Drosophila Genomics Resource Center) is a modified pUAST (see above); in addition to P-element ends, pTRW contains the selectable mini-white+ gene along with GAL4 UAS promoter sequences that drive expression of down stream cDNAs. Fly transformants expressing monomeric red fluorescent protein (mRFP)-fibrillarin, mRFP-RpS6, or mRFP-RpL26 were prepared as described above. To express GFP-RpL11 in transgenic flies, we amplified expressed sequence tag (EST) LD17235 (Drosophila Genomics Resource Center) and ligated the PCR product into pENTR/D-TOPO (Invitrogen) as described, and then we recombined the RpL11 cDNA into pTGW, a similar pUAST-based transformation plasmid engineered to express GFP-tagged fusions (Drosophila Genomics Resource Center) from GAL4 UASs.
To deplete NS1 by double-stranded RNAi expression, a 441-base pair segment of EST AT23067 that encodes the first 147 amino acids of NS1, was amplified using the forward primer (5′-ATAAGGATCCAGTAGATCTATGGCTTTAAAAAG-3′) containing BamHI and BglII restriction sites (underlined) positioned upstream of the initial ATG codon (italicized). The reverse primer (5′-GTATCCATGGTAGTTCTAGACACCTTGCGGAATTCCTTG-3′) contained underlined KpnI and XbaI sites. The resulting PCR product was first ligated into Drosophila transformation plasmid pUASp-Nba-CS2-BgX (Rorth, 1998; Zhu and Stein, 2004) by using KpnI and BamHI sites downstream of the GAL4 UASs but upstream of the CS2 gene intron. We ligated this first cDNA segment in reverse orientation with respect to the UAS sequences to prevent any possible translation of a truncated NS1 product that could potentially produce dominant negative effects. The second cDNA repeat was ligated down stream of the CS2 intron but in inverted manner with respect to the first element by using BglI and XbaI sites. Transformation was as described above. Seven upstream activation sequence (UAS)-RNAi lines were established. The D1 line was used for most experiments reported here. It maps to the third chromosome and is homozygous viable and fertile.
Microscopy
Preparation of whole mount tissues for immunofluorescence microscopy was performed as described by de Cuevas et al. (1996). In brief, formaldehyde-fixed tissues were probed overnight with the chicken anti-NS1 (described above) diluted 1/250–1/400 in 1× phosphate-buffered saline (PBS) containing 5% normal goat serum and 0.1% Triton (TX)-100. Tissues were washed and reprobed with an AlexaFluor 488-conjugated goat anti-chicken (Invitrogen) at 1/1000 for 4 h. Tissues were then washed in 1× PBS, 0.1%TX-100 as described and counterstained with 4′-6-diamidino-2-phenylindole dihydrochloride (DAPI; Polysciences, Warrington, PA) at 1.0 μg/ml before fluorescence microscopy.
Exogenous GFP-NS1 was expressed in larvae and adults by crossing the transgenic GFP-NS1 A1 line to the da-GAL4 driver line. Tissues were fixed in a formaldehyde-containing buffer as described previously (Cui and DiMario, 2007) before fluorescence microscopy. We used an acid-free chromosome preparation technique (DiMario et al., 2006) to directly visualize GFP-NS1 on spread salivary gland polytene chromosomes. We used an Axioskop (Carl Zeiss, Thornwood, NY) equipped with a SPOT SE digital camera and software (Diagnostic Instruments, Sterling Heights, MI) for phase contrast and conventional fluorescence microscopy. We used an inverted microscope (DMIRE2; Leica Microsystems, Deerfield, IL) equipped with TCS SP2 multiphoton hardware and software (Leica Microsystems) for confocal fluorescence microscopy.
Larval Malpighian tubules were prepared for routine transmission electron microscopy by using standard techniques of formaldehyde and glutaraldehyde fixation, glycine-containing buffer washes, osmium tetroxide postfixation, followed by ethanol dehydration and LR White embedding. We used a 100CX transmission electron microscope (JEOL, Tokyo, Japan) and Kodak negative film to capture images. Negatives were scanned at 1200 dpi, and electronic images were processed using Photoshop (Adobe Systems, Mountain View, CA).
RESULTS
Protein Sequence Conservation
NS1 in D. melanogaster is encoded by Conceptual Gene 3983 (CG3983) in cytological region 89E11 on the right arm of chromosome 3. The deduced protein contains 581 amino acids. Its predicted mass is 66 kDa, and its predicted pI is 9.8 (calculated using the EMBL IEP Service, http://www3.embl.de/cgi/pi-wrapper.pl). Full-length Drosophila NS1 and human NS (549 residues, 62 kDa, calculated pI = 9.5) share ∼33% identity and 67% similarity (Figure 1). GFP-NS1 localized well to HeLa cell nucleoli when expressed by transient transfection (data not shown).
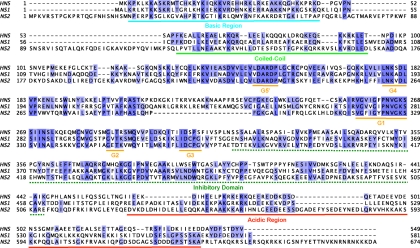
Figure 1.
Sequence comparisons between NS in Homo sapiens (HNS) and NS1 and NS2 in D. melanogaster. NS1 is 33% identical and 67% similar to human NS. Residues of identity are highlighted in blue. The blue line denoting the amino-terminal basic domain marks the region of homology used to categorize NS1 as an orthologue of NS rather than of GN3L3. The amino-terminal basic domain in HNS is ∼43% identical to the corresponding domain in NS1, but only 17% identical to the basic domain in NS2. The green line marks the coiled-coil domain within human NS. The greatest conservations between HNS, NS1, and NS2 occur within the circularly permutated GTP binding domains (G5′–G3, orange underlines). The green dashed line marks the inhibitory domain in HNS that prevents it from entering the nucleolus when GTP is not bound (Tsai and McKay, 2002). The red line marks the acidic carboxy terminal domain in HNS. NS1 has an 11 residue insertion in its carboxy-terminal domain compared with HNS, but it has substantial sequence homology at the beginning and end of the acidic domain with little in between.
GNL3L is another human GTP-binding nucleolar protein related to human NS, but in only the central GTP-binding regions. Although the amino-terminal basic domain of Drosophila NS1 (residues 3–49, blue underline in Figure 1) is ∼43% identical and ∼55% similar to the corresponding region in human NS, it shows no appreciable similarity to the amino terminal region of GNL3L. Therefore, we propose that Drosophila NS1 is more closely related to human NS than it is to human GNL3L. Sequence conservations between the amino-terminal basic domain in human NS and in Drosophila NS1 suggest conserved function(s). Tsai and McKay 2002,Tsai and McKay 2005 showed that the basic domain in human NS is required for nucleolar localization and for p53 interaction. Similarly, the amino-terminal basic domain in Drosophila NS1 is 46% identical and 70% similar to the amino-terminal basic domain in the yeast nucleolar GTPase Nug1p. We note that the basic domain in Nug1p targets it to the nucleolus, mediates its association with the 60S preribosomal particle, and binds RNA (Bassler et al., 2006).
Similar to all MMR_HSR1 GTP-binding proteins, Drosophila NS1 contains circularly permutated GTP-binding domains (orange underlines designated G5′, G4, G3, G2, and G1 in Figure 1). CG6501 in D. melanogaster encodes NS2 (also called Ngp1), a second nucleolar GTPase with permuted GTP-binding domains (Kaplan et al., 2008). BLAST alignments showed an overall 30% identity and 46% similarity between NS1 and NS2 (Figure 1); the greatest homologies were in the permuted GTP-binding motifs and within an inhibitory domain (dashed green underline in Figure 1) that normally retains human NS in the nucleoplasm when GTP is not bound to the up-stream GTP-binding domains (Tsai and McKay, 2005). The acidic carboxy region of human NS (red underline in Figure 1) functions with the inhibitory domain to release NS from the nucleolus (Tsai and McKay, 2005). This acidic region in human NS is more closely related to the carboxy domain of Drosophila NS1 than it is to the carboxy domain of NS2. This is particularly true for the last eight amino acids in NS. The carboxyl terminal domain of NS2 also differs from human NS and Drosophila NS1 in that it contains an additional 16 residue acidic insertion in the middle of the domain, and a basic 29 residue carboxy-terminal extension. The function of these additional sequences remains unknown.
Nucleotide Hydrolysis Assays
To characterize the intrinsic GTPase activity of NS1, purified NS1 was incubated with 10- to 5000-fold molar excess of GTP and the steady-state release of Pi was measured using the malachite green/molybdate colorimetric assay (Table 1). NS1 hydrolyzed GTP with kcat and Km values of 10.4 h−1 and 0.11 mM. Comparable intrinsic rates of hydrolysis have been reported for other circularly permuted GTPases such as E. coli and Bacillus subtilis YjeQ/RsgA/YloQ (Daigle et al., 2002; Himeno et al., 2004; Campbell et al., 2005) and yeast Nug1p (Bassler et al., 2006). NS1 demonstrated equal capacity to hydrolyze ATP with kcat and Km values of 12.7 h−1 and 0.15 mM. NS1 can also hydrolyze cytidine 5′-triphosphate (CTP) and uridine 5′-triphosphate (UTP), with significantly lower catalytic efficiency, as indicated by the kcat/Km values of 0.5 and 1.3 M−1 s−1 compared with 26.3 and 23.5 M−1 s−1 for GTP and ATP, respectively.
Table 1.
Steady-state kinetic parameters describing Drosophila nucleostemin-like proteina
| Substrate | Km (mM) | kcat (h−1) | kcat/Km(M−1 s−1) |
|---|---|---|---|
| GTP | 0.11 ± 0.03 | 10.4 ± 2.1 | 26.3 |
| ATP | 0.15 ± 0.06 | 12.7 ± 2.3 | 23.5 |
| CTP | 0.16 ± 0.04 | 0.3 ± 0.05 | 0.5 |
| UTP | 0.19 ± 0.13 | 0.9 ± 0.03 | 1.3 |
a Nucleotide hydrolysis by NS was assessed by measuring the release of free phosphate using the malachite green/molybdate assay (Lanzetta et al., 1979) as described in Materials and Methods. Values for Vmax, Km, and kcat were derived from a fit to the Michaelis–Menton equation V = (kcat/[E])[S]/(Km + [S]), by nonlinear regression curve fitting using Prism, version 4.0 (GraphPad Software) and are reported as average values with SDs.
Immunolabeling to Locate Native NS1
We expressed Drosophila NS1 in E. coli as a His-tagged fusion and then purified the protein by using nickel affinity columns. A chicken antibody was developed against the purified fusion; it labeled an endogenous protein of ∼72 kDa in extracts of cultured S2 cells (Schneider, 1972) (Figure 2A, lane 2). As a positive control, we prepared a transgenic fly line that expressed GFP-tagged NS1. Transgenic fly lysates contained an expected band at ∼96 kDa (72 kDa plus ∼27 kDa for the GFP tag; Figure 2A, lane 3). Although we failed to see endogenous NS1 in adult lysates, we could detect it in wild-type larval lysates by using similar immunoblotting techniques. We conclude that the chicken antiserum is specific for Drosophila NS1 but that endogenous NS1 may be too scarce in whole adult fly lysates to detect by the Western assays used here.
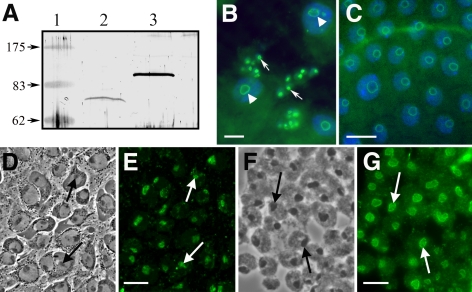
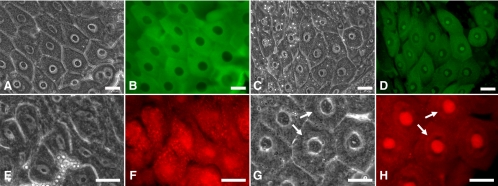
Figure 2.
Endogenous NS1 in Drosophila cells. (A) A Western blot probed with a chicken antiserum raised against Drosophila NS1 shows endogenous NS1 in whole extracts of Drosophila Schneider S2 culture cells at 72 kDa (lane 2) and exogenous GFP-NS1 at 96 kDa in whole extracts from transgenic adult flies (lane 3). Endogenous NS1 was not apparent in the whole fly extracts. (B) Standard immunofluorescence microscopy of a whole-mount midgut isolated from a wild-type, nontransfected third-instar larva. The tissue was dissected into a formaldehyde fixative and probed with the chicken anti-NS1 characterized in A, followed by an Alexa Fluor-conjugated rabbit anti-chicken secondary antibody, and then counterstained with DAPI. The merged image shows enrichment of NS1 within the nucleoli of diploid imaginal island cells (arrows) and within the peripheral (granular) regions of nucleoli of terminally differentiated, polyploid cells (arrowheads). Bar, 50 μm. (C) Adult ovarian follicle cells labeled with the chicken anti-NS1 serum and DAPI. The merged image shows the peripheral granular regions of the nucleoli. These follicle cells are terminally differentiated and polyploid, thus they contain large nucleoli. Bar, 50 μm. (D and E) Phase contrast and immunofluorescent images showing nucleoli in third-instar larval brain cells labeled by the chicken anti-NS1 serum. Black and corresponding white arrows are precisely aligned for comparative purposes. Bar, 50 μm. (F and G) Phase contrast and immunofluorescent images showing nucleoli in third-instar larval imaginal wing disk cells labeled by the chicken anti-NS1 serum. Peripheral granular regions of these nucleoli were preferentially labeled. Black and corresponding white arrows are precisely aligned for comparative purposes. Bar, 25 μm.
Immunofluorescence labeling using the chicken serum was more sensitive than the Western assays. We found native NS1 within the nucleoli of polyploid larval midgut cells (arrowheads in Figure 2B). These cells constitute the bulk of the larval midgut epithelium (Bodenstein, 1950). Smaller imaginal cells (arrows in Figure 2B) exist in clonally derived nests (islands) that are embedded between the larger polyploid cells. These island cells contain NS1 in relatively high concentrations within their smaller nucleoli. In response to ecdysone stimulation, the island cells proliferate during pupation as the larger polyploid cells degenerate (Bodenstein, 1950; Hartenstein and Jan, 1992). In this respect, the island cells are considered progenitors (Tepass and Hartenstein, 1995) that may require NS1 expression for their maintenance (see Figure 4).
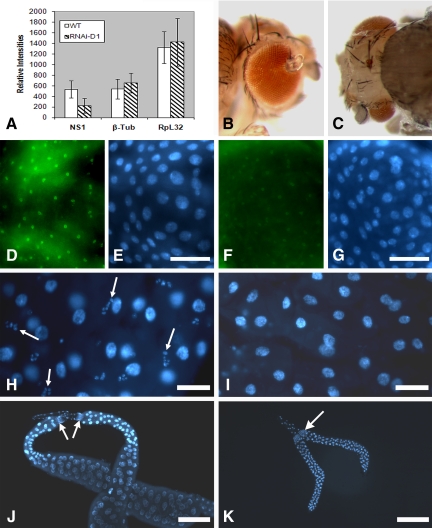
Figure 4.
RNAi-mediated depletion of NS1 in transgenic larvae and surviving adults. (A) Semiquantitative reverse transcription-PCR showed a 60% reduction in NS1 transcripts in RNAi-D1/da-GAL4 larvae versus control larvae. Transcripts encoding beta-tubulin and ribosomal protein L32 were not affected. (B and C) Two separate transgenic flies (RNAi-A2/da-GAL4) expressing RNAi to deplete NS1 displayed variable developmental defects to eyes and head. (D) The chicken anti-NS1 antibody characterized in Figure 2 localized endogenous NS1 in the proventriculus of control third-instar larvae larval. (E) DAPI staining of the tissue in (D). Bar, 200 μm. (F) The same anti-NS1 antibody labeled nucleoli weakly in the proventriculus isolated from RNAi-D1/da-GAL4 larvae. (G) DAPI staining showed the location of the nuclei in (F). Bar, 200 μm. (H) DAPI staining of a wild-type third-instar larval midgut just posterior to the proventriculus. Large polyploid nuclei in the principal midgut epithelial cells are clearly evident. Intermingled with the polyploid cells are nests of imaginal island cells (adult midgut precursors). Arrows point to their DAPI-stained nuclei. Bar, 200 μm. (I) DAPI staining of the midgut from a transgenic RNAi-D1/da-GAL4 larva again just posterior to the proventriculus. Nuclei in the principal midgut epithelial cells are again evident, but the imaginal island cells are missing. Bar, 200 μm. (J) Salivary glands from a wild-type third-instar larva stained with DAPI. Arrows point to the imaginal ring cells. Bar, 1 mm. (K) DAPI-stained salivary glands from a larva transheterozygous for the RNAi-expressing D1 transgene and a salivary gland-specific GAL4 driver (Bloomington stock 1824). The magnification is directly comparable to that in J. The number of nuclei (cells) remained constant, but the cells failed to grow in size. Arrow points to imaginal ring cells which are also comparable in number to wild type. Bar, 1 mm.
Mammalian NS resides in the peripheral granular regions of nucleoli, and as expected, the anti-NS1 serum labeled the peripheral regions of the large nucleoli in polyploid cells (arrowheads in Figure 2B). We observed similar peripheral nucleolar labeling in adult ovarian follicle cells, another terminally differentiated, polyploid cell type (Figure 2C). Besides larval midgut island cells, many other larval precursor cell types labeled with the antiserum. For example, nucleoli in larval brain cells (arrows in Figure 2, D and E) labeled well, and the antibody again preferentially labeled nucleolar peripheral granular regions in wing imaginal disk cells (Figure 2, F and G). Thus, the expression of NS1 is fairly ubiquitous in Drosophila, occurring in both precursor and terminally differentiated cell types. Supporting this claim is the recent finding that nucleostemin (NST-1) expression in C. elegans occurs in both proliferating cells and differentiated cells (Kudron and Reinke, 2008).
Exogenous NS1 Expression
To confirm that Drosophila NS1 localizes to nucleolar granular regions, we expressed both GPF-NS1 and mRFP-fibrillarin in transheterozygous larvae by using Hsp70 promoters, and we examined the location of these two proteins relative to each other in the giant nucleoli of third-instar larval salivary glands. We consider expression from the Hsp70 promoter to be relatively light compared with possible overexpressions achieved by crossing to various GAL4 driver lines. After a brief heat shock and recovery, we fixed the salivary glands, mounted them on microscope slides, and lightly squashed them. Confocal fluorescence microscopy located mRFP-fibrillarin and GFP-NS1 in individual giant nucleoli as shown in Figure 3, A–C (squashing often separated the nucleolus from the polytene chromosomes). Fibrillarin is the phylogenetically conserved nucleolar methyl-transferase associated with box C/D small nucleolar ribonucleoprotein (RNP). It is frequently used as a marker for the dense fibrillar components of nucleoli in which we know pre-rRNA methylation occurs (Tollervey et al., 1993; Cerdido and Medina, 1995). Ectopic mRFP-fibrillarin localized to the dense fibrillar components of the giant nucleolus as expected (Figure 3A). Arrows in Figure 3A denote the fibrillar centers that we know are devoid of fibrillarin. Conversely, GFP-NS1 enriched within distinctly separate subcompartments of the nucleolus, presumably the granular regions that typically surround the dense fibrillar components (Figure 3B). Merging the two images shows a distinct separation of fibrillarin and NS1 (Figure 3C). These localization patterns were similar to those observed by Politz et al. (2005) for fibrillarin and NS in mammalian nucleoli. We conclude that Drosophila NS1, like human NS, enriches specifically within the granular regions of Drosophila nucleoli. To our knowledge, GFP-NS1 is the first marker for the granular regions in Drosophila nucleoli.
Figure 3.
Exogenous GFP-NS1 in Drosophila cells. (A–C) Confocal fluorescence microscopy of a single giant nucleolus from a salivary gland cell of a third-instar transgenic larva that expressed both mRFP-fibrillarin (A) and GFP-NS1 (B) from Hsp70 promoters. Expression levels were considered mild. (A) mRFP-fibrillarin enriched within the dense fibrillar component of the nucleolus. The putative fibrillar centers (arrows) remaining unlabeled. (B) GFP-NS1 enriched within the granular components that typically surround the dense fibrillar components. (C) Merging images A and B clearly shows the separate nucleolar compartments. Bar, 25 μm. (D) Merged standard fluorescence microscopic images of an intact salivary gland nucleus from a third-instar larva that overexpressed GFP-NS1 from GAL4 UASs. The nucleolar dense fibrillar component and the granular regions were both enriched with GFP-NS1, whereas folded polytene chromosomes stained with DAPI. Small amounts of GFP-NS1 were observed in the nucleoplasm, free of the chromosome arms (arrows). Bar, 50 μm. (E) Small portions of salivary gland polytene chromosomes from a transgenic larva that overexpressed GFP-NS1 from GAL4 UASs. The merged image shows GFP-NS1 preferentially associated with one interband (transcriptionally active regions), whereas DAPI stained the condensed banded regions. A survey of the entire polytene chromosome set showed several interbands labeled with GFP-NS1. Bar, 25 μm. (F) A third-instar larva, transheterozygous for GFP-NS1 and da-GAL4 overexpressed GFP-NS1. The anterior is toward the upper left, and the posterior is toward the lower right. Melanotic masses accumulated in the proventriculus (top left arrow), gastric caeca (two arrowheads), and within the intestines (bottom right arrow).
Human NS shuttles between the nucleolus and the nucleoplasm (Tsai and McKay, 2005), but the functional associations between NS and other nuclear components under normal growth conditions remain largely unknown (Ma and Pederson, 2008b). To test whether NS1 has the potential to interact with nuclear components other than nucleoli, we overexpressed GFP-NS1 by crossing GFP-NS1 transgenic flies to transgenic daughterless (da)-GAL4 driver line. The da promoter has fairly ubiquitous activity in most tissues and throughout development. Consequently, transheterozygous progeny displayed copious amounts of GFP-NS1 in nearly all cell types. Figure 3D is a merged image of two conventional fluorescence micrographs showing a large polyploid nucleus from a third-instar larval salivary gland. We found overexpressed GFP-NS1 throughout the nucleolar dense fibrillar components and granular regions. What we interpret to be the fibrillar center contained less GFP-NS1. GFP-NS1 also accumulated within the nucleoplasm (arrows in Figure 3D), free of the DAPI-stained polytene chromosomes. When similar nuclei were squashed to spread the polytene chromosomes, GFP-NS1 seemed to associate with numerous interbands (transcriptionally active euchromatic regions). Figure 3E shows only a few representative interbands labeled by GFP-NS1. The observation suggests that NS1 has the potential to interact with polymerase (Pol) II-generated RNPs. Using the chicken polyclonal antibody, we searched for endogenous NS1 on polytene chromosomes from control glands (i.e., no exogenous GFP-NS1). A few loci labeled weakly, but not consistently, and further study is required to establish an association of endogenous NS1 with RNA Pol II-transcribed loci.
Three of seven transgenic lines contained multiple GFP-NS1 transgene insertions. Severe overexpression of GFP-NS1 in transheterozygous (with da-GAL4) progeny from these three lines showed delayed development compared with wild-type larvae, and they had varying degrees of larval and pupal lethality. Although overall nucleolar structure seemed normal in these larvae, many second- and third-instar larvae from two of the three lines developed melanotic masses within their proventriculus, gastric caeca, and intestines (blue arrows and arrowheads in Figure 3F). These tumor masses were restricted to the digestive tract and were not freely floating as described for many melanotic tumors in Drosophila (Sparrow, 1971).
Phenotypes Caused by the RNAi Depletion of NS1
Two copies of the first 441 bp of the NS1 cDNA coding sequence were ligated into pUASp-Nba-CS2-BgX as inverted repeats on either side of the CS2 intron (Zhu and Stein, 2004). pUASp contains multiple GAL4 UASs that drive transcription when exogenous GAL4 is provided in trans by genetic cross with various Drosophila GAL4 driver lines (Brand and Perrimon, 1993). Seven transgenic lines were recovered that expressed RNAi designed to deplete endogenous NS1 mRNA. We used the D1 line mostly, and the A2 line on occasion. Both lines map to the third chromosome, and both are homozygous viable and fertile.
We crossed the D1 and A2 RNAi lines to the homozygous da-GAL4 driver line (also on the third chromosome). Ninety percent of the RNAi-D1/da-GAL4 progeny die throughout the larval stages. Semiquantitative RT-PCR showed an ∼60% reduction in NS1 transcripts in RNAi-D1/da-GAL4 larvae compared with wild-type larvae (Figure 4A), whereas β-tubulin and RpL32 mRNAs levels were unaffected. RNAi-A2/da-GAL4 individuals died mostly during the larval stages, but a few survived to adulthood; many of the surviving flies displayed variable head, eye, and antennal/aristae defects (Figure 4, B and C). For example, the fly shown in Figure 4B displayed ectopic antennal segments growing out of the eye, whereas the fly in Figure 4C showed a small left eye, mispositioned left antennal segments and arista, missing median and left ocelli, and missing ocellar and interocellar bristles. These malformations were quite variable.
Immunofluorescence microscopy showed the substantial loss of NS1 in most tissues from RNAi-D1/da-GAL4 larvae. For example, the proventriculus isolated from a wild-type larva (Figure 4, D and E) showed ample antibody labeling, whereas the proventriculus isolated from a RNAi-D1/da-GAL4 larva (Figure 4, F and G) showed reduced labeling.
One of the most significant phenotypes observed in RNAi-D1/da-GAL4 larvae in terms of viability was the apparent loss of midgut imaginal island cells. These cells arise in embryonic stage 11 (5.20–7.20 h after fertilization at 25°C), they undergo limited cell division during the larval stages, but then upon ecdysone stimulation proliferate extensively during pupation to form the adult midgut (Skaer, 1993; Tepass and Hartenstein, 1995). DAPI staining displayed nuclei of both the polyploid epithelial cells and the nested imaginal island cells (arrows) in a region of wild-type larval midgut just posterior the proventriculus (Figure 4H). Island cells were missing or reduced in number in the same midgut region in RNAi-D1/da-GAL4 larvae (Figure 4I). Recall from Figure 2B that the nucleoli in these island cells labeled relatively intensely with anti-NS1, suggesting a robust expression of NS1 in these cells. The correlation of NS1 depletion with the loss of imaginal island cells in RNAi-D1/da-GAL4 larvae suggests that NS1 is required for the maintenance of these particular progenitor cells.
The loss of NS1 also caused reduced polyploid cell growth. We crossed the RNAi-D1 line to a salivary gland-specific GAL4 driver (P{GawB}AB1; Bloomington stock 1824) to deplete NS1 in only the salivary glands in transheterozygous larvae. DAPI-stained salivary glands from a wild-type third-instar larva are shown in Figure 4J. One gland overlaps the other in the image, and although one gland is shown in its entirety, the posterior portion of the other gland was cropped in the preparing the figure. Both glands are connected by a common anterior duct just above the arrows. Each gland has ∼125 polytene nuclei. Salivary glands arise from ectodermal infoldings during embryogenesis, 7 to 8 h after fertilization (Sonnenblick, 1950). From the time they first show up, no further mitoses occur in the cells that eventually become the large polytenic secretory cells. The arrows point directly to diploid imaginal ring cells that further proliferate during pupation to give rise to the adult salivary gland as the large polytenic cells degenerate. There are ∼150 ring cells per gland in a third-instar larva. These third-instar ring cells arise by mitotic division of ∼10 embryonic founder cells that proliferate at the second to third-instar larval molt (Curtiss and Heilig, 1995).
Salivary glands from a RNAi-D1/P{GawB}AB1 larva (Figure 4K) were significantly smaller when compared at the same magnification. The number of polytene nuclei (i.e., cells) in these glands was the same as in the wild-type glands. The number of ring cells in the RNAi-expressing glands (arrow in Figure 4K) also seems to be comparable with wild type. Thus, RNAi-mediated depletion of NS1 by using this particular GAL4 driver neither affected the number of polytenic cells nor the number of imaginal ring cells but it did retard the growth of the polytenic secretory cells. Larvae containing these NS1-depleted glands were viable due to the tissue-restricted expression of GAL4. The only obvious phenotype was the inability of resulting pupae to adhere to the walls of the vial. This was likely due to insufficient glue protein synthesis in the NS1-depleted salivary glands.
Loss of NS1 Blocks Nucleolar Release of the Large Ribosomal Subunit
Yeast GTPases Nug1p and Nog2p/Nug2p are required for the maturation and export of the large ribosomal subunit (Bassler et al., 2001; Saveanu et al., 2001). To test whether Drosophila NS1 is required for large ribosomal subunit maturation/transport, we first examined transgenic larvae that expressed GFP-tagged RpL11 (GFP-RpL11/+; da-GAL4/+). In these control larvae, GFP-RpL11 accumulated presumably within large ribosomal subunits in the cytoplasm of larval midgut epithelial (polyploid) cells (Figure 5, A and B). GFP labeling was barely detectable in the nucleoli, suggesting a minimum retention of GFP-RpL11 in the nucleoli during ribosome biogenesis. These larvae were perfectly healthy, indicating that exogenous expression of GFP-RpL11 had no detrimental effects. However, when we introduced RNAi-D1 on the third chromosome by genetic cross (GFP-RpL11/+; RNAi-D1/da-GAL4), GFP-RpL11 accumulated within the nucleoli (Figure 5, C and D) of the polyploid gut cells, and cytoplasmic labeling declined. Although GFP-RpL11 expression provided the clearest results, we also tested mRFP-RpL26. Control cells showed enhanced nuclear labeling and bright cytoplasmic foci (Figure 5, E and F), but when we introduced RNAi expression again by genetic cross, we observed similar nucleolar accumulations of mRFP-L26 (Figure 5, G and H). The nucleoli in these cells seemed hypertrophied, a common but not consistent phenotype associated with the RNAi-depletion of NS1. Besides mRFP-L26 accumulating in nucleoli, we observed a slight accumulation of mRFP-L26 just on the underside of the nuclear envelope (arrows in Figure 5H). Similar accumulations were not evident with GFP-RpL11. In a third effort, we used mRFP-S6 to monitor the export of the small ribosomal subunit, but we found no significant accumulation of mRFP-RpS6 in nucleoli when NS1 was depleted versus controls. Overall, the observations of Figure 5 indicate that NS1 is required for either the maturation or release of the large ribosomal subunits from the nucleolus to the nucleoplasm, and perhaps in the eventual transport of these subunits from the nucleus to the cytoplasm. RNAi-D1/da-GAL4 larvae are very slow to develop, and most die as pupae. A loss of cytoplasmic ribosomes and thus protein synthesis could explain this slow development and eventual lethality.
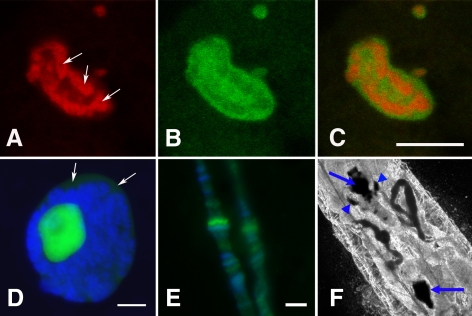
Figure 5.
RNAi-mediated depletion of NS1 causes large ribosomal subunit marker proteins GFP-RpL11 and mRFP-RpL26 to accumulate in nucleoli. (A and B) Phase contrast and fluorescence microscopy of midgut from control larvae (GFP-RpL11/+; +/da-GAL4) that expressed only GFP-RpL11. Heavy cytoplasmic labeling in the polyploid epithelial cells indicated GFP-RpL11 was incorporated into functional ribosomes. These larvae were perfectly healthy. (C and D) Phase contrast and fluorescence micrographs of midgut tissue from a larva (GFP-RpL11/+; RNAi-D1/da-GAL4) that expressed both GFP-RpL11 and RNAi specific for NS1 transcripts. Polyploid epithelial cells displayed increased accumulations of GFP-RpL11 within the nucleoli and reduced accumulations of GFP-RpL11 in the cytoplasm. Bars, 100 μm. (E and F) Midgut from a control larva (mRFP-RpL26/+; +/da-GAL4) that expressed only mRFP-RpL26. Fluorescence was brightest in the nuclei and regions of the cytoplasm, but nucleoli were indistinguishable from the rest of the nucleoplasm. (G and H) Midgut from a transgenic larva (mRFP-RpL26/+; RNAi-D1/da-GAL4) that expressed both mRFP-RpL26 and RNAi to deplete NS1. mRFP-RpL26 accumulated in nucleoli and on the underside of the nuclear envelope (arrows). Bars, 100 μm.
Ultrastructural Phenotypes
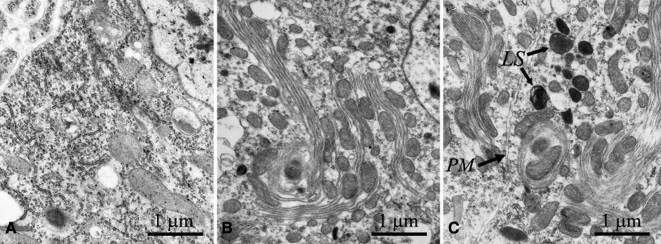
To verify the loss of cytoplasmic ribosomes, we examined Malpighian tubule cells from wild-type and RNAi-D1/da-GAL4 larvae at the ultrastructural level (Figure 6, A–C). The cytoplasm in normal Malpighian tubules (Figure 6A) showed an ample abundance of ribosomes. In contrast, variable numbers of Malpighian cells from RNAi-D1/da-GAL4 larvae showed an obvious reduction in cytoplasmic ribosomes, giving the cytoplasm a “washed out” appearance (Figure 6B). Nucleoli in these NS1-depleted cells showed no apparent morphological disruption at the ultrastructural level. In several cells, we observed extensive stacks of ribosome-free membranes that were in close proximity to clustered mitochondria (Figure 6, B and C). Figure 6C shows two mitochondria completely engulfed by several layers of membrane with several lysosomes in the vicinity. We interpret the stacked membranes, engulfed mitochondria, and concentrated lysosomes as indicators of autophagy (reviewed by McPhee and Baehrecke, 2009).
Figure 6.
Transmission electron microscopy of Malpighian tubules from wild-type control larvae (A) or RNAi-D1/da-GAL4 larvae (B and C). (A) The cytoplasm of control cells contained mitochondria and numerous ribosomes. The nucleus is in the upper right hand corner. (B) A similar region of a Malpighian tubule cell depleted of NS1 by RNAi showed a clear reduction in cytoplasmic ribosomes. Comparatively small mitochondria were found in close apposition to stacks of ribosome-free membrane. Formation of what we interpret to be a preautophagosome is nearly complete in the bottom left-hand quadrant of the image. The nucleus is again in the top right-hand corner. (C) Two adjacent Malpighian tubule cells from an RNAi-D1/da-GAL4 larva showing similar preautophagosome formation. The plasma membranes (PM) of the two cells are marked by the arrow. The cell on the right contains a fully formed autophagosome with two engulfed mitochondria. A cluster of lysosomes (LY) in close vicinity to the autophagosome indicates autophagy or type II programmed cell death as a stress response, possibly due to reduced protein synthesis.
DISCUSSION
Intrinsic GTPase Activity
Sequence analysis indicates that NS1 belongs to the Ylqf/YawG/HSR1_MMR1 class of GTPases characterized by a circularly permuted arrangement of their GTP-binding motifs (Leipe et al., 2002). The intrinsic GTPase activity of Drosophila NS1, kcat 10.4 h−1, is comparable with those reported for other circularly permuted GTPases: E. coli YjeQ (∼11 h−1) and B. subtilis YloQ (3.1 h−1), and to yeast Nog1p (6.1 h−1) (Daigle et al., 2002; Himeno et al., 2004; Campbell et al., 2005; Bassler et al., 2006). Nog1p, however, belongs to the OBG–HflX-like superfamily of GTPases, and its GTP-binding domains are not circularly permuted (Leipe et al., 2002). NS1 is able to hydrolyze ATP with similar efficiency, whereas lower hydrolysis rates were observed for CTP and UTP. This same broad substrate specificity was observed for YjeQ (Daigle et al., 2002; Himeno et al., 2004; Campbell and Brown, 2008). Interestingly, all of the circularly permuted GTPases studied thus far interact with the ribosome in a nucleotide-dependent manner; YjeQ and YloQ associate with bacterial 30S ribosomal subunits, YlqF binds to 50S ribosomal subunit, whereas yeast Nug1p, Nog2p, and Lsg1p associate with 60S ribosomal species (Saveanu et al., 2001; Kallstrom et al., 2003; Daigle and Brown, 2004; Campbell et al., 2005; Bassler et al., 2006; Matsuo et al., 2006). It is likely then that regulation of NS1 occurs, at least in part, through its association with the ribosome.
Endogenous NS1 and Its Depletion by RNAi Expression
We found endogenous NS1 in nearly all cell types examined in Drosophila larvae and adults (Figure 2). Highest steady-state concentrations of NS1 were within the peripheral regions of nucleoli. Our exogenous expression of GFP-NS1 and mRFP-fibrillarin clearly distinguished between the peripheral regions and the dense fibrillar regions. We believe NS1 represents the first Drosophila marker for the nucleolar granular region; a homologue to vertebrate B23 has not been identified in Drosophila. Although the precise functions of nucleostemin have yet to be determined, Romanova et al. (2009) showed that NS in HeLa cells is required for efficient processing of the 32S pre-rRNA intermediate into mature 28S rRNA. They further showed an association between NS and late pre-rRNA processing components Pes1 (a pre-60S component), DDX21 (a helicase required for both 18S and 28S processing), and EBP2 (predicted for 36S to 32S processing). Kudron and Reinke (2008) similarly showed that normal production of 18S and 26S rRNA requires nucleostemin (NST-1) in C. elegans and that loss of NST-1 causes larval arrest.
RNAi-mediated depletion of NS1 leads to variable losses of adult precursor (island) cells normally present within the larval midgut epithelium (Figure 4, H and I), whereas salivary ring cells were not adversely affected. This discrepancy may be a function of the particular GAL4 drivers used. The midgut island cells are considered precursors (Bodenstein, 1950; Tepass and Hartenstein, 1995); the ecdysone regulatory network induces these cells to proliferate during pupation to yield the adult midgut (Li and White, 2003). The island cells derive from the endoderm early in embryonic development (stage 11), but their cell fate specification depends on neurogenic genes (e.g., the Enhancer of split gene complex, Notch, Delta, and others) and proneural genes (e.g., achaete, scute, asense, and others) (Tepass and Hartenstein, 1995). Interestingly, mammalian NS is expressed in neuronal stem cells. The larval midgut island cells consistently labeled intensely with the NS1 antibody (Figure 2B), suggesting an enhanced expression of NS1 in these cells. We are currently tracking NS1 expression in wild-type island cells and salivary gland ring cells at earlier and later stages in development; a loss of NS1 in these cells could arrest cell cycle progression (Ma and Pederson 2007 Ma and Pederson 2008b). If so, the mechanism of cell cycle arrest due to NS1 depletion would be distinct from that in mammalian cells, because Drosophila lacks MDM2, the p53 E3 ubiquitin ligase, and p19Arf, the MDM2 inhibitor (Steller, 2000; Sogame et al., 2003; Brodsky et al., 2004).
Loss of NS1 in the terminally differentiated polyploid midgut cells caused significant accumulation of GFP-RpL11 or mRFP-RpL26 within nucleoli (Figure 5) and an apparent loss of cytoplasmic ribosomes (Figure 6B). Bassler et al. (2001) showed that yeast Nug1p is required for the nuclear export of the large ribosomal subunit. Saveanu et al. (2001) similarly showed that in the absence of Nog2p/Nug2p, the large ribosomal subunit leaves the nucleolus but accumulates in the nucleoplasm without transport to the cytoplasm. Kallstrom et al. (2003) then showed that the loss of yeast nuclear and nucleolar GTPase Nog1p causes the large ribosomal subunit to accumulate preferentially in the nucleolus. Yeast Nug1p and Nog2p/Nug2p, like Drosophila NS1, belong to the same family of nuclear GTPases that possess circular permuted GTP-binding domains. Our combined data suggest that NS1 functions in the nucleolar granular regions of most Drosophila cells, and its depletion perturbs large ribosomal subunit maturation, its release to the nucleoplasm, or perhaps in its export to the cytoplasm.
Possible Links between Nucleostemin and Stress Response
We found Drosophila NS1 in terminally differentiated cells, many of which are nondividing and polyploid with correspondingly large nucleoli. These cells afford us the opportunity to study NS1 in ribosome biogenesis and the responses these cells display when NS1 is lost. Ultrastructural examination showed that larval Malpighian tubule cells depleted for NS1 were variably deficient in cytoplasmic ribosomes (Figure 6B). Many of these same cells contained stacks of ribosome deficient membranes that surrounded and finally engulfed mitochondria (Figure 6, B and C). We interpret these structures as preautophagosomes. The proximity of lysosomes (Figure 6C) suggested autophagy or lysosome-dependent type II programmed cell death, a response known to occur in Drosophila under conditions of starvation and stress (reviewed by McPhee and Baehrecke, 2009).
The target of rapamycin (TOR) pathway (Wullschleger et al., 2006) could potentially link the loss of ribosome production with the onset of autophagy. Specifically, a link between TOR signaling and large ribosomal subunit maturation and export has been established with Nog1p, a GTPase in Saccharomyces cerevisiae that shuttles between the nucleolus and nucleoplasm (Honma et al., 2006). Nog1p is essential for both early and late-stage 60S ribosomal subunit biosynthesis. At restrictive temperatures, nog1-ts mutations cause accumulations of the 60S subunit (marked by RpL25-GFP) in the nucleoplasm rather than the nucleolus, suggesting Nog1p is required primarily for late-stage maturation of the 60S subunit in the nucleoplasm before the subunit's export to the cytoplasm. A block in TOR signaling by either nutrient starvation or rapamycin treatment entrapped Nog1p and its associated proteins within nucleoli. Honma et al. (2006) concluded that TOR signaling is necessary for the release of the Nog1p complex from the nucleolus perhaps after participating in a relatively early step in 60S subunit assembly. Interestingly, another yeast nuclear and nucleolar GTPase, Nog2p, also accumulated within nucleoli when TOR was inactivated, thus supporting the notion that activated TOR kinase regulates ribosome maturation and export. The finding that TOR signaling affects yeast Nog1p and Nog2p suggests a similar link may exist between TOR signaling and Drosophila NS1. Conversely, inactivation of TOR kinase did not enhance nucleolar retention of the permuted GTPase Nug1p, or that of Bsm1, the nuclear and nucleolar factor required for 40S subunit assembly (Honma et al., 2006). Our future efforts will test the hypothesis that TOR signaling regulates ribosome biosynthesis and/or nuclear export by modulating NS1 activity either directly or indirectly.
ACKNOWLEDGMENTS
This work partially fulfills the requirements for R. R.'s Ph.D. thesis in the Department of Biological Sciences, Louisiana State University, Baton Rouge. R. R. was supported by the National Science Foundation's Bridge to Doctorate Program. E. R. was supported by a 2008 summer stipend from the Louisiana Biomedical Research Network. This work was supported by the National Science Foundation grant MCB-0234245 and by a 2007–2008 Council on Research Faculty Research Grant from the Office of Research and Economic Development, Louisiana State University, Baton Rouge.
Abbreviations used:
- GFP
green fluorescent protein
- mRFP
monomeric red fluorescent protein
- NS
nucleostemin
- RNAi
RNA interference
- RNP
ribonucleoprotein
- Rp
ribosomal protein
- TOR
target of rapamycin
- UAS
upstream activation sequence.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-06-0592) on August 26, 2009.
REFERENCES
- Bassler J., Grandi P., Gadal O., Leßmann T., Petfalski E., Tollervey D., Lechner J., Hurt E. Identification of a pre-ribosomal 60S particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- Bassler J., Kallas M., Hurt E. The Nug1 GTPase reveals an N-terminal RNA binding domain that is essential for association with 60S pre-ribosomal particles. J. Biol. Chem. 2006;281:24737–24744. doi: 10.1074/jbc.M604261200. [DOI] [PubMed] [Google Scholar]
- Beekman C., Nichane M., De Clercq S., Maetens M., Floss T., Wurst W., Bellefroid E., Marine J.-C. Evolutionarily conserved role of nucleostemin: controlling proliferation of stem/progenitor cells during early vertebrate development. Mol. Cell. Biol. 2006;26:9291–9301. doi: 10.1128/MCB.01183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenstein D. The postembryonic development of Drosophila. In: Demerec M., editor. Biology of Drosophila. New York: John Wiley & Sons; 1950. pp. 275–367. [Google Scholar]
- Brand A. H., Perrimon N. Targeted gene expression as a means of altering fates and generating phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brodsky M. H., Weinert B. T., Tsang G., Rong Y. S., McGinnis N. M., Golic K. G., Rio D. C., Rubin G. M. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol. Cell Biol. 2004;24:1219–1231. doi: 10.1128/MCB.24.3.1219-1231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell T. L., Daigle D. M., Brown E. D. Characterization of the Bacillus subtilis GTPase YloQ and its role in ribosome function. Biochem. J. 2005;389:843–852. doi: 10.1042/BJ20041873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell T. L., Brown E. D. Genetic interaction screens with ordered overexpression and deletion clone sets implicate the Escherichia coli GTPase YjeQ in late ribosome biogenesis. J. Bacteriol. 2008;190:2537–2545. doi: 10.1128/JB.01744-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdido A., Medina F. J. Subnuclear location of fibrillarin and variation in its levels during the cell cycle and during differentiation of plant cells. Chromosoma. 1995;103:625–634. doi: 10.1007/BF00357689. [DOI] [PubMed] [Google Scholar]
- Cui Z., DiMario P. J. RNAi knockdown of Nopp140 induces Minute-like phenotypes in Drosophila. Mol. Biol. Cell. 2007;18:2179–2191. doi: 10.1091/mbc.E07-01-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtiss J., Heilig J. S. Establishment of Drosophila imaginal precursor cells is controlled by the Arrowhead gene. Development. 1995;121:3819–3825. doi: 10.1242/dev.121.11.3819. [DOI] [PubMed] [Google Scholar]
- Dai M.-S., Sun X.-X., Lu H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol. Cell Biol. 2008;28:4365–4376. doi: 10.1128/MCB.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle D. M., Rossi L., Berghuis A. M., Aravind L., Koonin E. V., Brown E. D. YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with circularly permuted G-motifs and marked burst kinetics. Biochemistry. 2002;41:11109–11117. doi: 10.1021/bi020355q. [DOI] [PubMed] [Google Scholar]
- Daigle D. M., Brown E. D. Studies of the interaction of Escherichia coli YjeQ with the ribosome in vitro. J. Bacteriol. 2004;186:1381–1387. doi: 10.1128/JB.186.5.1381-1387.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cuevas M., Lee J. K., Spradling A. C. α-Spectrin is required for germline cell division and differentiation in the Drosophila ovary. Development. 1996;122:3959–3968. doi: 10.1242/dev.122.12.3959. [DOI] [PubMed] [Google Scholar]
- DiMario P. J., Rosby R., Cui Z. Direct visualization of GFP-fusion proteins on polytene chromosomes. Dros. Infor. Serv. 2006;89:115–118. [Google Scholar]
- Du X., Subba Rao M.R.K., Chen X.-Q., Wu W., Mahalingam S., Balasundaran D. The homologous putative GTPases Grn1p from fission yeast and the human GNL3L are required for growth and play a role in processing of nucleolar pre-rRNA. Mol. Biol. Cell. 2006;17:460–474. doi: 10.1091/mbc.E05-09-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy J. B. GAL4 system in Drosophila: a geneticist's Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- Grisendi S., Mecucci C., Falini B., Pandolfi P. P. Nucleophosmin and cancer. Nat. Rev. Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- Hartenstein V., Jan Y. N. Studying Drosophila embryogenesis with PlacZ enhancer trap lines. Roux's Arch. Dev. Biol. 1992;201:194–220. doi: 10.1007/BF00188752. [DOI] [PubMed] [Google Scholar]
- Hedges J., West M., Johnson A. W. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 2005;24:567–579. doi: 10.1038/sj.emboj.7600547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H., et al. A novel GTPase activated by the small subunit of ribosome. Nucleic Acids Res. 2004;32:5303–5309. doi: 10.1093/nar/gkh861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma Y., Kitamura A., Shioda R., Maruyama H., Ozaki K., Oda Y., Mini T., Jenö P., Maki Y., Yonezawa K., Hurt E., Ueno M., Uritani M., Hall M. N., Ushimaru T. TOR regulates late steps of ribosome maturation in the nucleoplasm via Nog1 in response to nutrients. EMBO J. 2006;25:3832–3842. doi: 10.1038/sj.emboj.7601262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. W. Nuclear export of ribosomal subunits. In: Olson M.O.J., editor. The Nucleolus. New York: Kluwer Academic/Plenum Publishers; 2003. pp. 286–301. [Google Scholar]
- Kallstrom G., Hedges J., Johnson A. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm respectively. Mol. Cell. Biol. 2003;23:4344–4355. doi: 10.1128/MCB.23.12.4344-4355.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D. D., Zimmermann G., Suyama K., Meyer T., Scott M. P. A nucleostemin family GTPase, NS3, acts in serotonergic neurons to regulate insulin signaling and control body size. Genes Dev. 2008;22:1877–1893. doi: 10.1101/gad.1670508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart D. P., Crawford J. M., Montague R. A. Quantitative microinjection of Drosophila embryos. In: Sullivan W., Ashburner M., Hawley R. S., editors. Drosophila Protocols. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 345–359. [Google Scholar]
- Kudron M. M., Reinke V. C. elegans nucleostemin is required for larval growth and germline stem cell division. PLoS Genet. 2008;4:e1000181. doi: 10.1371/journal.pgen.1000181. doi: 10.1371/journal.pgen.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lanzetta P. A., Alvarez L. J., Reinarch P. S., Candia O. A. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- Leipe D. D., Wolf Y. I., Koonin E. V., Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- Li T.-R., White K. P. Tissue-specific gene expression and ecdysone-regulated genomic networks in Drosophila. Dev. Cell. 2003;5:59–72. doi: 10.1016/s1534-5807(03)00192-8. [DOI] [PubMed] [Google Scholar]
- Ma H., Pederson T. Depletion of the nucleolar protein nucleostemin causes G1 cell cycle arrest via the p53 pathway. Mol. Biol. Cell. 2007;18:2630–2635. doi: 10.1091/mbc.E07-03-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Pederson T. Nucleostemin is a binding partner of nucleophosmin in human osteosarcoma cells. Mol. Biol. Cell. 2008a;19:2870–2875. doi: 10.1091/mbc.E08-02-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Pederson T. Nucleostemin: a multiplex regulator of cell-cycle progression. Trends Cell Biol. 2008b;18:575–579. doi: 10.1016/j.tcb.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Matsuo Y., Morimoto T., Kuwano M., Loh P. C., Oshima T., Ogasawara N. The GTP-binding protein YlqF participates in the late step of 50S ribosomal subunit assembly in Bacillus subtilis. J. Biol. Chem. 2006;281:8110–8117. doi: 10.1074/jbc.M512556200. [DOI] [PubMed] [Google Scholar]
- McPhee C. K., Baehrecke E. H. Autophagy in Drosophila melanogaster. Biochim. Biophys. Acta. 2009;1793:1452–1460. doi: 10.1016/j.bbamcr.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Yasumoto H., Tsai R.Y.L. Multiple controls regulate nucleostemin portioning between nucleolus and nucleoplasm. J. Cell Sci. 2006;119:5124–5136. doi: 10.1242/jcs.03292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Zhu Q., Tsai R.Y.L. Nucleolar trafficking of nucleostemin family proteins: common versus protein-specific mechanisms. Mol. Cell Biol. 2007;27:8670–8682. doi: 10.1128/MCB.00635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Going in GTP cycles in the nucleolus. J. Cell Biol. 2005;168:177–178. doi: 10.1083/jcb.200412038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normile D. Common control for cancer, stem cells. Science. 2002;298:1869. doi: 10.1126/science.298.5600.1869. [DOI] [PubMed] [Google Scholar]
- Politz J.C.R., Polena I., Trask I., Bazett-Jones D. P., Pederson T. A nonribosomal landscape in the nucleolus revealed by the stem cell protein nucleostemin. Mol. Biol. Cell. 2005;16:3401–3410. doi: 10.1091/mbc.E05-02-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud E. G., Andrade M. A., Bonneau F., Bach T., Ly N., Knop M., Scheffzek K., Pepperkok R. Human Lsg1 defines a family of essential GTPases that correlates with the evolution of compartmentalization. BMC Biol. 2005;3:21. doi: 10.1186/1741-7007-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova L., Grand A., Zhang L., Rayner S., Katoku-Kikyo N., Kellner S., Kikyo N. Critical role of nucleostemin in pre-rRNA processing. J. Biol. Chem. 2009;284:4968–4977. doi: 10.1074/jbc.M804594200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P. Gal4 in the Drosophila female germline. Mech. Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- Saveanu C., Bienvenu D., Namane A., Gleizes P-E., Gas N., Jacquier A., Fromont-Racine M. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 2001;20:6475–6484. doi: 10.1093/emboj/20.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- Skaer H. The alimentary canal. In: Bate M., Martinez-Arias A., editors. The Development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 941–1012. [Google Scholar]
- Sogame N., Kim M., Abrams J. M. Drosophila p53 preserves genomic stability by regulating cell death. Proc. Natl. Acad. Sci. USA. 2003;100:4696–4701. doi: 10.1073/pnas.0736384100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenblick B. P. The early embryology of Drosophila melanogaster. In: Demerec M., editor. Biology of Drosophila. New York: John Wiley & sons; 1950. pp. 62–167. [Google Scholar]
- Sparrow J. C. Melanotic “tumours.”. In: Ashburner M., Wright T.R.F., editors. The Genetics and Biology of Drosophila. Vol. 2b. New York: Academic Press; 1971. pp. 277–313. [Google Scholar]
- Spradling A. C. P element-mediated transformation. In: Roberts D. B., editor. Drosophila: A Practical Approach. Oxford, United Kingdom: IRL Press; 1986. pp. 175–197. [Google Scholar]
- Steller H. Drosophila p53, meeting the grim reaper. Nat. Cell Biol. 2000;2:E100–E102. doi: 10.1038/35014093. [DOI] [PubMed] [Google Scholar]
- Szebeni A., Hingorani K., Negi S., Olson M.O.J. Role of protein kinase CK2 phosphorylation in the molecular chaperone activity of nucleolar protein B23. J. Biol. Chem. 2003;278:9107–9115. doi: 10.1074/jbc.M204411200. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U., Hartenstein V. Neurogenic and proneural genes control cell fate specification in the Drosophila endoderm. Development. 1995;121:393–405. doi: 10.1242/dev.121.2.393. [DOI] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen H., Jansen R., Kern H., Hurt E. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- Tsai R.Y.L., McKay R. D. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16:2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai R.Y.L., McKay R. D. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J. Cell Biol. 2005;168:179–184. doi: 10.1083/jcb.200409053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M. N. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Zhu X., Stein D. RNAi-mediated inhibition of gene function in the follicle cell layer of the Drosophila ovary. Genesis. 2004;40:101–108. doi: 10.1002/gene.20070. [DOI] [PubMed] [Google Scholar]