Abstract
Deleted in Liver Cancer 1 (DLC1) is a GTPase-activating protein (GAP) with specificity for RhoA, RhoB, and RhoC that is frequently deleted in various tumor types. By inactivating these small GTPases, DLC1 controls actin cytoskeletal remodeling and biological processes such as cell migration and proliferation. Here we provide evidence that DLC1 binds to phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2) through a previously unrecognized polybasic region (PBR) adjacent to its RhoGAP domain. Importantly, PI(4,5)P2-containing membranes are shown to stimulate DLC1 GAP activity in vitro. In living cells, a DLC1 mutant lacking an intact PBR inactivated Rho signaling less efficiently and was severely compromised in suppressing cell spreading, directed migration, and proliferation. We therefore propose that PI(4,5)P2 is an important cofactor in DLC1 regulation in vivo and that the PBR is essential for the cellular functions of the protein.
INTRODUCTION
In recent years, Deleted in Liver Cancer 1 (DLC1) has emerged as an important tumor suppressor gene that may be deleted almost as frequently as p53 (Lahoz and Hall, 2008; Xue et al., 2008). DLC1 was first isolated as a candidate tumor suppressor gene in primary human hepatocellular carcinoma, and loss of expression has subsequently been shown in other tumor types, including colon, breast, prostate, and lung (Durkin et al., 2007b). Transfection of the DLC1 cDNA into different carcinoma cell lines lacking DLC1 expression was then demonstrated to inhibit cell growth and tumorigenicity in nude mice (Ng et al., 2000; Yuan et al., 2003, 2004; Zhou et al., 2004). Comparison of breast cancer sublines by transcriptional profiling revealed that DLC1 expression is linked to their metastatic potential, with down-regulation favoring the formation of pulmonary metastases in athymic mice (Goodison et al., 2005). Recent studies using RNA interference-based approaches provide further proof for a tumor suppressor function of DLC1. Its down-regulation in the breast carcinoma cell lines MCF7 and MDAMB436 promoted enhanced wound closure and Transwell migration via a Dia1-dependent pathway, demonstrating that DLC1 loss is sufficient for the acquisition of a more migratory phenotype (Holeiter et al., 2008). A tumor suppressor function of DLC1 was moreover confirmed in vivo, where DLC1 down-regulation was shown to cooperate with myc overexpression in p53 null cells to promote liver tumorigenesis in mice (Xue et al., 2008). The structurally related genes DLC2 and DLC3 are thought to serve a similar tumor suppressive function, as they are also frequently lost in various tumor types and their reexpression in cancer cells was shown to inhibit proliferation, colony formation, and growth in soft agar (Ching et al., 2003; Durkin et al., 2007a).
DLC1 is a GTPase-activating protein (GAP) protein with in vitro activity for the small GTPases RhoA, RhoB, and RhoC, and to a lesser extent Cdc42 (Wong et al., 2003; Healy et al., 2008). The Rho family of GTPases are important regulators of diverse biological responses, including actin cytoskeletal rearrangements, gene transcription, cell cycle regulation, apoptosis, and membrane trafficking (Jaffe and Hall, 2005; Ridley, 2006). Rho proteins cycle between a GTP-bound active state to interact with effector proteins, modulating their activity and localization, and an inactive GDP-bound state. Activation of Rho proteins is controlled by the guanine nucleotide exchange factors (GEFs), which promote the release of bound GDP and facilitate GTP binding. GAP proteins, on the other hand, are negative regulators that increase the intrinsic GTPase activity of Rho GTPases to accelerate the return to the inactive state (Jaffe and Hall, 2005; Ridley, 2006; Bos et al., 2007). RhoA, RhoB, and RhoC are posttranslationally modified by prenylation of a conserved carboxy-terminal cysteine. The prenyl group anchors the GTPase into membranes, and this modification is essential for activity. Although RhoA and RhoC are geranylgeranylated, RhoB is also farnesylated, which accounts for differences in localization (Wheeler and Ridley, 2004). Another layer of complexity is added by the guanine nucleotide dissociation inhibitors, which sequester Rho GTPases in the cytoplasm by binding to the prenyl group (Jaffe and Hall, 2005; Ridley, 2006).
Active RhoA, RhoB, and RhoC promote the formation of actin stress fibers and focal adhesions (Wheeler and Ridley, 2004). In accordance with its RhoGAP function, microinjection of p122, the rat homolog of DLC1, suppressed the formation of lysophosphatidic acid–induced stress fibers and focal adhesions (Sekimata et al., 1999). Furthermore, stable expression of human DLC1 in hepatocellular and breast carcinoma cell lines was shown to reduce cell motility and invasiveness, consistent with the inhibition of Rho signaling (Goodison et al., 2005; Wong et al., 2005). Regulation of GAP proteins is achieved by several mechanisms such as protein or lipid interactions and posttranslational modification (Bernards and Settleman, 2005). However, little is known about the molecular regulation of DLC1. Recent reports have provided evidence that interaction with tensin proteins is important for the recruitment of DLC1 to focal adhesions (Yam et al., 2006; Qian et al., 2007; Liao et al., 2007). DLC1 mutants deficient in tensin binding and thus focal adhesion localization lose their ability to suppress colony formation, indicating that DLC1 location to these sites is linked to biological activity (Qian et al., 2007; Liao et al., 2007). On the other hand, the phosphorylation-dependent interaction with 14-3-3 adaptor proteins retains DLC1 in the cytoplasm and inhibits its cellular functions (Scholz et al., 2009). Negative regulation is also achieved by direct binding of p120RasGAP to the GAP domain of DLC1, which was shown to impair the growth-suppressing properties of DLC1 (Yang et al., 2009).
DLC1 is a multidomain protein that contains an amino-terminal sterile α motif (SAM) and a StAR (steroidogenic acute regulatory)-related lipid transfer (START) domain at its carboxy-terminus. SAM domains mediate homo- and heterotypic interactions with other SAM domains, but they were also shown to interact with other proteins, lipids, and RNA (Kim and Bowie, 2003). START domains are typically found in lipid transfer proteins and form a hydrophobic pocket to accommodate a single lipid molecule that is shuttled between membranes (Alpy and Tomasetto, 2005). Protein and/or lipid interactions via SAM and START domains are thus potential mechanisms by which DLC localization and/or activity may be regulated. Because the active forms of Rho GTPases are associated with cellular membranes, it is here where the DLC proteins are expected to exert their function. Therefore, we argued that DLC1 may be recruited to and activated at membrane proximal sites not only by protein but also by lipid interaction to inactivate target Rho GTPases. Here we identify a novel interaction of DLC1 with negatively charged phospholipids, with highest affinity ob served for phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2), that is independent of SAM and START domains. Instead, the interaction is mediated by a polybasic region (PBR) directly preceding the GAP domain that is conserved within the DLC protein family. In vitro assays with prenylated RhoA show that the presence of PI(4,5)P2-containing membranes stimulate DLC1's GAP activity. We further demonstrate that, in a cellular context, an intact PBR is essential for DLC1 inhibition of Rho signaling and, accordingly, suppression of cell proliferation and directed migration. To our knowledge this is the first report of a RhoGAP protein being activated by PI(4,5)P2, thus establishing a novel role for this lipid in Rho GTPase regulation.
MATERIALS AND METHODS
Antibodies and Reagents
Antibodies used were as follows: mouse anti-GFP mAb (Roche Applied Science, Mannheim, Germany), mouse anti-paxillin mAb (Becton Dickinson, Heidelberg, Germany), mouse anti-tubulin mAb (Sigma Aldrich, St. Louis, MO), goat anti-GST pAb (GE Healthcare, Piscataway, NJ), IRDye 800-labeled goat anti-GST IgG (Rockland Biosciences, Gilbertsville, PA). Horseradish peroxidase (HRP)-labeled anti-mouse IgG antibody was from GE Healthcare, HRP-labeled anti-goat IgG antibody was from Santa Cruz Biotechnology (Santa Cruz, CA), Alexa Fluor 546–labeled anti-mouse IgG antibody, Alexa Fluor 546–labeled phalloidin, and 1-hexadecanoyl-2-(1-pyrenedecanoyl)-sn-glycero-3-phosphocholine (pyrene-PC) were from Molecular Probes (Eugene, OR). Phosphatidylserine (PS), phosphatidic acid (PA), phosphatidylcholine (PC), and PI(4,5)P2 were purchased from Sigma Aldrich, phosphatidylinositol-4-phosphate (PI(4)P) and phosphatidylinositol-3,4,5-trisphosphate (PI(3,4,5)P3) were from Biomol (Hamburg, Germany), and phosphatidylinositol (PI) and porcine brain lipids were from Avanti Polar Lipids (Alabaster, AL). All chemicals, if not stated otherwise, were purchased from Roth (Karlsruhe, Germany).
DNA Cloning
The plasmids encoding full-length human DLC1 (GenBank accession no. AAK07501.1) pEGFPC1-DLC1 and pEFrPGKpuroFlag-DLC1 and the truncated variants pEGFPC1-DLC1 ΔSAM and pEGFPC1-DLC1 ΔSTART were described previously (Holeiter et al., 2008; Scholz et al., 2009). DLC1 ΔNlong (aa 596-1091) and DLC1 ΔNshort (aa 637-1091) were amplified by PCR using pEGFPC1-DLC1 as a template with primers containing BamHI restriction sites. Forward primers used were 5′-CGC GGA TCC AAA TAC TCA CTC CTA AAG CTA ACG G-3′ for DLC1 ΔNlong and 5′-CGC GGA TCC AGT GTG TTT GGG GTC CCA C-3′ for DLC1 ΔNshort. The reverse primer used was 5′-CGC GGA TCC TCA CCT AGA TTT GGT GTC TTT GG-3′. DLC1 K626A/R627G and ΔPBR (Δ623–631) mutants were generated by QuikChange site-directed PCR mutagenesis using pEGFPC1-DLC1 as a template according to the manufacturer's instructions (Stratagene, La Jolla, CA). The forward primers used were as follows: DLC1 K626A/R627G 5′-CGT GCC CAA GTT CAT GGC GGG GAT CAA GGT TCC AGA C-3′ and DLC1 ΔPBR 5′-GCT GGG CCG TGC CCG ACT ACA AGG ACC GG-3′. DLC1 constructs were subcloned into pEFrPGKpuro-Flag and pGEX6-P3 vectors by BamHI restriction. The DLC1 ΔPBR cDNA was also subcloned into the pcDNA5/FRT/TO-GFP vector, which we described recently (Scholz et al., 2009). The human DLC3β cDNA, a shorter form lacking the SAM domain, was amplified by PCR using clone IRATp970E0455D (imaGenes, Berlin, Germany) as a template with forward primer 5′-CCG GAA TTC TAC CTT GAA TAA TTG TGC CTC GAT G-3′ and reverse primer 5′-CCG GAA TTC TTC ACA GCT TTG TCT CAG GGC-3′ and cloned into the pEGFPC1 vector as an EcoRI fragment. The 5′ region encoding the SAM domain present in DLC3α was amplified by PCR using cDNA derived from HeLa cells as a template with the forward primer 5′-CCG GAA TTC TCC TCT GCT GGA CGT TTT CTG-3′ and reverse primer 5′-TTC TGA GTC TTC ATT CTG CTT GC-3′. The PCR product and pEGFPC1-DLC3β were digested with EcoRI and BpiI, and the DLC3 fragments were ligated with EcoRI-digested pEGFPC1, generating pEGFPC1-DLC3α used in this study. All amplified cDNAs were confirmed by sequencing. Oligonucleotides were purchased from MWG Biotech (Ebersberg, Germany).
Cell Culture and Transfection
All cell lines used were cultured in RPMI media (Invitrogen, Karlsruhe, Germany) supplemented with 10% fetal calf serum (FCS; PAA Laboratories, Karlsruhe, Germany) in a humidified atmosphere of 5% CO2 at 37°C. Flp-In T-Rex HEK293 cells (Invitrogen) were grown in RPMI containing 10% FCS, 100 μg/ml zeocin, and 15 μg/ml blasticidin. These cells stably express the Tet repressor and contain a single Flp recombination target (FRT) site and were used to generate the HEK293 Flp-In DLC1 lines. Cells were cotransfected with pcDNA5/FRT/TO-GFP-DLC1 WT and ΔPBR and the Flp recombinase expression plasmid pOG44 at a ratio of 1:10 and then selected with 100 μg/ml hygromycin. HEK293 cells were transfected using TransIt reagent (Mirus, Madison, WI), and MCF7 cells were transfected using Lipofectamine 2000 (Invitrogen) or by nucleofection (Amaxa, Köln, Germany).
Purification of Recombinant Proteins from E. coli
pGEX6-P3 DLC1 ΔNlong and ΔNshort vectors or empty vector were transformed into BL21 bacteria. Production cultures were inoculated from overnight cultures (1/25) and grown for 3 h at 37°C. Cultures were then shifted to 30°C, induced with 0.5 mM IPTG (Fermentas, St. Leon-Rot, Germany), and protein production continued for 3–4 h. Bacteria were harvested by centrifugation and resuspended in PBS with PMSF and Complete protease inhibitors (Roche). Bacteria were lysed by sonification (2 × 10 cycles, 50% duty cycle), debris was removed by centrifugation, and the supernatant was incubated with glutathione beads (Roche) for 2 h at 4°C. Beads were washed with PBS, and protein was eluted with 30 mM glutathione in 50 mM Tris, pH 7.4, and 100 mM NaCl buffer. Purity and concentration of the protein was determined by SDS-PAGE and Coomassie staining. Purified proteins were stored at −80°C. Glutathione S-transferase (GST)-RhoA was purified in the same manner, but all buffers were supplemented with 5 mM MgCl2 and 1 μM GDP. In vitro prenylation of GST-RhoA was performed in a buffer containing 50 mM Tris·HCl, pH 8.0, 100 mM NaCl, 20 mM KCl, 5 mM MgCl2, 1 mM DTE, 50 μM GDP, 5 μM ZnCl2, and 0.5% NP40. A reaction mixture of 20 μM GST-RhoA, 25 μM yeast GST-GGTaseI, and 50 μM of geranylgeranyl pyrophosphate (Sigma Aldrich) was incubated at 4°C for 14 h. Completeness of GST-RhoA prenylation was verified by mass spectrometry.
Hypotonic Cell Lysis
HEK293T cells transiently expressing the different GFP-tagged DLC1 variants were lysed in 50 mM Tris·HCl, pH 7.4, containing Complete protease inhibitors, 1 mM PMSF, 5 mM ß-glycerophosphate, and 5 mM sodium fluoride. Lysates were passed through a 0.45-mm needle, and nuclei and debris were removed by centrifugation (16,000 × g, 4°C, 10 min). The total protein concentration, and the amount of green fluorescent protein (GFP)-tagged proteins were determined by Bradford assay (Bio-Rad, Munich, Germany) and measurement of GFP fluorescence (excitation 466 nm, emission 512 nm), respectively, using a Tecan Infinite 200M reader (Crailsheim, Germany).
Lipid ELISA
Phospholipids dissolved in ethanol were spotted onto polystyrol 96-well plates (2 μg per well; Greiner Bio-One, Frickenhausen, Germany) and left to dry. The wells were blocked with PBS containing 0.05% Tween-20 (PBS-T) and 5% lipid-free bovine serum albumin (BSA) for 1 h, incubated with hypotonic cell lysate (150 μg total protein) containing equal amounts of GFP-tagged proteins for 40 min and then washed three times with PBS-T. The wells were then incubated with anti-GFP antibody in PBS-T for 2 h, followed by HRP-conjugated secondary antibody in PBS-T for 1 h. ABTS (2,2′-azino-di (3-ethylbenzthiazoline-6-sulfonate) peroxidase substrate at 1 mg/ml in ABTS buffer (Roche) was added, and absorption at 405 nm was recorded with a Tecan Infinite 200M plate reader after 30 min.
Lipid Vesicle Preparation
The indicated amount of lipid in chloroform was dried under a nitrogen stream. For SUV (small unilamellar vesicle) preparations, buffer containing 20 mM HEPES, pH 7.2, 5 mM EDTA, and 100 mM NaCl was added, and lipids were rehydrated by sonification (10 min, 20% duty cycle) on ice. For membrane interaction assays, SUVs were prepared in the presence of pyrene-PC (10 mol%). For MLV preparations, buffer containing 20 mM Tris·HCl, pH 7.5, 25 mM NaCl, and 4 mM EDTA was added, and after incubation for 1 h on ice, lipids were rehydrated by vigorous vortexing.
Membrane Interaction Assay
Protein binding to vesicles was evaluated by measuring FRET from tryptophan residues to the pyrene fluorophore of pyrene-PC as follows. Emission scans were recorded in the absence of protein (wavelength, 310–500 nm; excitation, 290 nm; slit widths, 4 nm) at 20°C for samples containing 30 μl pyrene-PC-containing SUVs (16 μM lipid) in 1,8 ml buffer (20 mM HEPES, pH 7.2, 5 mM EDTA, 100 mM NaCl) using an Aminco Bowman series 2 spectrofluorometer (SLM Instruments, Rochester, NY). Then, equal amounts of recombinant GST-tagged DLC1 ΔNlong or ΔNshort were added and the emission spectrum was recorded again. FRET/tryptophan ratios were calculated using the mean peak values (FRET: 376–379 nm; tryptophan: 336–338 nm) after subtraction of values in the absence of protein. Ratios for BL only were set as 1.
Multilamellar Vesicle Flotation Assay
Multilamellar vesicle (MLV; 1 mM total lipid) were prepared using bovine brain lipids with or without 10 mol% PI(4,5)P2. 100 μl MLVs, 25 μl HEPES buffer (20 mM HEPES, pH 7.2, 5 mM EDTA, and 100 mM NaCl), and 25 μl GST-tagged DLC1 ΔNlong, ΔNshort or GST as a control (∼1 mg/ml) were incubated at room temperature (RT) for 30 min. The suspension was adjusted to 30% sucrose by addition of 100 μl 75% sucrose, overlayed with 200 μl 25% sucrose in buffer, and 50 μl sucrose-free buffer. Samples were centrifuged at 240,000 × g for 1 h. The bottom (250 μl) and top (100 μl) fractions were collected and analyzed by Western blotting using an IRDye-labeled (Rockland Immunochemicals, Gilbertsville, PA) α-GST antibody, a Licor Odyssey scanner and the Licor Odyssey software (Bad Homburg, Germany) for quantification. Bound-to-unbound protein ratios were calculated and the BL only ratios of each protein were set to 100%.
In Vitro RhoGAP Activity Assay
GST-RhoA (∼10 μg) was preloaded with γ32P-GTP (Hartmann Analytic, Braunschweig, Germany) in 60 μl loading buffer (20 mM Tris·HCl, pH 7.5, 25 mM NaCl, 4 mM EDTA, 0.1 mM DTT, 0.125 μCi γ32P-GTP) at 30°C for 5 min. Preloading of GST-RhoAGG (∼10 μg) was performed in the presence of 20 μg MLVs. Preloading was stopped by adding 10 mM MgCl2. Ten microliters of the loading mix was added to 40 μl reaction mix (20 mM Tris·HCl, pH 7.5, 0.1 mM DTT, 1 mM GTP, and 1 mg/ml BSA) on ice, with or without DLC1 Δlong or ΔNshort (∼1 μg) and shifted to 20°C to start the reaction. At 0 and 10 min, 10 μl of the reaction mix was added to 1 ml of ice-cold wash buffer (20 mM Tris·HCl, pH 7.5, and 5 mM MgCl2), and samples were filtered through nitrocellulose membranes (Millipore, Schwalbach, Germany), retaining RhoA-bound γ32P-GTP. After washing, the amount of γ32P-GTP was measured by scintillation counting (Cherenkov radiation, 1600TR scintillation analyzer; Packard Instrument, Dreieich, Germany).
Raichu-RhoA Biosensor Measurements
HEK293T cells were transiently transfected with plasmids encoding the Raichu-RhoA biosensor (Yoshizaki et al., 2003) and Flag-DLC1 WT, K626A/R627G and ΔPBR. Cells were lysed in 50 mM Tris, pH 7.5, 100 mM NaCl, and 0.5% Triton X-100, and debris was removed by centrifugation at 16,000 × g for 5 min. Lysates were transferred to white Nunc 96-well plates (Wiesbaden-Biebrich, Germany) and emission ratios (fluorescence resonance energy transfer/cyan fluorescent protein [FRET/CFP]) were determined by measuring CFP and yellow fluorescent protein (YFP) fluorescence after background subtraction at 475 and 530 nm, respectively, using a Tecan Infinite 200M plate reader (excitation, 433 nm). Expression levels of the biosensor were monitored by measuring YFP fluorescence (excitation, 480 nm) and those of the DLC1 variants by immunoblotting of lysates.
Luciferase Assays
HEK293T cells were grown on collagen-coated 24-well plates (Greiner) and transfected with 50 ng each of the 3DA.Luc firefly luciferase reporter containing three serum response factor (SRF)-binding elements, pRL-TK, a Renilla luciferase plasmid under the control of the thymidine kinase promoter, and pEGFPC1-DLC1 wild type (WT), K626A/R627G and ΔPBR, respectively. After serum starvation over night, cells were stimulated with 15% FCS for 6 h. Cells were lysed with 300 μl passive lysis buffer (Promega, Heidelberg, Germany), and luciferase activities in 10 μl lysate were measured by addition of 50 μl firefly substrate (470 μM D-luciferin, 530 μM ATP, 270 μM CoA, 33 mM DTT, 20 mM tricine, 2.67 mM MgSO4, and 0.1 mM EDTA, pH 7.8), followed by addition of 100 μl Renilla substrate (0.7 μM coelenterazine, 2.2 mM Na2EDTA, 0.44 mg/ml BSA, 1.1 M NaCl, 1.3 mM NaN3, and 0.22 M potassium phosphate buffer, pH 5.0). Luminescence was measured with a Tecan Infinite 200M plate reader.
Immunofluorescence Microscopy
Cells grown on collagen-coated glass coverslips were transfected with the indicated expression plasmids, and after 24 h cells were fixed in 4% PFA for 10 min, washed, and incubated with PBS containing 0.1 M glycine for 15 min. Cells were permeabilized with PBS containing 0.1% Triton for 5 min and blocked with 5% goat serum (Invitrogen) in PBS containing 0.1% Tween-20 for 30 min. Cells were then incubated with paxillin-specific antibody diluted in blocking buffer for 2 h, followed by incubation with secondary Alexa Fluor 546–coupled antibody diluted in blocking buffer for 1 h. F-actin staining was done by incubating fixed cells with Alexa Fluor 546–coupled phalloidin diluted in blocking buffer for 1 h. Coverslips were mounted in Fluoromount G (Southern Biotechnology, Birmingham, AL). Focal adhesion localization was analyzed with a Zeiss Axiovision system (Oberkochen, Germany) equipped with an Apotome using a Plan-Apochromat 63×/1.40 NA oil DIC M27 objective and the Axiovision Software. F-actin staining was analyzed with a confocal laser scanning microscope (TCS SL, Leica, Wetzlar, Germany) using 488- and 543-nm excitation, an HCX PL APO CS 100.0× 1.40 NA oil UV objective lens and Leica Software. For signal quantification, four to five random pictures were taken with a 40.0×/1.25 NA HCX PL APO oil objective lens, and mean fluorescence intensities were measured for the GFP and phalloidin channels using the ImageJ software (http://rsb.info.nih.gov/ij/).
Live Cell Imaging
Noninduced HEK293 FlpIn cells and cells induced to express DLC1 WT and ΔPBR (100 ng/ml doxycycline for 24 h) were harvested and resuspended in phenol red-free RPMI (Invitrogen) containing 10% FCS and plated onto collagen-coated glass bottom dishes (World Precision Instruments, Berlin, Germany). After 5 min, recording was started by acquiring bright-field images every 30 s with a Zeiss Axiovision system equipped with a heated incubation chamber using a Plan-Apochromat 20×/0.8 NA M27 objective and the Axiovision software.
Migration Assays
For random migration, MCF7 cells were transfected using the Amaxa nucleofector (Kit V; program P-020). The next day, 105 cells were seeded into the upper well of a Transwell (8.0 μm; Costar, Cambridge, MA) and allowed to migrate for 24 h. Both the upper and the lower camber contained medium supplemented with 10% FCS. Cells on the upper side of the membranes were removed using a cotton swab, and cells on the underside were fixed in 4% PFA and stained with 0.1% crystal violet. Cells were counted in five independent microscopic fields at a 20-fold magnification. For directed migration, HEK293T Flp-In cells were induced with 100 ng/ml doxycycline for 24 h before the experiment, noninduced cells were used as controls. Transwells were coated with 2.5 μg/ml collagen on the underside and 5 × 104 cells in medium containing 0.5% FCS were added to the top chamber. The bottom chamber was supplemented with 10% FCS. Cells were allowed to migrate for 4 h and then analyzed as described above.
MTT Assays
MCF7 cells were transfected using the Amaxa nucleofector (Kit V; program P-020), and 1000 cells were plated into 96-well cell culture plates. At each time point, 10 μl 3-(4,5-dimethylthiazol-2yl-)2,5-diphenyl tetrazolium bromide (MTT) solution (5 mg/ml) was added, and the cells were incubated for 2 h. The medium was aspirated, and the cells were lysed in 100 μl 50% dimethylformamide containing 10% SDS. Absorbance at 570 nm was determined with background subtraction at 630 nm using a Tecan Infinite 200M plate reader.
Statistical analysis
P values were determined by unpaired student's t test, and significance is indicated as follows: *** p ≤ 0.001, ** p ≤ 0.01, * p ≤ 0.05, and n.s. is p > 0.05.
RESULTS
DLC1 Interacts with Negatively Charged Phospholipids Independently of Its SAM and START Domains
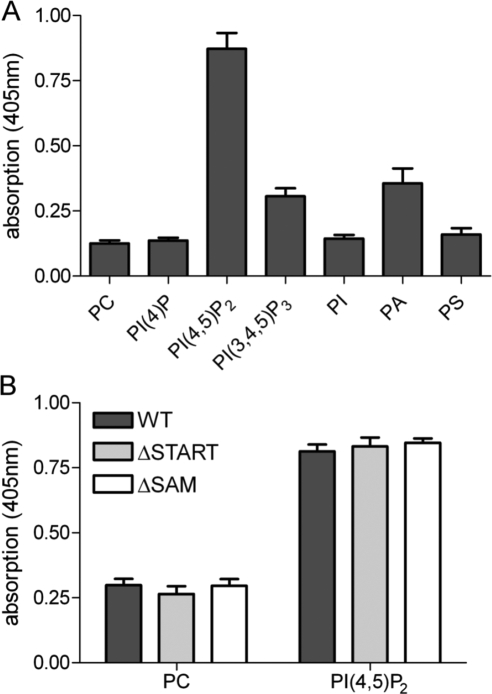
To investigate the ability of DLC1 to bind to different lipid species, we performed lipid ELISA assays. Here, lipids spotted onto 96-well plates were incubated with hypotonic lysates of HEK293T cells transiently expressing GFP-tagged DLC1. Bound DLC1 was then detected with a GFP-specific primary and HRP-labeled secondary antibody, followed by incubation with ABTS peroxidase substrate (Figure 1A). Using this assay, DLC1 was found to strongly bind to PI(4,5)P2, and modestly to PA, and PI(3,4,5)P3, while binding to PC, PI, PS, and PI(4)P) was negligible (Figure 1A). Interestingly, PI(4,5)P2-binding of DLC1 variants lacking the amino-terminal SAM (DLC1 ΔSAM) and carboxy-terminal START domains (DLC1 ΔSTART) were identical to that of the WT protein (Figure 1B), indicating that the interaction was independent of these domains. Of note, lipid ligands of the START domain are unlikely to be identified in this assay, because lipid uptake into the hydrophobic pocket requires administration in solution.
Figure 1.
DLC1 binding to phospholipids. (A) In 96-well plates, immobilized egg PC plus 15 mol% of the indicated lipids was incubated with hypotonic lysate from HEK293T cells transiently expressing GFP-tagged DLC1. Bound DLC1 was detected with GFP-specific antibody, followed by HRP-conjugated secondary antibody, and visualized with ABTS peroxidase substrate. Absorption was measured after 30 min at 405 nm. The mean of two independent experiments performed with triplicate samples is shown; error bars, SEM. (B) Immobilized PI(4,5)P2 (15 mol% in PC) was incubated with hypotonic lysate from HEK293T cells transiently expressing GFP-tagged DLC1 wild-type (WT), ΔSAM, and ΔSTART proteins. Detection of bound proteins was done as described in A. A representative experiment with triplicate samples is shown. Error bars, SEM.
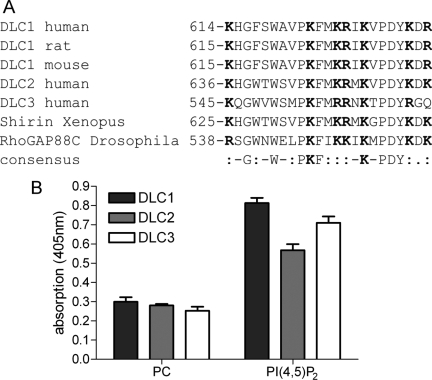
DLC Proteins Possess a Highly Conserved Polybasic Region Adjacent to the GAP Domain
Binding of negatively charged phospholipids such as PI(4,5)P2 can be mediated by short sequences enriched in basic amino acids (Fivaz and Meyer, 2003). A ScanProsite motif search identified such a PBR in close proximity of the DLC1 RhoGAP domain. Sequence comparison of human, mouse and rat DLC1 proteins including the Drosophila and Xenopus homologues revealed conservation of this region among the different species. In addition, the PBR was present in DLC2 and DLC3 (Figure 2A). Lipid ELISA analysis showed that DLC2 and DLC3 were also able to interact with PI(4,5)P2 (Figure 2B), raising the possibility that lipid binding is a general mechanism of the DLC protein family mediated by a conserved PBR.
Figure 2.
The DLC protein family contains a conserved polybasic region and interacts with PI(4,5)P2. (A) Sequence comparison of DLC1 from different species and human DLC2 and DLC3. Basic amino acids are highlighted in bold. (B) GFP-tagged DLC1, DLC2 and DLC3 were tested for their ability to bind to immobilized PI(4,5)P2 as described in Figure 1. A representative experiment with triplicate samples is shown. Error bars, SEM.
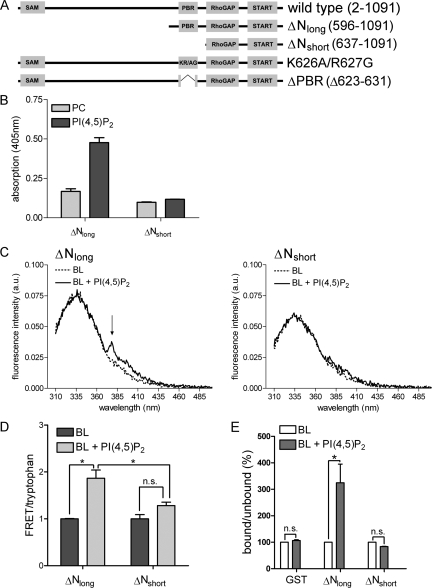
DLC1 Interaction with PI(4,5)P2 Is Mediated by the Polybasic Amino Acid Cluster
To investigate the importance of the identified PBR for DLC1 interaction with negatively charged phospholipids, we generated carboxy-terminal constructs containing the GAP and START domains, either comprising the PBR (DLC1 ΔNlong) or lacking this region (DLC1 ΔNshort; Figure 3A). Lipid ELISA analysis showed that the PBR was indeed responsible for mediating the interaction with PI(4,5)P2 (Figure 3B). The amount of DLC1 ΔNlong bound by immobilized PI(4,5)P2 was significantly higher than that bound by PC alone, whereas binding of DLC1 ΔNshort was not enhanced by PI(4,5)P2. To assess if the PBR also recognizes PI(4,5)P2 when incorporated into membranes, we used a FRET-based assay, in which the aromatic amino acid tryptophan served as the FRET donor and pyrene-labeled PC as the FRET acceptor. Pyrene-PC was incorporated into SUVs made of porcine brain lipids (BL), with or without the addition of 10 mol% PI(4,5)P2. These vesicles were then incubated with recombinant GST-tagged DLC1 ΔNlong and ΔNshort purified from E. coli. In the case of DLC1 ΔNlong, a clear FRET peak was visible in the presence of vesicles comprised of BL and PI(4,5)P2, indicating the close proximity of the donor and acceptor (Figure 3C, left, FRET marked with an arrow). When DLC1 ΔNshort was incubated with PI(4,5)P2-containing vesicles no FRET peak was observed (Figure 3C, right). FRET-tryptophan ratios are quantified in Figure 3D. In the case of DLC1 ΔNshort there was only a small nonsignificant difference between the FRET-tryptophan ratios for vesicles with and without PI(4,5)P2 (Figure 3D). To independently test whether DLC1 associates with PI(4,5)P2-containing membranes in a PBR-dependent manner, we performed a lipid flotation assay. Here, MLVs composed of porcine brain lipids were incubated with GST-DLC1 ΔNlong, ΔNshort, and GST alone. MLV-bound protein was then separated from unbound protein by sucrose gradient centrifugation and quantified by Western blotting. Addition of 10 mol PI(4,5)P2 lead to a significant enrichment of GST-ΔNlong in the vesicle-containing fraction, whereas GST or GST-DLC1 ΔNshort were not enriched (Figure 3E). Collectively, our results provide evidence that DLC1 interacts with PI(4,5)P2-containing membranes via a region rich in basic amino acids that directly precedes its GAP domain.
Figure 3.
The DLC1 polybasic region mediates PI(4,5)P2interaction. (A) Schematic representation of DLC1 WT, ΔNlong, ΔNshort, K626A/R627G, and ΔPBR. (B) GFP-DLC1 ΔNlong and ΔNshort were tested for their ability to bind to immobilized PI(4,5)P2 as described in Figure 1. (C) Recombinant GST-DLC1 ΔNlong (left panel) or ΔNshort proteins (right panel) were incubated with SUVs comprised of porcine brain lipids (BL) and pyrene-PC, with or without the addition of 10 mol% PI(4,5)P2. The samples were excited at 290 nm, and emission scans were recorded from 310 to 500 nm (a.u., arbitrary units). Background spectra were recorded before protein addition. The arrow indicates pyrene fluorescence emission (376–379 nm) caused by FRET. (D) FRET-tryptophan ratios were calculated as described in Materials and Methods. Data shown are the mean of two experiments in the case of BL and three experiments in the case of BL + PI(4,5)P2; error bars, SEM. (E) Recombinant GST-DLC1 ΔNlong, ΔNshort or GST alone were mixed with MLVs composed of the indicated lipids. MLV-bound protein was separated from unbound protein by density gradient centrifugation, and the amount of bound and unbound protein was analyzed by Western blotting using an IRDye-labeled α-GST antibody and a Licor Odyssey scanner. Bound-unbound ratios were determined. Data shown are the mean of two experiments; error bars, SE.
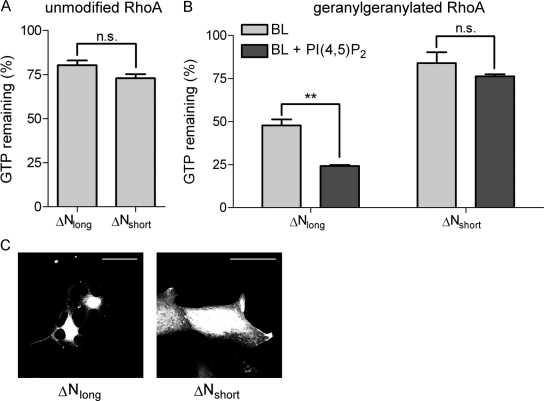
DLC1 Activity Is Stimulated by PI(4,5)P2-containing Membranes
Because of the close proximity of the PI(4,5)P2-binding motif to the DLC1 GAP domain, we speculated that lipid binding may modulate DLC1 GAP activity. We first determined in vitro GAP activities of GST-tagged DLC1 ΔNlong and ΔNshort in the absence of membranes using recombinant GTP-loaded RhoA as a substrate. As shown in Figure 4A, both DLC1 ΔNlong and ΔNshort displayed similar in vitro GAP activity for RhoA, proving that the deletion of the PBR per se does not impact on the DLC1 GAP domain. Of note, the addition of PI(4,5)P2-containing vesicles did not affect the rate of RhoA-GTP hydrolysis in this assay (data not shown). In the cell, RhoA is posttranslationally modified by geranylgeranylation, which targets it to membranes. Importantly, prenylation has been previously shown to alter the susceptibility of Rho GTPases toward GAP proteins (Ligeti et al., 2004). Thus, to investigate the impact of membranes on DLC1 GAP activity and the role of PI(4,5)P2-binding, we set up an in vitro assay with prenylated recombinant RhoA (RhoAGG) that mimics more closely the cellular environment. To compare for the different Rho preparations, we normalized the results for RhoA and RhoAGG to their intrinsic GTPase activity (Figure 4, A and B). Interestingly, the presence of MLVs composed of brain lipids (BL) strongly increased the GAP activity of DLC1 ΔNlong for RhoGG (from 80 to 48% remaining GTP), compared with nonprenylated RhoA (Figure 4, A and B). Addition of PI(4,5)P2 to these MLVs stimulated GAP activity even further (approximately twofold; Figure 4B, left). In contrast to DLC1 ΔNlong, DLC1 ΔNshort activity toward RhoAGG in a membrane environment was comparable to that observed for unprenylated RhoA and was not stimulated by incorporation of PI(4,5)P2 into vesicles (Figure 4B, right).
Figure 4.
Lipid interaction stimulates in vitro DLC1 RhoGAP activity. (A) GST-RhoA (10 μg) was loaded with γ32P-GTP and incubated with equal amounts of GST-DLC1 ΔNlong and ΔNshort at 20°C. GTP hydrolysis was determined after 0 and 10 min by measuring protein-bound γ32P-GTP. The intrinsic RhoA GTPase activity was set to 100%. The graph shows a representative experiment performed with triplicate samples; error bars, SEM. (B) GST-RhoA was geranylgeranylated in vitro as described in Materials and Methods. Ten micrograms GST-RhoAGG was loaded with γ32P-GTP in the presence of 10 μg MLVs and incubated with equal amounts of GST-DLC1 ΔNlong and ΔNshort at 20°C. GTP hydrolysis was determined after 0 and 10 min by measuring protein-bound γ32P-GTP. The intrinsic RhoA GTPase activity was set to 100%. The graph shows a representative experiment performed with triplicate samples; error bars, SEM. (C) MCF7 cells were grown on collagen-coated coverslips and transfected with expression plasmids encoding GFP-DLC1 ΔNlong and ΔNshort. After 24 h cells were fixed and photographed. The images shown are stacks of three to four confocal sections taken at 0.5-μm intervals; scale bar, 50 μm.
To confirm these activity differences in intact cells, we ectopically expressed GFP-tagged variants of DLC1 ΔNlong and ΔNshort in MCF7 breast epithelial cells. DLC1 overexpression has been shown to induce cell rounding and leads to the development of neurite-like extensions (Sekimata et al., 1999; Wong et al., 2005). Expression of an isolated GAP domain is known to be sufficient for the induction of such effects. Indeed, MCF7 cells expressing DLC1 ΔNlong showed the typical “neurite-like” morphology (Figure 4C). By contrast, cells expressing DLC1 ΔNshort displayed a normal flat morphology (Figure 4C). Taken together, these results strongly suggest that the PBR-mediated lipid interaction is important for DLC1 RhoGAP function in vitro and in vivo.
The PBR in DLC1 Is Required for Inactivation of Rho Signaling
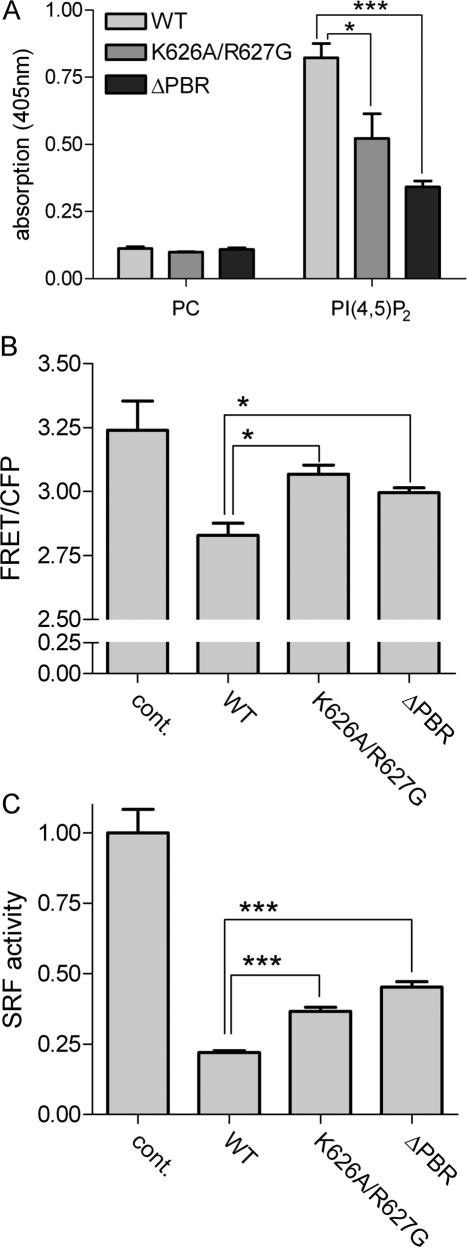
To study the role of the identified lipid binding region in the context of full-length DLC1, we generated mutants deficient in PI(4,5)P2-binding by 1) exchanging two basic amino acids in the core of the PBR (K626A/R627G) and 2) deleting nine amino acids (623–631), generating a DLC1 ΔPBR mutant (see Figure 2A). These mutations are expected to have a minimal impact on the overall structure of the protein. Lipid ELISA analysis confirmed that both mutations decreased PI(4,5)P2-binding, with the ΔPBR deletion being more potent (Figure 5A). To determine the effects of these DLC1 variants on cellular Rho activity, we made use of a previously described Raichu-RhoA biosensor, whose FRET-CFP emission ratios reflect endogenous Rho-GTP levels (Yoshizaki et al., 2003). Compared with the WT, DLC1 K626A/R627G and ΔPBR were less efficient at reducing the emission ratio of Raichu-RhoA (Figure 5B), indicating reduced GAP activities of the mutant proteins. To independently determine the contribution of the PBR to DLC1 RhoGAP function, we performed SRF luciferase reporter assays, which are responsive to signaling by active Rho. Here, cells were transfected with an SRF reporter, serum-starved over night, and then restimulated with serum. Compared with the WT protein, which suppressed SRF-dependent transcription by 75%, the DLC1 K626A/R627G and ΔPBR mutants were less potent at inhibiting transcription (Figure 5C).
Figure 5.
The DLC1 PBR is required for RhoGAP activity in intact cells. (A) GFP-tagged DLC1 WT, K626A/R627G, and ΔPBR were tested for their ability to bind immobilized PI(4,5)P2 as described in Figure 1. (B) HEK293T cells were cotransfected with plasmids encoding the Raichu-RhoA biosensor and Flag-DLC1 WT, K626A/R627G, ΔPBR, or empty vector (cont.). The next day, the emission ratio of Raichu-RhoA was determined by measuring YFP (FRET) and CFP fluorescence (excitation 433 nm) in cell lysates. One representative experiment out of three is shown; error bars, SEM. (C) HEK293T cells were transiently transfected with the 3DA.Luc reporter, a plasmid encoding for Renilla luciferase and empty vector (cont.), pEGFPC1 DLC1 WT, K626A/R627G, or ΔPBR, respectively. Cells were serum-starved overnight and then restimulated with 15% serum for 6 h. Firefly luciferase activity was determined and normalized by Renilla luciferase activity. The control was set to 1. Equal expression of the DLC1 variants was verified by measuring GFP fluorescence in the lysates with a fluorescence spectrometer. One representative experiment out of three is shown, each performed with triplicate samples; error bars, SEM.
PBR Deletion Does Not Alter DLC1 Localization But Activity in MCF7 Cells
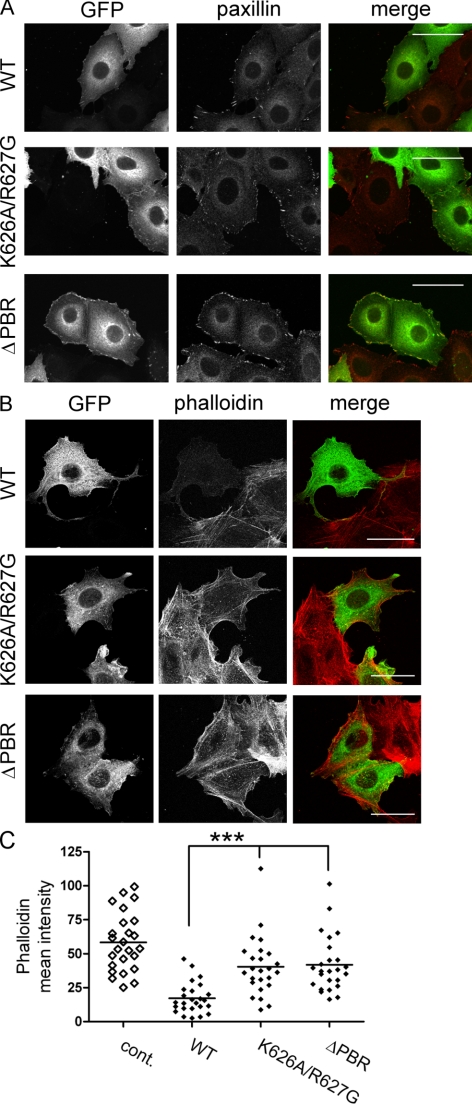
To investigate whether the PBR may be involved in DLC1 subcellular localization, we ectopically expressed DLC1 WT, K626A/R627G and ΔPBR in MCF7 cells. Consistent with previous observations (Holeiter et al., 2008), full-length DLC1 localized to focal adhesions as determined by paxillin costaining (Figure 6A). Because DLC1 overexpression leads to focal adhesion dissolution, only cells with very low expression levels that retained their normal morphology were selected for the analysis. DLC1 K626A/R627G and ΔPBR were also recruited to paxillin-positive sites but appeared to have a reduced effect on focal adhesion dissolution (Figure 6A). DLC1 has also been reported to localize to the leading edge of migrating cells (Healy et al., 2008). However, membrane localization of DLC1 WT, K626A/R627G, and ΔPBR proteins were comparable as judged microscopically and by fractionation assays (Supplemental Figure S1), suggesting that mutation or deletion of the PBR does not alter DLC1 subcellular localization.
Figure 6.
The PBR is required for DLC1-mediated actin cytoskeletal changes. (A and B) MCF7 cells grown on collagen-coated coverslips were transfected with expression plasmids encoding GFP-DLC1 WT, K626A/R627G, and ΔPBR and fixed after 24 h. (A) Focal adhesions were visualized with a paxillin-specific primary and Alexa Fluor 546–coupled secondary antibody (red). (B) Actin stress fibers were stained with Alexa Fluor 546–conjugated phalloidin (red). The images shown are stacks of five to six confocal sections taken at 0.5-μm intervals. Scale bars, 50 μm. (C) The mean phalloidin intensity of cells expressing similar levels of the different GFP-DLC1 variants (n > 25) was quantified. Nontransfected cells were used as a control (cont.).
Rho activity is known to correlate with stress fiber formation, which is suppressed upon DLC1 overexpression (Sekimata et al., 1999; Wong et al., 2005). Phalloidin staining revealed a reduced capacity of DLC1 K626A/R627G and ΔPBR to inhibit stress fiber formation compared with the WT protein (Figure 6B). Quantification of the mean intensity of the phalloidin signal confirmed that mutation or deletion of the PBR partially restored the F-actin content of DLC1-expressing cells compared with control cells (Figure 6C), indicating diminished cellular RhoGAP activities of these mutants, which is in line with the results obtained in Figure 5, B and C.
The PBR Is Required for DLC1 Cellular Functions
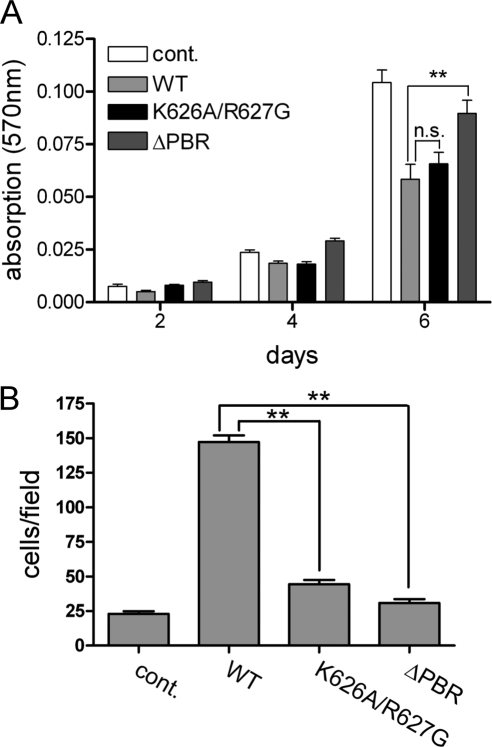
We previously showed that DLC1 expression inhibits growth of MCF7 cells (Scholz et al., 2009). The DLC1 K626A/627G mutant inhibited cell growth almost as potently as the WT protein as measured by MTT assay, indicating that the exchange of the two basic amino acids within the PBR is not sufficient to suppress the antiproliferative effects of DLC1. However, the DLC1 ΔPBR mutant was notably impaired at inhibiting cell growth (Figure 7A). DLC1 expression is further known to affect cell motility in a GAP-dependent manner (Kim et al., 2008). We therefore analyzed the impact of PBR mutation on DLC1 function using Transwell assays. Interestingly, expression of the DLC1 WT protein dramatically stimulated random motility of MCF7 cells (Figure 7B). Both PBR mutation (K626A/R627G) and deletion (ΔPBR) almost completely abrogated this effect (Figure 7B). Taken together, our results show that an intact PBR is required for effects of DLC1 on cell proliferation and motility.
Figure 7.
The PBR is required for DLC1 cellular functions. (A and B) MCF7 cells were transfected by nucleofection with GFP-DLC1 WT, K626A/R627G, ΔPBR, and vector alone as a control (cont.). (A) Proliferation of the cells was measured by MTT assay. A representative experiment is shown. Data are the mean of nine wells, normalized to the absorbance at day 0; error bars, SEM. (B) Cell motility was measured in Transwells assays, in which lower and upper chambers contained medium supplemented with 10% FCS. Cells that had migrated across the filter after 24 h were fixed and stained. The number of migrated cells was determined by counting five independent microscopic fields (20-fold magnification). Data shown are the mean of duplicate wells and are representative of two independent experiments.
Inhibition of Cell Spreading and Directed Migration by DLC1 Requires an Intact PBR
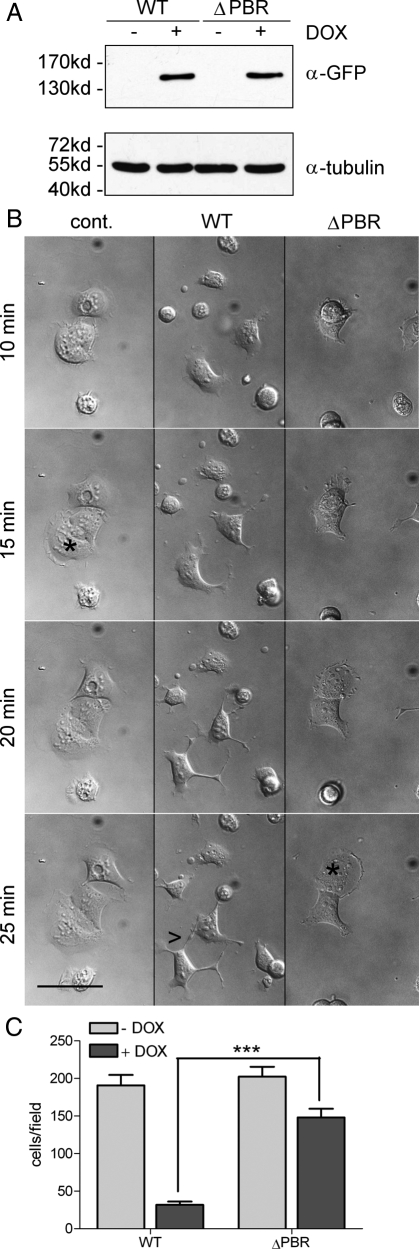
To study in greater detail the contribution of the newly identified lipid interaction motif to DLC1's effects on cytoskeletal remodeling, we generated stable HEK293 Flp-In cell lines inducibly expressing either GFP-tagged WT DLC1 or DLC1 ΔPBR. Because of targeted genomic integration of the cDNA, selected cells contained a single DLC1 copy that can be switched on by the addition of doxycycline. This is advantageous over high expression levels obtained in transient transfections and the growth suppressing and adaption effects encountered when generating stable cell lines. Figure 8A shows that equal amounts of GFP-tagged DLC1 WT and ΔPBR were expressed upon doxycycline induction of these stable HEK293 Flp-In lines. To study the effect of DLC1 expression on cell morphology and actin cytoskeletal remodeling, we plated cells onto collagen-coated dishes and recorded cell spreading by live cell imaging. Compared with noninduced cells, expression of the DLC WT protein did not alter cellular adhesion properties but severely impacted on cell spreading (Figure 8B). Cells expressing WT DLC1 failed to form stable lamellipodia, which were seen in control cells by 15 min after plating (Figure 8B, asterisk), but instead developed abnormal cellular projections (Figure 8B, see arrowhead). These cells furthermore displayed a high degree of rapidly changing protrusive activity (see Supplemental Movies), which was previously described for HEK293 cells expressing a DLC1 variant lacking the SAM domain (Kim et al., 2008). By contrast, DLC1 ΔPBR expressing cells spread normally in a manner comparable to the control (Figure 8B and Supplemental Movies).
Figure 8.
A DLC1 mutant deficient in PI(4,5)P2 binding fails to inhibit cell spreading and directed migration. (A) HEK293 Flp-In DLC1 WT and ΔPBR cells were left untreated (−) or treated (+) with 100 ng/ml doxycycline overnight to induce DLC1 expression. Whole cell extracts were separated by SDS-PAGE and analyzed by Western blotting with GFP-specific (top) and tubulin-specific. antibodies (bottom) as a loading control. (B) HEK293 Flp-In DLC1 WT and ΔPBR cells were treated with 100 ng/ml doxycycline overnight. Noninduced HEK293 Flp-In DLC1 WT cells were used as a control. Cells were harvested and plated onto collagen-coated glass bottom dishes. After 5 min, bright-field images were taken every 30 s for 90 min. The figure shows snapshots of the movies (see Online Supplemental files) at the indicated time points; asterisks mark cells with lamellipodia; arrowheads point to projections. Scale bar, 50 μm. (C) HEK293 Flp-In DLC1 WT, and ΔPBR cells were left untreated or treated with 100 ng/ml doxycycline. Cells, 5 × 104, were seeded in medium containing 0.5% FCS into the upper chamber of a Transwell. The lower well contained medium supplemented with 10% FCS. Cells that had migrated across the filter after 4 h were fixed and stained. The number of migrated cells was determined by counting five independent microscopic fields (20-fold magnification). Data shown are the mean of duplicate wells and are representative of two independent experiments. Error bars, SEM.
Transient expression of DLC1 ΔSAM in HEK293 cells was reported to increase the velocity but impair the directionality of migration (Kim et al., 2008). We therefore investigated the impact of DLC1 WT and DLC1 ΔPBR on directed cell migration by performing Transwell filter assays. Cells were induced with doxycycline and seeded onto collagen-coated Transwell cell culture inserts, and migration was stimulated with a serum gradient (0.5–10%). DLC1 WT expression led to a severe decrease in the number of migrated cells, whereas DLC1 ΔPBR expression had only a slight impact on the migratory of cells (Figure 8C). We thus conclude that lipid interaction via the PBR is essential for DLC1 activity and its control of Rho-dependent cellular processes.
DISCUSSION
Here we have identified an evolutionarily conserved lipid interaction module within the DLC protein family that is necessary for cellular GAP function. Membrane binding is a common feature of many GTPases that often contain carboxy-terminal lipidation motifs. These can be accompanied by polybasic sequences that aid in localization and specificity to phospholipid membranes (Williams, 2003; Heo et al., 2006). Similarly, effector proteins are often targeted to lipids, due to the necessity of interacting with membrane-localized upstream GTPases (Papayannopoulos et al., 2005; Takahashi and Pryciak, 2007). Finally, GEFs and GAPs, which are generally multidomain proteins physically associate with their cognate GTPases and thus often contain different membrane targeting sequences, such as PH, PX, C1, C2, FYVE, ENTH, and FERM domains (Hurley, 2006; Bos et al., 2007; Gureasko et al., 2008). Despite the abundance of these membrane targeting sequences, there are only a limited number of reports on lipid regulation of GAP proteins. While a role for phosphoinositides in the PH domain-dependent activation of several ArfGAP proteins such as ASAP1, ACAP1/2 and ARAP1/3 has been established (Brown et al., 1998; Jackson et al., 2000; Krugmann et al., 2002; Miura et al., 2002), no such activation mechanisms have been described for any of the ∼70 mammalian RhoGAP proteins. In yeast, the Cdc42p-specific GAP proteins Rga1/2p were reported be stimulated by PE and PS and inhibited by PI(4,5)P2 via an unknown mechanism (Saito et al., 2007). Within the RhoGAP family, the DLC proteins are of particular interest to due their established tumor suppressor function, making the understanding of their regulation especially relevant. Because DLC proteins do not possess a specific membrane-targeting domain but rather contain a PBR, the necessity of lipid binding for their GAP activity has remained unrecognized until now.
Polybasic sequences generally contain multiple positively charged amino acids that can interact with acidic phospholipids through electrostatic interaction. They further often contain hydrophobic amino acids, present also in the PBR of the DLC proteins, that partition into the hydrophobic core of the lipid bilayer and prevent recognition as nuclear targeting sequences (Heo et al., 2006). The DLC1 PBR from amino acids 614–636 is interspersed by two prolines but may adopt an amphipathic helix in its core section of alternating basic and hydrophobic residues (KFMKRIKV). We show that the DLC1 PBR mediates binding of negatively charged PI(4,5)P2 in an immobilized form and when incorporated into lipid vesicles. PI(4,5)P2 is a phospholipid species enriched in the plasma membrane. Consistent with a previous report (Healy et al., 2008), we observed DLC1 recruitment to the leading edge of migrating cells in addition to localization to focal adhesions, suggesting that Rho inactivation occurs locally at these sites. However, negatively charged phospholipids such as PI(4,5)P2 are unlikely to be the sole determinant of DLC1 plasma membrane localization. Fusion of a peptide encompassing the DLC1 PBR to GFP led to GFP relocation from the nucleus to the cytoplasm, but was not sufficient to promote plasma membrane association (data not shown). Because lipid binding affinities of PBRs are generally rather low, they often act in concert with other interaction domains to promote efficient membrane targeting (Fivaz and Meyer, 2003). This provides an explanation for the similar subcellular distributions of DLC1 ΔNlong and ΔNshort and DLC1 WT and ΔPBR proteins. Although SAM and START domains did not contribute to PI(4,5)P2 binding in lipid ELISA assays, these domains may well cooperate in DLC1 membrane localization. It was recently shown that the DLC1 SAM domain associates with membrane ruffles in fibroblast growth factor-stimulated cells (Zhong et al., 2009) and our own experiments with the isolated START domain suggest that this domain strongly interacts with membranes in vitro and in vivo (data not shown). Furthermore, protein interactions are expected to be critically involved in DLC1 plasma membrane recruitment and activation, perhaps via binding of the Rho GTPase itself. Such activation models have been established for the Ras exchange factor SOS and the Cdc42 effector N-WASP. In both cases, PI(4,5)P2 and Ras and Cdc42 binding, respectively, act in a cooperative manner to promote full activation (Papayannopoulos et al., 2005; Gureasko et al., 2008).
Most reported biochemical studies on GAP proteins including those on DLC1 have been conducted with bacterially produced unmodified GTPases. It is known, however, that the lipid environment and prenylation state of the small GTPase are important factors that can modify the substrate specificity of GAPs. For example, p190RhoGAP is a GAP protein that displays in vitro activity for Rho and Rac. Interestingly, PS, PI and PI(4,5)P2 inhibited its RhoGAP activity but activated its RacGAP activity when using small GTPases derived from insect cells as substrates (Ligeti et al., 2004). This is in line with our observation that the stimulating effect of PI(4,5)P2-containing membranes on DLC1 GAP activity is only evident for prenylated RhoA. Our results further clarify discrepancies relating to the boundaries of the DLC1 GAP domain. According to SwissProt database information the GAP domain spans amino acids 641–847. The DLC1 GAP domain was recently proposed to extend amino-terminally beyond amino acid 641 because a fragment comprising amino acids 629-1075 failed to produce morphological changes upon expression in cells (Kim et al., 2008). Based on our data, this is attributable to the deletion of a functional PBR motif, because DLC1 ΔNshort spanning amino acids 637-1091 lacked in vivo activity, while retaining full GAP activity in vitro toward unmodified RhoA (Figure 4A). Rat DLC1 was originally reported to bind and activate phospholipase C δ1 (PLC-δ1) by an unknown mechanism involving the carboxy-terminal half of the protein (Homma and Emori, 1995; Sekimata et al., 1999). Human DLC1 was not found to stimulate phospholipid hydrolysis activity of PLC-δ1 in overexpression assays (Healy et al., 2008), but it cannot be ruled out that DLC1-bound PI(4,5)P2 is utilized locally as a substrate by PLC-δ1. We have recently shown that DLC1 associates with the lipid phosphatase PTEN (Heering et al., 2009). Along these lines it is tempting to speculate that locally produced PI(4,5)P2 by the action of PTEN may stimulate DLC1 activity.
To examine the importance of the PBR to DLC1 cellular function, we generated DLC1 K626A/R627G and ΔPBR mutants. Although PI(4,5)P2 binding of these mutants was only partially abolished and Rho signaling was only moderately restored, their biological activities were strongly reduced. This proves that subtle changes in Rho-GTP levels can have a significant impact on cellular responses. Indeed, proliferation of MCF7 cells expressing DLC1 ΔPBR was partially restored compared with control cells and the ability of the DLC1 PBR mutants to stimulate random cell motility was almost abrogated compared with the WT protein. To study the role of the DLC1 PBR on actin cytoskeletal remodeling, we made use of a doxycycline-inducible cellular system. In HEK293 Flp-In cells expressing the WT protein, cell spreading associated with lamellipodia and focal adhesion formation and directed migration were severely compromised. In contrast to this, the cellular actin architecture and migration characteristics of cells expressing DLC1 ΔPBR were more comparable to parental cells, proving that the PBR is required for DLC1 to suppress Rho-dependent biological processes.
Many proteins that are crucial to the assembly of the migration machinery are regulated by PI(4,5)P2. The formation of focal adhesions is dependent on localized PI(4,5)P2 generation, and PI(4,5)P2 is enriched at the leading edge (Janmey and Lindberg, 2004; Ling et al., 2006). The establishment and maintenance of local PI(4,5)P2 pools is controlled by the balanced action of phosphatidylinositol phosphate kinases and enzymes that consume PI(4,5)P2, such as phospholipases and phosphatidylinositol-3-kinase as well as PI(4,5)P2 sequestering proteins (Janmey and Lindberg, 2004; Ling et al., 2006). It is conceivable that PI(4,5)P2-dependent DLC1 activation is part of a negative feedback because Rho signaling contributes to PI(4,5)P2 production by activating phosphatidylinositol phosphate kinases (Oude Weernink et al., 2004). We attempted to investigate whether modulation of cellular PI(4,5)P2 levels may influence DLC1 GAP activity and thus RhoA-GTP levels. However, PI(4,5)P2 depletion by activation of phospholipases with 3m3FBS did not restore RhoA-GTP levels in cells expressing DLC1, as determined with Raichu-RhoA biosensor experiments (data not shown). However, such changes may not be seen in overexpression systems and are likely to require analysis by imaging at the single cell level. A challenge for the future is thus the identification of cooperating factors, both lipids and proteins, in DLC1 recruitment to membrane proximal sites in combination with the spatiotemporal analysis of DLC1-regulated Rho signaling events during cell spreading and migration.
Supplementary Material
ACKNOWLEDGMENTS
We wish to thank Irene Ng (University of Hong Kong, Hong Kong, China) for the plasmid encoding GFP-tagged DLC2, Michiyuki Matsuda (Osaka University, Osaka, Japan) for the Raichu-RhoA biosensor, Guido Posern (Max Planck Institute of Biochemistry, Martinsried, Germany) for the luciferase reporter 3DA.luc, Simone Schoenwaelder (Monash University, Melbourne, Australia) for pGEX-RhoA, and Kirill Alexandrov and Roger Goody (Max Planck Institute of Molecular Physiology, Dortmund, Germany) for recombinant GGTase. We are grateful to Peter Müller for helpful discussions and Klaus Pfizenmaier for critical reading of the manuscript. The laboratory of Monilola A. Olayioye is funded by grants of the Deutsche Forschungsgemeinschaft (DFG; SFB 495-Junior Research Group) and the Deutsche Krebshilfe (OM-106708 and -107545). Thomas G. Pomorski is supported by a Heisenberg grant of the DFG.
Abbreviations used:
- DLC
deleted in liver cancer
- FRET
fluorescence resonance energy transfer
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor
- PA
phosphatidic acid
- PBR
polybasic region
- PC
phosphatidylcholine
- PI
phosphatidylinositol
- PI(3,5)P3
phosphatidylinositol-3,4,5-trisphosphate
- PI(4)P
phosphatidylinositol-4-phosphate
- PI(4)P2
phosphatidylinositol-4,5-bisphosphate
- PLC-δ1
phospholipase C δ1
- PS
phosphatidylserine
- SAM
sterile α motif
- START
StAR-related lipid transfer
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-03-0247) on August 26, 2009.
REFERENCES
- Alpy F., Tomasetto C. Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J. Cell Sci. 2005;118:2791–2801. doi: 10.1242/jcs.02485. [DOI] [PubMed] [Google Scholar]
- Bernards A., Settleman J. GAPs in growth factor signalling. Growth Factors. 2005;23:143–149. doi: 10.1080/08977190500130480. [DOI] [PubMed] [Google Scholar]
- Bos J. L., Rehmann H., Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Brown M. T., Andrade J., Radhakrishna H., Donaldson J. G., Cooper J. A., Randazzo P. A. ASAP1, a phospholipid-dependent arf GTPase-activating protein that associates with and is phosphorylated by Src. Mol. Cell. Biol. 1998;18:7038–7051. doi: 10.1128/mcb.18.12.7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching Y. P., Wong C. M., Chan S. F., Leung T. H., Ng D. C., Jin D. Y., Ng I. O. Deleted in liver cancer (DLC) 2 encodes a RhoGAP protein with growth suppressor function and is underexpressed in hepatocellular carcinoma. J. Biol. Chem. 2003;278:10824–10830. doi: 10.1074/jbc.M208310200. [DOI] [PubMed] [Google Scholar]
- Durkin M. E., Ullmannova V., Guan M., Popescu N. C. Deleted in liver cancer 3 (DLC-3), a novel Rho GTPase-activating protein, is downregulated in cancer and inhibits tumor cell growth. Oncogene. 2007a;26:4580–4589. doi: 10.1038/sj.onc.1210244. [DOI] [PubMed] [Google Scholar]
- Durkin M. E., Yuan B. Z., Zhou X., Zimonjic D. B., Lowy D. R., Thorgeirsson S. S., Popescu N. C. DLC-1, a Rho GTPase-activating protein and tumour suppressor. J. Cell Mol. Med. 2007b;11:1185–1207. doi: 10.1111/j.1582-4934.2007.00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fivaz M., Meyer T. Specific localization and timing in neuronal signal transduction mediated by protein-lipid interactions. Neuron. 2003;40:319–330. doi: 10.1016/s0896-6273(03)00634-2. [DOI] [PubMed] [Google Scholar]
- Goodison S., Yuan J., Sloan D., Kim R., Li C., Popescu N. C., Urquidi V. The RhoGAP protein DLC-1 functions as a metastasis suppressor in breast cancer cells. Cancer Res. 2005;65:6042–6053. doi: 10.1158/0008-5472.CAN-04-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gureasko J., Galush W. J., Boykevisch S., Sondermann H., Bar-Sagi D., Groves J. T., Kuriyan J. Membrane-dependent signal integration by the Ras activator Son of sevenless. Nat. Struct. Mol. Biol. 2008;15:452–461. doi: 10.1038/nsmb.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy K. D., Hodgson L., Kim T. Y., Shutes A., Maddileti S., Juliano R. L., Hahn K. M., Harden T. K., Bang Y. J., Der C. J. DLC-1 suppresses non-small cell lung cancer growth and invasion by RhoGAP-dependent and independent mechanisms. Mol. Carcinog. 2008;47:326–337. doi: 10.1002/mc.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heering J., Erlmann P., Olayioye M. A. Simultaneous loss of the DLC1 and PTEN tumor suppressors enhances breast cancer cell migration. Exp. Cell Res. 2009;315:2505–2514. doi: 10.1016/j.yexcr.2009.05.022. [DOI] [PubMed] [Google Scholar]
- Heo W. D., Inoue T., Park W. S., Kim M. L., Park B. O., Wandless T. J., Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holeiter G., Heering J., Erlmann P., Schmid S., Jahne R., Olayioye M. A. Deleted in liver cancer 1 controls cell migration through a Dia1-dependent signaling pathway. Cancer Res. 2008;68:8743–8751. doi: 10.1158/0008-5472.CAN-08-0984. [DOI] [PubMed] [Google Scholar]
- Homma Y., Emori Y. A dual functional signal mediator showing RhoGAP and phospholipase C-delta stimulating activities. EMBO J. 1995;14:286–291. doi: 10.1002/j.1460-2075.1995.tb07002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J. H. Membrane binding domains. Biochim. Biophys. Acta. 2006;1761:805–811. doi: 10.1016/j.bbalip.2006.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson T. R., Brown F. D., Nie Z., Miura K., Foroni L., Sun J., Hsu V. W., Donaldson J. G., Randazzo P. A. ACAPs are arf6 GTPase-activating proteins that function in the cell periphery. J. Cell Biol. 2000;151:627–638. doi: 10.1083/jcb.151.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A. B., Hall A. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Janmey P. A., Lindberg U. Cytoskeletal regulation: rich in lipids. Nat. Rev. Mol. Cell. Biol. 2004;5:658–666. doi: 10.1038/nrm1434. [DOI] [PubMed] [Google Scholar]
- Kim C. A., Bowie J. U. SAM domains: uniform structure, diversity of function. Trends Biochem. Sci. 2003;28:625–628. doi: 10.1016/j.tibs.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Kim T. Y., Healy K. D., Der C. J., Sciaky N., Bang Y. J., Juliano R. L. Effects of structure of Rho GTPase-activating protein DLC-1 on cell morphology and migration. J. Biol. Chem. 2008;283:32762–32770. doi: 10.1074/jbc.M800617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krugmann S., et al. Identification of ARAP3, a novel PI3K effector regulating both Arf and Rho GTPases, by selective capture on phosphoinositide affinity matrices. Mol. Cell. 2002;9:95–108. doi: 10.1016/s1097-2765(02)00434-3. [DOI] [PubMed] [Google Scholar]
- Lahoz A., Hall A. DLC1, a significant GAP in the cancer genome. Genes Dev. 2008;22:1724–1730. doi: 10.1101/gad.1691408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y. C., Si L., DeVere White R. W., Lo S. H. The phosphotyrosine-independent interaction of DLC-1 and the SH2 domain of cten regulates focal adhesion localization and growth suppression activity of DLC-1. J. Cell Biol. 2007;176:43–49. doi: 10.1083/jcb.200608015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligeti E., Dagher M. C., Hernandez S. E., Koleske A. J., Settleman J. Phospholipids can switch the GTPase substrate preference of a GTPase-activating protein. J. Biol. Chem. 2004;279:5055–5058. doi: 10.1074/jbc.C300547200. [DOI] [PubMed] [Google Scholar]
- Ling K., Schill N. J., Wagoner M. P., Sun Y., Anderson R. A. Movin' on up: the role of PtdIns(4,5)P(2) in cell migration. Trends Cell Biol. 2006;16:276–284. doi: 10.1016/j.tcb.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Miura K., Jacques K. M., Stauffer S., Kubosaki A., Zhu K., Hirsch D. S., Resau J., Zheng Y., Randazzo P. A. ARAP1, a point of convergence for Arf and Rho signaling. Mol. Cell. 2002;9:109–119. doi: 10.1016/s1097-2765(02)00428-8. [DOI] [PubMed] [Google Scholar]
- Ng I.O., Liang Z. D., Cao L., Lee T. K. DLC-1 is deleted in primary hepatocellular carcinoma and exerts inhibitory effects on the proliferation of hepatoma cell lines with deleted DLC-1. Cancer Res. 2000;60:6581–6584. [PubMed] [Google Scholar]
- Oude Weernink P. A., Schmidt M., Jakobs K. H. Regulation and cellular roles of phosphoinositide 5-kinases. Eur. J. Pharmacol. 2004;500:87–99. doi: 10.1016/j.ejphar.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Papayannopoulos V., Co C., Prehoda K. E., Snapper S., Taunton J., Lim W. A. A polybasic motif allows N-WASP to act as a sensor of PIP(2) density. Mol. Cell. 2005;17:181–191. doi: 10.1016/j.molcel.2004.11.054. [DOI] [PubMed] [Google Scholar]
- Qian X., Li G., Asmussen H. K., Asnaghi L., Vass W. C., Braverman R., Yamada K. M., Popescu N. C., Papageorge A. G., Lowy D. R. Oncogenic inhibition by a deleted in liver cancer gene requires cooperation between tensin binding and Rho-specific GTPase-activating protein activities. Proc. Natl. Acad. Sci. USA. 2007;104:9012–9017. doi: 10.1073/pnas.0703033104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A. J. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006;16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Saito K., Fujimura-Kamada K., Hanamatsu H., Kato U., Umeda M., Kozminski K. G., Tanaka K. Transbilayer phospholipid flipping regulates Cdc42p signaling during polarized cell growth via Rga GTPase-activating proteins. Dev. Cell. 2007;13:743–751. doi: 10.1016/j.devcel.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Scholz R. P., Regner J., Theil A., Erlmann P., Holeiter G., Jahne R., Schmid S., Hausser A., Olayioye M. A. DLC1 interacts with 14-3--3 proteins to inhibit RhoGAP activity and block nucleocytoplasmic shuttling. J. Cell Sci. 2009;122:92–102. doi: 10.1242/jcs.036251. [DOI] [PubMed] [Google Scholar]
- Sekimata M., Kabuyama Y., Emori Y., Homma Y. Morphological changes and detachment of adherent cells induced by p122, a GTPase-activating protein for Rho. J. Biol. Chem. 1999;274:17757–17762. doi: 10.1074/jbc.274.25.17757. [DOI] [PubMed] [Google Scholar]
- Takahashi S., Pryciak P. M. Identification of novel membrane-binding domains in multiple yeast Cdc42 effectors. Mol. Biol. Cell. 2007;18:4945–4956. doi: 10.1091/mbc.E07-07-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler A. P., Ridley A. J. Why three Rho proteins? RhoA, RhoB, RhoC, and cell motility. Exp. Cell Res. 2004;301:43–49. doi: 10.1016/j.yexcr.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Williams C. L. The polybasic region of Ras and Rho family small GTPases: a regulator of protein interactions and membrane association and a site of nuclear localization signal sequences. Cell Signal. 2003;15:1071–1080. doi: 10.1016/s0898-6568(03)00098-6. [DOI] [PubMed] [Google Scholar]
- Wong C. M., Lee J. M., Ching Y. P., Jin D. Y., Ng I. O. Genetic and epigenetic alterations of DLC-1 gene in hepatocellular carcinoma. Cancer Res. 2003;63:7646–7651. [PubMed] [Google Scholar]
- Wong C. M., Yam J. W., Ching Y. P., Yau T. O., Leung T. H., Jin D. Y., Ng I. O. Rho GTPase-activating protein deleted in liver cancer suppresses cell proliferation and invasion in hepatocellular carcinoma. Cancer Res. 2005;65:8861–8868. doi: 10.1158/0008-5472.CAN-05-1318. [DOI] [PubMed] [Google Scholar]
- Xue W. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev. 2008;22:1439–1444. doi: 10.1101/gad.1672608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam J. W., Ko F. C., Chan C. Y., Jin D. Y., Ng I. O. Interaction of deleted in liver cancer 1 with tensin2 in caveolae and implications in tumor suppression. Cancer Res. 2006;66:8367–8372. doi: 10.1158/0008-5472.CAN-05-2850. [DOI] [PubMed] [Google Scholar]
- Yang X. Y., Guan M., Vigil D., Der C. J., Lowy D. R., Popescu N. C. p120Ras-GAP binds the DLC1 Rho-GAP tumor suppressor protein and inhibits its RhoA GTPase and growth-suppressing activities. Oncogene. 2009 doi: 10.1038/onc.2008.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki H., Ohba Y., Kurokawa K., Itoh R. E., Nakamura T., Mochizuki N., Nagashima K., Matsuda M. Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. J. Cell Biol. 2003;162:223–232. doi: 10.1083/jcb.200212049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B. Z., Jefferson A. M., Baldwin K. T., Thorgeirsson S. S., Popescu N. C., Reynolds S. H. DLC-1 operates as a tumor suppressor gene in human non-small cell lung carcinomas. Oncogene. 2004;23:1405–1411. doi: 10.1038/sj.onc.1207291. [DOI] [PubMed] [Google Scholar]
- Yuan B. Z., Zhou X., Durkin M. E., Zimonjic D. B., Gumundsdottir K., Eyfjord J. E., Thorgeirsson S. S., Popescu N. C. DLC-1 gene inhibits human breast cancer cell growth and in vivo tumorigenicity. Oncogene. 2003;22:445–450. doi: 10.1038/sj.onc.1206064. [DOI] [PubMed] [Google Scholar]
- Zhong D., Zhang J., Yang S., Soh U. J., Buschdorf J. P., Zhou Y. T., Yang D., Low B. C. The SAM domain of the RhoGAP DLC1 binds EF1A1 to regulate cell migration. J. Cell Sci. 2009;122:414–424. doi: 10.1242/jcs.027482. [DOI] [PubMed] [Google Scholar]
- Zhou X., Thorgeirsson S. S., Popescu N. C. Restoration of DLC-1 gene expression induces apoptosis and inhibits both cell growth and tumorigenicity in human hepatocellular carcinoma cells. Oncogene. 2004;23:1308–1313. doi: 10.1038/sj.onc.1207246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.