Abstract
Functional interactions of the translational activator Mss51 with both the mitochondrially encoded COX1 mRNA 5′-untranslated region and with newly synthesized unassembled Cox1 protein suggest that it has a key role in coupling Cox1 synthesis with assembly of cytochrome c oxidase. Mss51 is present at levels that are near rate limiting for expression of a reporter gene inserted at COX1 in mitochondrial DNA, and a substantial fraction of Mss51 is associated with Cox1 protein in assembly intermediates. Thus, sequestration of Mss51 in assembly intermediates could limit Cox1 synthesis in wild type, and account for the reduced Cox1 synthesis caused by most yeast mutations that block assembly. Mss51 does not stably interact with newly synthesized Cox1 in a mutant lacking Cox14, suggesting that the failure of nuclear cox14 mutants to decrease Cox1 synthesis, despite their inability to assemble cytochrome c oxidase, is due to a failure to sequester Mss51. The physical interaction between Mss51 and Cox14 is dependent upon Cox1 synthesis, indicating dynamic assembly of early cytochrome c oxidase intermediates nucleated by Cox1. Regulation of COX1 mRNA translation by Mss51 seems to be an example of a homeostatic mechanism in which a positive effector of gene expression interacts with the product it regulates in a posttranslational assembly process.
INTRODUCTION
The largest subunit of mitochondrial cytochrome c oxidase, Cox1, is encoded in the mitochondrial DNA (mtDNA) of all eukaryotic species that have been examined (Gray et al., 2004), and it is synthesized by their organellar genetic systems. Cox1 is highly hydrophobic, spanning the inner mitochondrial membrane 12 times, and it is complexed with several metal ions and two heme A moieties that participate directly in electron transport (Tsukihara et al., 1996). It is assembled into the core of cytochrome c oxidase, largely surrounded by subunits encoded by nuclear genes. The processes by which Cox1 is assembled with the other subunits and cofactors into an active enzyme are highly complex, requiring at least 30 genes in Saccharomyces cerevisiae (Herrmann and Funes, 2005; Khalimonchuk and Rodel, 2005; Cobine et al., 2006; Fontanesi et al., 2006; Barrientos et al., 2009). The assembly pathway is not understood in detail. In mammals, analysis of mutant and drug-treated cell lines indicates that Cox1 is a component of the earliest assembly intermediates (Nijtmans et al., 1998; Williams et al., 2004), and similar analysis in yeast is consistent with this idea (Horan et al., 2005).
An important function of this assembly process may be to prevent incompletely assembled components of cytochrome c oxidase from generating damaging reactive oxygen species, before they are contained by the holoenzyme. Indeed, mutations in several yeast genes required for cytochrome c oxidase assembly cause hypersensitivity to hydrogen peroxide (Pungartnik et al., 1999; Williams et al., 2005; Banting and Glerum, 2006), and a key component of the reactive prooxidant species is Cox1 (Khalimonchuk et al., 2007). One feature of the assembly process that is likely to play a role in minimizing the level of such prooxidant species is the coupling of Cox1 synthesis to assembly of cytochrome c oxidase (Barrientos et al., 2004).
Translation of S. cerevisiae mitochondrially coded mRNAs within the organelle is tightly controlled by nuclearly encoded mRNA-specific translational activators that, in most cases, recognize the 5′-untranslated regions (UTRs) of their target mRNAs (reviewed in Fox, 1996; Towpik, 2005). For example, Pet309 recognizes the leader of the COX1 mRNA and specifically activates synthesis of the Cox1 protein (Manthey and McEwen, 1995). Furthermore, Pet309 also interacts with the activators of COX2 and COX3 mRNA translation to colocalize synthesis of the three core subunits of cytochrome c oxidase, promoting efficient assembly (Sanchirico et al., 1998; Naithani et al., 2003).
Mss51 is the second known COX1 mRNA-specific translational activator (Decoster et al., 1990; Siep et al., 2000). It is of particular interest because dominant MSS51 missense mutations can suppress the leaky cytochrome c oxidase assembly defect caused by a shy1Δ mutation in yeast (Barrientos et al., 2002). Mutations affecting the human homologue of SHY1, SURF1 (Mashkevich et al., 1997), cause a similar cytochrome oxidase deficiency associated with Leigh syndrome (Tiranti et al., 1998; Zhu et al., 1998). PSI-BLAST comparisons indicate that both the human and mouse genomes encode a possible orthologue of yeast Mss51 (NP_001019764 and XP_912342) (Saccharomyces Genome Database), although the function of this mammalian protein is unknown.
Yeast Mss51 has two genetically distinct activities that make it an excellent candidate for a regulatory protein coupling Cox1 synthesis to cytochrome c oxidase assembly (Perez-Martinez et al., 2003). First, Mss51 is required to translate an mRNA encoding a reporter gene, ARG8m, inserted at the COX1 locus in place of the COX1 protein coding sequence, demonstrating that Mss51 has a target in either the 5′- or 3′-UTRs of the COX1 mRNA (or both). This activity may resemble that of other known translational activators. However, Mss51 (but not Pet309) is also required to express a chimeric mRNA bearing the untranslated regions of the COX2 mRNA flanking either the COX1 coding sequence or a COX1::ARGm translational fusion gene, demonstrating that Mss51 has a second genetically defined target mapping in the COX1 coding sequence itself (Perez-Martinez et al., 2003). Furthermore, immune precipitation of epitope-tagged Mss51 efficiently coprecipitates newly synthesized, unassembled Cox1 (Perez-Martinez et al., 2003; Barrientos et al., 2004; Mick et al., 2007). This strongly suggests that the second target of Mss51 action is Cox1 itself. This protein–protein interaction seems to be necessary for Cox1 synthesis (Perez-Martinez et al., 2003) and is also likely to be required for early steps in the cytochrome c oxidase assembly pathway (Mick et al., 2007; Pierrel et al., 2007; Zambrano et al., 2007).
Evidence indicating a coupling of Cox1 synthesis to assembly emerged from a systematic study demonstrating that most yeast mutations which disrupt cytochrome c oxidase assembly reduce, but do not eliminate, in vivo pulse labeling of Cox1 (Barrientos et al., 2004), a phenomenon observed previously in a few mutants (Poutre and Fox, 1987; Calder and McEwen, 1991). These observations suggest the existence of an assembly–feedback control system similar to that discovered previously in the chloroplast of Chlamydomonas reinhardtii (Choquet et al., 1998). Overproduction of Mss51 in several of the yeast assembly mutants largely reversed their Cox1 synthesis reductions (Barrientos et al., 2004). Interestingly, one cytochrome c oxidase assembly mutation, cox14Δ, did not reduced Cox1 pulse labeling, indicating that Cox14 is required for the feedback. Coprecipitation experiments revealed that Cox14 interacts both with newly synthesized Cox1 and with Mss51. Based on these findings and the dual activities of Mss51 (Perez-Martinez et al., 2003), Barrientos et al. (2004) proposed a model in which sequestration of Mss51 in assembly intermediates containing Cox1 and Cox14 could limit Cox1 synthesis (Barrientos et al., 2004).
In this article, we present evidence that strongly supports a regulatory role for Mss51 in controlling Cox1 translation and demonstrates dynamic interactions among newly synthesized Cox1, Mss51, and Cox14 that couple Cox1 synthesis to early steps in cytochrome c oxidase assembly.
MATERIALS AND METHODS
Strains, Media, and Genetic Methods
The S. cerevisiae strains used in this study are listed in Table 1. Standard genetic methods and media recipes were as described previously (Rose et al., 1988; Fox et al., 1991). Complete fermentable media were YPD or YPRaf (containing 2% glucose or 2% raffinose). Nonfermentable medium was YPEG (3% glycerol and 3% ethanol) or 2% lactate. Minimal media contained 0.67% yeast nitrogen base, 2% glucose, and Complete Supplement Mixtures (CSMs) purchased from Bio 101 (Vista, CA). Sequences encoding 3xHA or 3xMyc epitope tags were added to the 3′ ends of nuclear gene coding sequences, without altering mRNA flanking sequences, by pop-in pop-out transformation as described previously (Schneider et al., 1995). Mitochondrial transformation, and integration of altered genes into rho+ mtDNA, was as described previously (Perez-Martinez et al., 2003; Bonnefoy et al., 2007). Plasmids (see below) bearing cox1Δ::ARG8m-1, cox1Δ::ARG8m-2, and COX1::3xHA chimeric genes were transformed into the rho0 strain NAB69 by high-velocity microprojectile bombardment. Mitochondrial transformants were identified by their ability to rescue arginine growth when mated with a rho+ cox3Δ::arg8m-1 mutant (Bonnefoy and Fox, 2000) or by respiratory growth when mated with a rho+ strain carrying a cox1-D369N mutation, L45 (Meunier et al., 1993). Altered mitochondrial genes in the transformants were integrated by homologous recombination into rho+ mtDNA by isolating cytoductants issued from crosses of the transformants to either NB40-36a followed by selection for Arg+ growth, or to XPM10b followed by selection for respiratory growth, as appropriate.
Table 1.
Yeast strainsa used in this study
| Strain | Genotype | Reference |
|---|---|---|
| CAB267 | MATα, ade2, ura3Δ, cox14Δ::URA3, MSS51::3xHA [ρ+] | This study |
| CAB268 | MATα, ade2, ura3Δ, cox14Δ::URA3 [ρ+] | This study |
| CAB299 | MATα, ura3, leu2-3,112, lys2, his4-519, MSS51::3xHA, COX14::3×MYC [ρ+, cox1Δ::ARG8m, cox2Δ::COX1b, COX2] | This study |
| CAB300 | MATα, ura3, leu2-3,112, lys2, his4-519, MSS51::3xHA, COX14::3×MYC [ρ+ ΔΣaI, ΔΣbI] | This study |
| CAB302 | MATα, ura3, leu2-3,112, lys2, his4-519, MSS51::3xHA, COX14::3×MYC, pet309::URA3[ρ+ ΔΣaI, ΔΣbI] | This study |
| CAB306 | MATα, ura3, leu2-3,112, lys2, his4-519, MSS51::3xHA, COX14::3×MYC, cbs2::URA3,[ρ+ ΔΣaI, ΔΣbI] | This study |
| CAB312 | MATα, ura3Δ::KanMX3, leu2-3,112, his4-519, MSS51::3xMYC, [ρ+] | This study |
| CAB313 | MATα, ura3-52, leu2-3,112, lys2, his4-519, arg8::hisG, MSS51::3xMYC, [ρ+ COX1::3xHA ΔΣaI, ΔΣbI] | This study |
| CAB315 | MATα, ura3-52, leu2-3,112, lys2, his4-519, arg8::hisG, MSS51::3xMYC, mss2Δ::LEU2[ρ+ COX1::3xHA ΔΣaI, ΔΣbI] | This study |
| DAU1 | MATα, ade2, ura3Δ [ρ+] | Costanzo and Fox (1988) |
| NAB69 | MATa, ade2-101, arg8::hisG, ura3-52, kar1-1 [ρ0] | Perez-Martinez et al. (2003) |
| NB40-36a | MATα, lys2, leu2-3,112, arg8::hisG, ura3-52 [ρ+] | Perez-Martinez et al. (2003) |
| SB7 | MATα, ade2, ura3Δ, MSS51::3xHA [ρ+] | Perez-Martinez et al. (2003) |
| TF258 | MATα, ura3, leu2-3,112, lys2, his4-519, MSS51::3xHA, COX14::3xMYC [ρ+] | This study |
| TF259 | MATα, ura3, leu2-3,112, lys2, his4-519, MSS51::3xHA, COX14::3xMYC [ρ+ cox1Δ] | This study |
| TF266 | MATα, ura3, leu2-3,112, lys2, his4-519, MSS51::3xHA, COX14::3xMYC [ρ+ cox2-N15I] | This study |
| XPM10b | MATα, lys2, leu2-3,112, arg8::hisG, ura3-52 [ρ+, cox1Δ::ARG8m] | (Perez-Martinez et al., 2003) |
| XPM34 | MATα, lys2, leu2-3,112, arg8::hisG, ura3-52 [ρ+ cox2-N15I] | This study |
| XPM63a | MATα, lys2, leu2-3,112, arg8::hisG, ura3-52, mss51Δ::LEU2 [ρ+, cox1Δ::ARG8m] | (Perez-Martinez et al., 2003) |
| XPM270a | MATα, ade2, ura3Δ, MSS51::3xHA arg8::hisG, [ρ+, cox1Δ::ARG8m, cox2Δ::COX1b, COX2] | This Study |
| XPM271a | MATα, lys2, leu2-3,112, arg8::hisG, ura3-52 [ρ+, cox1Δ::ARG8m-1] | This study |
| XPM275a | MATα, lys2, leu2-3,112, arg8::hisG, ura3-52, mss51Δ::LEU2 [ρ+, cox1Δ::ARG8m-1] | This study |
| XPM286 | MATα/MATa, ade2/ADE2, lys2/LYS2, leu2-3,112/LEU2, arg8::hisG/arg8Δ::URA3, ura3-52/ura3Δ [ρ+, cox1Δ::ARG8m] | This study |
| XPM287 | MATα/MATa, pet309::URA3/PET309, ade2/ADE2, lys2/LYS2, leu2-3,112/LEU2, arg8::hisG/arg8Δ::URA3, ura3-52/ura3Δ [ρ+, cox1Δ::ARG8m] | This study |
| XPM288 | MATα/MATa, mss51Δ::LEU2/MSS51, ade2/ADE2, lys2/LYS2, leu2-3,112/LEU2, arg8::hisG/arg8Δ::URA3, ura3-52/ura3Δ [ρ+, cox1Δ::ARG8m] | This study |
| XPM291 | MATα/MATa, mss51Δ::LEU2/MSS51, pet309::URA3/PET309, ade2/ADE2, lys2/LYS2,leu2-3,112/LEU2, arg8::hisG/arg8Δ::URA3, ura3-52/ura3Δ [ρ+, cox1Δ::ARG8m] | This study |
| XPM304b | MATα, lys2, leu2-3,112, arg8::hisG, ura3-52 [ρ+, cox1Δ::ARG8m-2] | This study |
| XPM312a | MATα, lys2, leu2-3,112, arg8::hisG, ura3-52, mss51Δ::LEU2 [ρ+, cox1Δ::ARG8m-2] | This study |
a All strains are congenic or isogenic to D273-10B, except for NAB69. Mitochondrial genotypes are shown in brackets. ΔΣaI ΔΣbI indicate intronless mtDNA (Labouesse, 1990).
b Chimeric cox2Δ::COX1 gene inserted ectopically upstream of COX2 (Perez-Martinez et al., 2003).
Construction of Chimeric Mitochondrial Genes
Chimeric genes were generated by the fusion polymerase chain reaction technique (Ho et al., 1989) using Pfu polymerase (Stratagene, La Jolla, CA) or Taq polymerase (Invitrogen, Carlsbad, CA), as described previously (Perez-Martinez et al., 2003). The cox1Δ::ARG8m-1 construct (pXPM76) consists of 395 base pairs of the COX1 5′-UTR sequence, followed by ARG8m, and 119 base pairs encoding the COX2 3′-UTR replacing 525 base pairs corresponding to the COX1 3′-UTR. After the COX2 3′-UTR sequence, there are 465 base pairs of downstream COX1 flanking sequence to allow integration at the COX1 locus. The cox1Δ::ARG8m-2 construct (pXPM80) consists of 568 base pairs of upstream COX1 flanking sequence, followed by 73 base pairs encoding the COX2 promoter and 5′-UTR, ARG8m, and 990 base pairs of downstream COX1 flanking sequence encoding its 3′-UTR. The COX1::3xHA construct (pXPM63) consists of the intronless COX1 coding sequence (Labouesse, 1990) with 90 base pairs encoding three HA epitopes inserted upstream of the stop codon, flanked by 395 base pairs of upstream sequence and 990 base pairs of downstream sequence.
Analysis of Mitochondrial Proteins
Yeast cells were grown in 10 ml of complete raffinose medium until late log phase. Cells were disrupted by vortexing with glass beads, and crude mitochondria were obtained as described by Diekert et al. (2001), except that protease inhibitor mini-tablets (Roche Diagnostics, Indianapolis, IN) were added instead of phenylmethylsulfonyl fluoride. Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) (Laemmli, 1970). For Western blots, proteins were transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore, Billerica, MA), or nitrocellulose where indicated, and probed with an anti-Arg8 antibody (Steele et al., 1996), anti-Cox1 (MitoSciences, Eugene, OR), anti-hemagglutinin (HA) (Roche Diagnostics), or anti-Myc (Roche Diagnostics). Immune complexes were detected with either goat anti-rabbit immunoglobulin (Ig)G or anti-mouse IgG conjugated to horseradish peroxidase (Bio-Rad Laboratories, Hercules, CA) and the enhanced chemiluminescence (ECL) kit (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom), or when using anti-Cox1, the ECL Plus kit (GE Healthcare). Alternatively, for the experiment of Figure 2A and similar experiments, the secondary antibody was AlexaFluor488-conjugated goat anti-mouse IgG (Invitrogen). Fluorophore signals were detected using a Storm PhosphorImager 840 (GE Healthcare). The signals were quantitated by subtracting the sum of pixel intensities of a background area from the sum of pixel intensities of an equal sample area as described (Demlow and Fox, 2003). For immunoprecipitation, mitochondria were solubilized in 1% digitonin and incubated with anti-HA agarose (Roche Diagnostics) as described previously (Herrmann et al., 2001; Perez-Martinez et al., 2003). Total proteins were precipitated from solubilized extracts and immune supernatants using StrataClean resin (Stratagene, San Diego, CA) (Ziegler et al., 1997). Mitochondrial translation products were radiolabeled in 300–700 μg of highly purified mitochondria for 30 min at 25°C in the presence of [35S]methionine, as described previously (Westermann et al., 2001; Perez-Martinez et al., 2003).
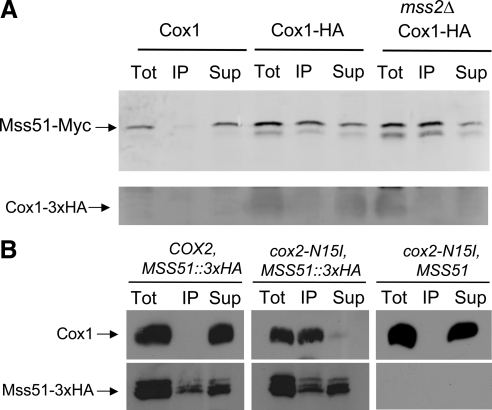
Figure 2.
A large fraction of total Mss51-3xMyc coimmune precipitates with Cox1-3xHA from mitochondrial extracts. (A) Mitochondria were isolated from three strains that each contained Mss51-3xMyc encoded by a modified chromosomal MSS51 gene. CAB312 (Cox1) was otherwise wild-type; CAB313 (Cox1-HA) has a modified intronless mitochondrial COX1 gene encoding Cox1-3xHA; CAB315 (mss2Δ Cox1-HA) had both Cox1-3xHA and a nuclear mss2Δ mutation. Mitochondria were solubilized with digitonin. Half of each extract was analyzed as total protein (Tot). The other half was subjected to immunoprecipitation with anti-HA antibody, yielding a precipitate (IP) and supernatant (Sup). The three fractions from each strain were subjected to SDS gel electrophoresis and Western blotting with anti-Myc antibody. Immune complexes were visualized by phosphorimaging after reaction with AlexaFluor coupled anti-mouse-IgG (see Materials and Methods). (B) Mitochondria were isolated from two strains that contained Mss51-3xHA encoded by a modified chromosomal MSS51 gene; TF258 (COX2) contained wild-type mtDNA; TF266 (cox2-N15I) had a missense substitution mutation that prevents processing of the pre-Cox2 precursor. In addition, the control strain XPM34 contained unmodified Mss51 and the cox2-N15I mutation. Mitochondria were extracted and subjected to immune precipitation as described in A. The Western blots were probed with anti-Cox1 antibody and subsequently with anti-HA antibody, then visualized by ECL detection (see Materials and Methods). The cox2-N15I blots probed with anti-Cox1 were overexposed relative to the others (see text).
RESULTS
Mss51 Acts on the COX1 mRNA 5′-UTR and Is Present at Levels near Rate Limiting for Expression
Mss51 is required posttranscriptionally for expression of a chimeric mitochondrial mRNA composed of the COX1 5′- and 3′-UTRs flanking the mitochondrial reporter gene ARG8m, demonstrating that one of its activities is mediated through untranslated COX1 mRNA sequences (Perez-Martinez et al., 2003). Other well studied mitochondrial translational activators target exclusively mRNA 5′-UTRs (Towpik, 2005). However, the unusual nature of Mss51's dual activities raised the possibility that it might target the 3′-UTR, the mRNA region mediating many other cases of translational control (Kuersten and Goodwin, 2003). To determine whether the 5′- or 3′-UTR could be a sole target of Mss51 in vivo, we created strains in which COX2 5′- and 3′-UTRs individually replaced the COX1 5′- or 3′-UTRs flanking ARG8m by inserting chimeric genes into mtDNA at the COX1 locus (see Materials and Methods). We deleted the 525 base pairs encoding the COX1 3′-UTR, replacing them with 118 base pairs encoding the COX2 3′-UTR. In a separate strain, we deleted the 505 base pairs encoding the COX1 5′-UTR, replacing them with 73 base pairs encoding the COX2 promoter and COX2 mRNA 5′-UTR, which contains the target of the COX2-specific translational activator Pet111 (Dunstan et al., 1997; Green-Willms et al., 2001) (Figure 1A). In strains containing a wild-type nuclear genome both chimeric mitochondrial mRNAs supported Arg+ growth. In strains containing the mss51Δ mutation, the chimeric ARG8m mRNA bearing the COX1 5′-UTR and COX2 3′-UTR failed to support Arg+ growth. However, the mss51Δ strain containing the chimeric mRNA bearing the COX2 5′-UTR and COX1 3′-UTR grew well in the absence of arginine. As expected, expression of this ARG8m mRNA bearing the COX2 5′-UTR was also independent of the COX1-specific translational activator Pet309 (Manthey and McEwen, 1995; Perez-Martinez et al., 2003) but was dependent upon the COX2-specific translational activator Pet111 (data not shown). Thus, Mss51 has a translational activation target in the COX1 mRNA 5′-UTR and may act there together with Pet309. This target could correspond to a region of the COX1 mRNA 5′-UTR that interacts with Mss51 in the yeast three-hybrid system (Zambrano et al., 2007).
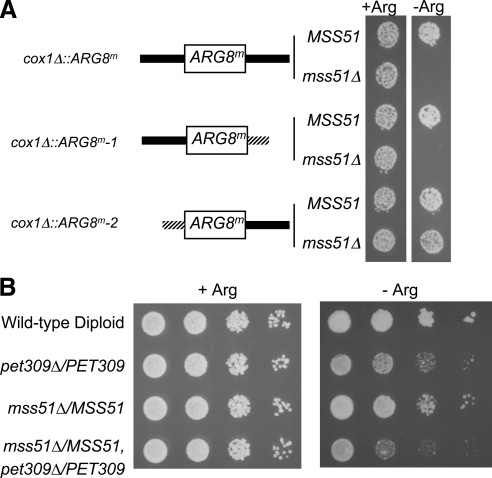
Figure 1.
Mss51 acts on the COX1 mRNA 5′-UTR and is rate limiting, along with Pet309, for mRNA translation. (A) Cells grown in liquid YPD were spotted on CSM raffinose medium containing (+Arg) or lacking (−Arg) arginine and incubated for 4 d at 30°C. The cells contained cox1Δ::ARG8m genes at the COX1 locus in mtDNA encoding chimeric mRNAs: black bars, COX1 untranslated regions; stippled bars, COX2 untranslated regions. cox1Δ::ARG8m (top diagram) has ARG8m flanked by native COX1 5′- and 3′-UTRs, with (XPM10b) or without (XPM63a) MSS51. cox1Δ::ARG8m-1 (middle diagram) has ARG8m flanked by the native COX1 5′-UTR, and the COX2 3′-UTR (525 base pairs of COX1 downstream sequence replaced by 118 base pairs of COX2 downstream sequence) with (XPM271a) or without (XPM275a) MSS51. cox1Δ::ARG8m-2 (bottom diagram) has ARG8m flanked by the COX2 5′-UTR (505 base pairs of COX1 upstream sequence replaced by 73 base pairs of COX2 upstream sequence), and the native COX1 3′-UTR with (XPM304b) or without (XPM312a) MSS51. (B) Diploid cells containing the cox1Δ::ARG8m reporter in mtDNA were grown in liquid YPD. Ten-fold serial dilutions of diploid cells were spotted on CSM glucose media containing (+Arg) or lacking (−Arg) arginine, and incubated for 4 d at 30°C. The indicated relevant nuclear genotypes correspond to the following strains (Table 1): wild type, XPM286; pet309Δ/PET309, XPM287; mss51Δ/MSS51, XPM288; and pet309Δ/PET309 mss51Δ/MSS51, XPM291.
For mRNA-specific translational activation through the COX1 5′-UTR to play a role in modulating Cox1 synthesis, Mss51 and/or Pet309 should be present at or near rate-limiting levels in mitochondria. To test whether this is so, we examined expression of ARG8m, inserted in place of the COX1 codons, in diploid strains lacking one copy of MSS51, PET309, or both. Expression of the reporter in heterozygous diploids relative to homozygous wild type was assayed by growth on medium lacking arginine (Figure 1B). In both cases, reduced gene dosage decreased the rate of growth on medium lacking arginine, with pet309Δ/PET309 having a stronger effect than mss51Δ/MSS51. A diploid heterozygous for both nuclear mutations was more strongly affected than either single mutant. We conclude that the levels of Mss51 and Pet309 are at or near rate limiting for translational activation through the COX1 5′-UTR.
A Significant Fraction of Total Mss51-3xMyc Interacts with Cox1-3xHA
Mick et al. (2007) have shown that affinity purification of Mss51 from unlabeled wild-type cells yields cytochrome c oxidase assembly intermediate complexes that contain unassembled Cox1. If intermediate complexes containing Cox1 bind a significant fraction of total Mss51, then the levels of assembly intermediates could affect the availability of rate-limiting Mss51 for translational activation through the COX1 mRNA 5′-untranslated leader (UTL). To ask whether such complexes contain a significant fraction of total cellular Mss51, we examined the efficiency with which Mss51 would coimmune precipitate with Cox1 from digitonin solubilized extracts of mitochondria. We were unable to immune precipitate Cox1 with the commercially available anti-Cox1 antibody (MS418; MitoSciences). We therefore modified the COX1 gene in mtDNA by the addition of sequences encoding three HA-epitopes at the Cox1 C terminus (see Materials and Methods). Otherwise wild-type cells containing this mtDNA grew normally on nonfermentable carbon sources. To allow detection of Mss51 in fractions derived from this strain, we added sequences encoding three Myc-epitopes at the C terminus of Mss51 without altering sequences flanking MSS51 (Schneider et al., 1995). These epitopes did not affect respiratory growth of otherwise wild-type strains.
Mitochondria were isolated from log-phase cells containing both Cox1-3xHA and Mss51-3xMyc, as well as a control strain lacking the tag on Cox1, and were solubilized with digitonin. One-half of each extract was immune precipitated with anti-HA coupled to agarose beads. Total proteins were collected from the other half of each extract, and from the immune supernatants, by precipitation with StrataClean resin (Stratagene). The precipitated fractions were analyzed by semiquantitative Western blotting using anti-Myc primary antibody and fluorescein-labeled secondary antibody. Signals were detected digitally by phosphorimaging (see Materials and Methods). The average percent of total of Mss51-3xMyc recovered in the anti-HA immune precipitates from four trials was 41% (SD = 22). One such experiment is shown in Figure 2A.
The average percentage of total Cox1-3xHA precipitated by anti-HA was 24% (SD = 10). This relatively low efficiency of direct immune-precipitation suggests that the three HA-epitopes may be sterically blocked in a high fraction of fully assembled cytochrome c oxidase complexes. Thus, the immune precipitates may be enriched for assembly intermediates. These data are consistent with the hypothesis that a significant fraction of Mss51 may be sequestered with Cox1 in assembly intermediates and thus unavailable for translational activation through the COX1 mRNA 5′-UTL.
If cytochrome c oxidase assembly is blocked at a step downstream in the pathway, then the fraction of Mss51-3xMyc present in assembly intermediates could be higher than in wild type. We tested this by measuring coimmune precipitation from a strain lacking MSS2, a nuclear gene required for export of the Cox2 C-tail domain to the intermembrane space (Broadley et al., 2001). However, in this case the average percentage of Mss51-3xMyc precipitating with Cox1-3xHA was 50 in four trials (SD = 35) (Figure 2A), not significantly higher than in wild type. In addition, the epitope tag apparently stabilized unassembled Cox1 in the mss2 mutant. Although these data do not provide supporting evidence for increased sequestration of Mss51 with Cox1 in the assembly defective mutant, it must be noted that the measurements are inherently imprecise, and the physiologically relevant differences between wild type and mutant could be small.
We also compared wild type to an assembly defective strain by asking what fraction of total unmodified Cox1 protein would coimmune precipitate with tagged Mss51-3xHA. Here, we disrupted cytochrome c oxidase assembly using the mitochondrial cox2-N15I mutation, which prevents processing of the pre-Cox2 leader peptide (Saracco, 2003). Solubilized extracts of mitochondria were subjected to immune precipitation with immobilized anti-HA, and the precipitates were analyzed by Western blotting (Figure 2B). Owing to the weak immune reaction of the available anti-Cox1 monoclonal antibody with Cox1, we had to use for the Westerns an enhanced chemiluminescence detection system that does not allow quantitative analysis of signal strength. The steady-state level of Cox1 in extracts of COX2 mitochondria was far higher than that in the cox2-N15I mutant extracts, as expected. We therefore overexposed the anti-Cox1 blot from the mutant relative to wild type, to achieve comparable signal strength. We also probed the blots with anti-HA to detect Mss51-3xHA and exposed these wild-type and mutant blots equally. (It seems that the 3xHA epitope was partially destroyed by proteolysis during the immune precipitation, producing multiple bands and apparent incomplete recovery of total Mss51-3xHA in the precipitate plus supernatant fractions.) Importantly, the residual Cox1 present in the cox2-N15I mutant was highly enriched in the anti-HA immune precipitate (in an epitope-dependent manner), in contrast to wild-type where most of the Cox1, which is assembled into cytochrome oxidase, remained in the soluble extract after immune precipitation (Figure 2B). Thus, when assembly was disrupted the steady-state level of Cox1 was greatly reduced, but virtually all of the Cox1 present was associated with Mss51.
Coimmune Precipitation of Newly Synthesized Cox1 with Mss51-3xHA Depends upon Cox14
Cox14 is a short mitochondrial protein, encoded in the nucleus, that interacts with newly synthesized Cox1 and with Mss51 (Barrientos et al., 2004). Cox14 is required for assembly of active cytochrome c oxidase and accumulation of Cox1, but unlike most other assembly-defective mutants, cox14 mutants do not exhibit assembly feedback inhibition of COX1 mRNA translation as judged by pulse labeling (Barrientos et al., 2004).
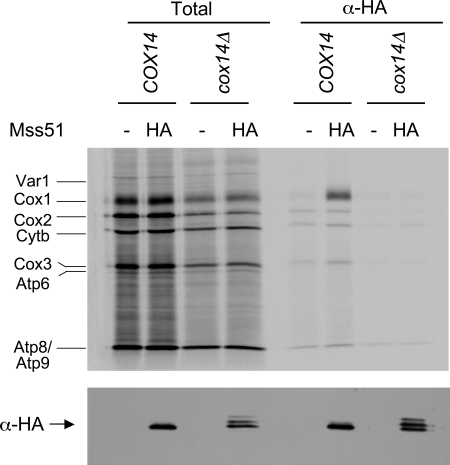
Stable interaction of Mss51 with newly synthesized Cox1 could be dependent on Cox14. To test this idea, we asked whether Cox1, newly synthesized in mitochondria from a cox14Δ mutant, would coimmune precipitate with Mss51-3xHA. Mitochondria were isolated from COX14 and cox14Δ strains that contained either wild-type Mss51 or the functional Mss51-3xHA. After mitochondrial translation in the presence of [35S]methionine and solubilization, anti-HA antibody was added to the mitochondrial lysates and immune precipitates were analyzed by SDS gel electrophoresis and autoradiography. Mss51-3xHA selectively coprecipitated Cox1 from the wild-type COX14 strain but failed to do so from the cox14Δ strain (Figure 3), contrary to results reported previously (Barrientos et al., 2004). Probing of a Western blot of these lysates with anti-HA antibody confirmed that Mss51-3xHA was present in the absence of Cox14. These results strongly suggest that Cox14 is required for Mss51 to form complexes with newly synthesized Cox1 that are stable under our solubilization conditions.
Figure 3.
The interaction between Mss51 and newly synthesized Cox1 is greatly reduced in the absence of Cox14. Mitochondria were purified from four strains whose relevant genotype was either COX14 or cox14Δ and contained either Mss51 or Mss51-3xHA, as indicated. The mitochondria were allowed to synthesize mitochondrially coded proteins in the presence of [35S]methionine (see Materials and Methods) and then solubilized with digitonin. 10% of the extract was subjected to SDS-PAGE (Total), whereas 90% was immunoprecipitated with anti-HA antibody and then subjected to SDS-PAGE (α-HA). The gel was blotted to a nitrocellulose membrane, which was autoradiographed (top). Labeled translation products are indicated as follows: cytochrome c oxidase subunit 1, Cox1; subunit 2, Cox2; subunit 3, Cox3; cytochrome b, Cytb; subunit 6 of ATPase, Atp6; subunit 8, Atp8; subunit 9, Atp9; and the ribosomal protein, Var1. After autoradiography, the membrane was probed with anti-HA antibody and visualized by ECL detection to confirm precipitation of Mss51-3xHA (bottom). The strains, from left to right, were DAU1, SB7, CAB268, and CAB267 (see Table 1).
It is impossible to test reciprocally whether the interaction of Cox14 with newly synthesized Cox1 depends upon Mss51, because in the absence of Mss51, Cox1 cannot be synthesized.
Interaction between Mss51 and Cox14 Depends upon Cox1 Synthesis
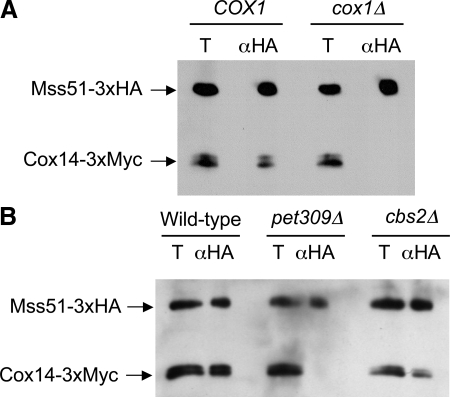
Mss51 and Cox14 have been shown by coimmunoprecipitation to interact physically (Barrientos et al., 2004). These nuclearly coded proteins could interact in a stable complex, or associate dynamically in response to synthesis of mitochondrially encoded Cox1. We first tested this by constructing strains containing Mss51-3xHA and Cox14-3xMyc, with either wild-type mtDNA, or mtDNA bearing a cox1Δ mutation that eliminates the coding sequence, as well as 787 base pairs of upstream, and 525 base pairs of downstream sequence. Mitochondria were isolated from both strains and solubilized with digitonin. The soluble extracts were immunoprecipitated with an anti-HA antibody, and the precipitates were analyzed by Western blot probed with both anti-HA and anti-Myc (Figure 4A). As expected, Mss51-3xHA coimmunoprecipitated Cox14-3xMyc from the extract of the COX1 mitochondria. However, Cox14-3xMyc was not coprecipitated from the extract of the cox1Δ mitochondria.
Figure 4.
Cox1 synthesis is necessary for the interaction between Mss51 and Cox14. (A) Mitochondria were purified from cells containing Mss51-3xHA and Cox14-3xMyc, and either wild-type mtDNA (COX1) or a cox1 deletion (cox1Δ) in mtDNA (strains TF258 and TF259). The mitochondria were solubilized with digitonin and the extracts were incubated with anti-HA antibody (αHA). The precipitates were analyzed by Western blot probed with anti-Myc and anti-HA antibodies. The total fractions (T) corresponds to 10% of the mitochondrial extract, the αHA precipitates were from the remaining 90%. (B) Mitochondria were purified from cells containing Mss51-3xHA, Cox14-3xMyc, and intronless mtDNA, that were otherwise wild type (Wild-type), pet309::URA3 (pet309), or cbs2::URA3 (cbs2) (strains CAB300, CAB302, and CAB306). Analysis was as described in A.
To ask whether the Cox1 protein or the COX1 mRNA was necessary for the interaction between Mss51-3xHA and Cox14-3xMyc, we tested for coimmunoprecipitation in a pet309Δ strain whose mtDNA does not contain introns. Strains of this genotype contain near wild-type levels of mature COX1 mRNA but do not translate it (Manthey and McEwen, 1995). Consistent with the hypothesis that Cox1 synthesis is necessary for the interaction between Mss51 and Cox14, the pet309Δ mutation prevented coimmune precipitation of Cox14-3xMyc by Mss51-3xHA (Figure 4B). As a control, we tested whether a cbs2Δ mutation, which produces a respiratory negative phenotype by specifically preventing translation of apo-cytochrome b from the COB mRNA (Rödel, 1986), would prevent the Mss51–Cox14 interaction. It did not, confirming the specific requirement for Cox1 synthesis of the Mss51–Cox14 interaction (Figure 4B).
Mss51 Can Interact with Newly Synthesized Cox1 and with Cox14 Independently of the COX1 mRNA 5′-UTR
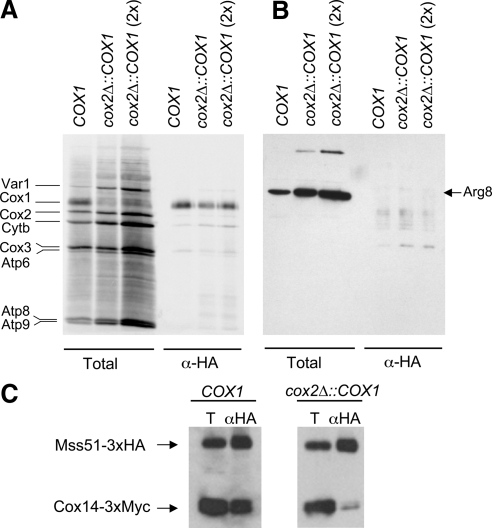
Cox1 protein can be synthesized and assembled into functional cytochrome oxidase from a chimeric mRNA bearing the COX1 coding sequence flanked by COX2 5′- and 3′-UTRs, transcribed from an ectopic cox2Δ::COX1 locus in mtDNA. Translation of this chimeric mRNA is reduced relative to wild type, but nevertheless dependent upon MSS51 function, demonstrating a second function for Mss51 protein, distinct from its interaction with the COX1 mRNA 5′-UTR (Perez-Martinez et al., 2003). If the interaction between Mss51 and newly synthesized Cox1 corresponds to this second function, then this physical interaction also should be independent of the COX1 5′-UTR on the mRNA. To test this, we radiolabeled newly synthesized Cox1 in mitochondria isolated from strains containing Mss51-3xHA and either the COX1 gene in wild-type mtDNA, or the ectopic cox2Δ::COX1 locus in mtDNA with cox1Δ::ARG8m at the COX1 locus. After solubilization with digitonin and addition of anti-HA antibody, immune precipitates were isolated and analyzed by SDS gel electrophoresis and autoradiography of a gel-blot (Figure 5A). Newly synthesized radiolabeled Cox1 was selectively precipitated with Mss51-3xHA from extracts of mitochondria bearing either the wild-type COX1 mRNA or the chimeric cox2Δ::COX1 mRNA, demonstrating that the Mss51-Cox1 physical interaction does not require the COX1 mRNA 5′-UTR in cis to the coding sequence. As a further control, we also probed the blot with anti-Arg8 antibody. As expected, no coimmune precipitation of Arg8 protein with Mss51-3xHA was detectable despite its translation from an mRNA bearing the COX1 mRNA 5′-UTL (Figure 5B).
Figure 5.
Mss51 interaction with newly synthesized Cox1 is not dependent on the COX1 mRNA UTRs. (A) Mitochondria were purified from two strains containing Mss51-3xHA, with either wild-type mtDNA (SB7) or modified mtDNA bearing the cox1Δ::ARG8m deletion and cox2Δ::COX1, an ectopic chimeric gene encoding an mRNA with the COX1 codons flanked by COX2 5′- and 3′-UTRs (XPM270a). The mitochondria were allowed to synthesize labeled proteins, and Total and α-HA precipitated proteins from wild-type (COX1) and the ectopic chimeric (cox2Δ::COX1) mRNAs were analyzed as described in the experiment in Figure 3. The third lane in each series had twice the amount of mitochondria (700 μg), as indicated. (B) After autoradiography, the membrane was probed with anti-Arg8 antibody and visualized by ECL detection. (C) Extracts of unlabeled mitochondria from strains containing Mss51-3xHA and Cox14-3xMyc, and either wild-type mtDNA (COX1) (TF258) or modified mtDNA the encoding the ectopic chimeric mRNA with COX1 codons flanked by COX2 5′- and 3′-UTRs (CAB299) were analyzed by immune precipitation and Western blot as described in Figure 4.
We also tested by coimmune precipitation whether the Cox1-translation-dependent interaction between Mss51 and Cox14 requires prior interaction of Mss51 with the COX1 mRNA 5′-UTR. Cox14-3xMyc was coprecipitated with Mss51-3xHA from extracts of mitochondria containing the chimeric cox2Δ::COX1 mRNA, albeit at lower efficiency than from mitochondria with the wild-type COX1 mRNA (Figure 5C). Thus, the interaction between Mss51 and Cox14 can occur in the absence of the COX1 mRNA 5′-UTR in cis to the coding sequence. The lower efficiency of Cox14 coprecipitation may reflect the lower level of Cox1 synthesis observed from the chimeric mRNA (Perez-Martinez et al., 2003).
DISCUSSION
Mss51 has functional interactions with both the COX1 mRNA and with newly synthesized unassembled Cox1 protein. Its involvement in both gene regulation and assembly of the protein whose synthesis it regulates suggests a role for Mss51 in homeostatic coupling of Cox1 synthesis to its assembly into cytochrome c oxidase (Perez-Martinez et al., 2003). Our data strongly support this hypothesis.
By manipulation of the mtDNA sequences flanking the COX1 coding sequences in rho+ mtDNA, we genetically mapped the Mss51 target in the COX1 mRNA to the 5′-UTR. This A+U-rich 450 nucleotide mRNA leader also contains the target of the COX1 mRNA-specific translational activator Pet309 (Manthey and McEwen, 1995). Although neither of these targets have been further localized genetically, the downstream 245 nucleotides of the COX1 mRNA 5′-UTR interact in a yeast three-hybrid assay with an N-terminal portion of Mss51 (but not full-length Mss51), suggesting that the biological target of Mss51 is relatively close to the protein coding sequence (Zambrano et al., 2007). Although the mechanism(s) by which these, and other, mitochondrial translational activators function is unknown, it is interesting to note the overall similarities to the control of cytoplasmic translation of mRNAs bearing internal ribosome entry sites (IRES) in their 5′-UTRs by IRES-transacting factors (Komar and Hatzoglou, 2005).
mRNA-specific translational activation is known to be rate limiting for expression of reporter genes inserted into the COX2 and COX3 mitochondrial loci in yeast (Steele et al., 1996; Cohen and Fox, 2001; Green-Willms et al., 2001), and we found that PET309 gene dosage is similarly limiting for expression of COX1. Interestingly, Mss51 is approximately fourfold more abundant than Pet309 in S288c-related cells grown on glucose (Ghaemmaghami et al., 2003) and approximately eightfold more abundant in D273–10B-related cells grown on raffinose (unpublished data). Despite this relative abundance of Mss51, we nevertheless detected reduced expression of the ARG8m reporter inserted into the COX1 locus in diploid cells containing a single MSS51 nuclear gene, relative to homozygous wild-type diploids. Thus, Mss51 is present at levels that are near rate limiting for translational activation of the COX1 mRNA, consistent with the possibility that it has a role in regulating the level of Cox1 synthesis in mitochondria. Reduced gene dosage of both PET309 and MSS51 together had a stronger negative effect on reporter gene expression than reduced dosage of either alone. This suggests that when both Mss51 and Pet309 levels are lowered, relatively few mRNAs are simultaneously occupied by both necessary factors. However, this result does not distinguish whether the Mss51 and Pet309 have distinct functions in 5′-UTR–dependent translational activation, or work together to execute a single activity.
Barrientos et al. (2004) observed that that the synthesis of Cox1 within mitochondria, as measured by pulse labeling in vivo, was reduced in several mutant strains unable to assemble cytochrome c oxidase. However, this assembly–feedback regulation was not observed in assembly-defective cox14 mutants, nor in double mutants lacking both COX14 and other genes necessary for assembly. Thus, the 70-amino acid Cox14 protein is required both for assembly and for feedback regulation. Furthermore, Cox14 coprecipitated with both newly synthesized Cox1 and with Mss51. Based on these data, Barrientos et al. (2004) proposed that reduced synthesis of Cox1 in most assembly-defective mutants could be due to Cox14-dependent sequestration of Mss51 in assembly intermediates, although they reported that the association of Mss51 with newly synthesized Cox1 was not dependent upon Cox14.
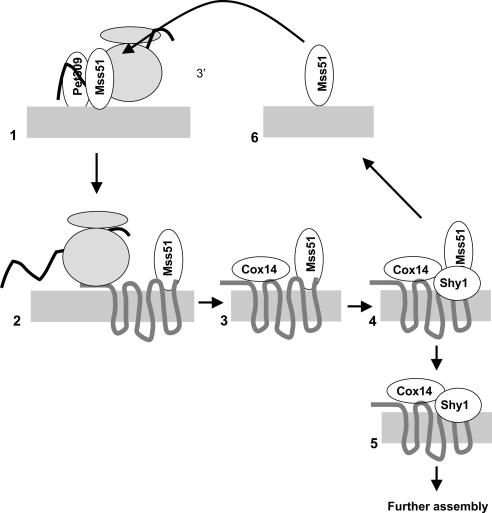
Our findings, together with those of previous studies, support sequestration of Mss51 in early assembly intermediates as a mechanism for coupling Cox1 synthesis and assembly, in a sequence of events depicted in Figure 6. First, on the inner surface of the inner membrane (Siep et al., 2000; Tavares-Carreon et al., 2008) Mss51 and Pet309 activate translation of the COX1 mRNA (Manthey and McEwen, 1995; Perez-Martinez et al., 2003) through functional interactions with a site or sites in its 5′-UTR. Here, it is important to note that the levels of both Pet309 and Mss51 are at or near rate limiting for translational activation of chimeric COX1 mRNA encoding the reporter ARG8m. Thus, this is likely to be a point of regulation for Cox1 synthesis, as well as its localization (Naithani et al., 2003).
Figure 6.
Dual activities of Mss51, and the stabilization of Mss51–Cox1 interaction by Cox14, couple Cox1 synthesis to assembly (see Discussion). 1) Mss51 and Pet309 activate COX1 mRNA through the 5′-UTR. 2) Mss51 interacts with newly synthesized Cox1 and allows completed translation through an unknown mechanism (Perez-Martinez et al., 2003). 3) Cox14 enters a complex that stabilizes the binding of Mss51 to newly synthesized Cox1. 4) Shy1 associates with the complex shown in 3 and facilitates dissociation of an early assembly intermediate (5) and Mss51 (6). Released Mss51 is available to activate another round of COX1 mRNA translation. Not shown: the protein Coa1 enters the pathway in step 3 or 4 (Pierrel et al., 2007), and Coa2 enters the assembly complex downstream of step 4 (Pierrel et al., 2008). Cox14 and Shy1 remain associated with assembled cytochrome oxidase supercomplexes (Mick et al., 2007).
Second, as the completed or nearly completed Cox1 polypeptide emerges, it interacts with Mss51 (Figure 6, step 2). This step is inferred from the fact that Mss51 is required for Cox1 synthesis even when the COX2-mRNA specific translational activator Pet111 carries out the upstream activation function on a COX2 mRNA 5′-UTR fused to the COX1 coding sequence in a chimeric mRNA (Perez-Martinez et al., 2003). It presumably precedes involvement of Cox14, because Cox1 synthesis is robust in cox14Δ mutants (Barrientos et al., 2004). This inferred interaction must be weak because we did not detect coimmune precipitation of Cox1 with Mss51 from solubilized extracts in the absence of Cox14 (Figure 3). The role of Mss51 at this step is unknown. However, it may function by antagonizing a late Cox1 translation elongation arrest, a regulatory mechanism that has been documented in bacteria (Perez-Martinez et al., 2003; Woolhead et al., 2006).
Third, a complex containing at least Mss51 and Cox14 assembles dynamically with newly synthesized Cox1 (Figure 6, step 3). It is possible that Mss51 molecules interacting with the COX1 mRNA 5′-UTR normally transit in cis directly to a nascent Cox1 polypeptide translated from that mRNA molecule. This would generate a requirement for another Mss51 molecule to interact with the COX1 mRNA to activate the next round of translation. However, we found that Mss51 does interact physically with newly synthesized Cox1 translated from the chimeric COX1 mRNA bearing the COX2 mRNA 5′-UTR. Thus, Mss51 is capable of binding to nascent Cox1 (and to Cox14) in the absence of a COX1 mRNA 5′-UTR in cis to the coding sequence. The Cox1–Mss51–Cox14 complex may also contain the assembly factor Coa1 (Pierrel et al., 2007), not depicted in Figure 6.
The interaction between Mss51 and Cox14 must be bridged dynamically by newly synthesized Cox1, because Mss51 and Cox14 do not coimmune precipitate from extracts of mitochondria from either cox1Δ mutant, or a pet309 mutant, that specifically fail to synthesize Cox1. This finding is consistent with the observation of high molecular weight complexes containing Mss51 that are absent after treatment of wild-type cells with chloramphenicol or in a pet309 mutant (Pierrel et al., 2008). The assembly intermediate complex(es) containing Mss51, Cox14 and newly synthesized Cox1 contain roughly half of total Mss51, based on our coimmune precipitation with epitope-tagged Cox1. This finding is consistent with the observation that ∼50 of Mss51 is associated with Cox14 (Barrientos et al., 2004). Interestingly, these complexes seem to contain >0.1 of total cellular Cox1 based on data reported by Mick et al. (2007), indicating a significant pool of Cox1 is present in early assembly intermediates containing Mss51.
Fourth, Shy1 associates with Cox1-containing complexes, probably after Mss51, Cox14, and Coa1, although the evidence for this order is not conclusive (Figure 6, step 4) (Mick et al., 2007; Pierrel et al., 2007). Both Shy1 and Cox14 are associated physically with Mss51, with downstream assembly intermediates lacking Mss51, and with fully assembled respiratory supercomplexes containing active cytochrome c oxidase (complex IV) and the cytochrome bc1 complex (complex III) (Mick et al., 2007). We found that a cox1 deletion mutation in mtDNA prevented coimmune precipitation of Shy1 with Mss51 (unpublished data), consistent with the dynamic assembly of Shy1-containing complexes nucleated by newly synthesized Cox1. Mutations inactivating Shy1 in yeast, and its orthologues in humans and bacteria, decrease but do not eliminate cytochrome c oxidase activity, apparently by decreasing the efficiency of heme a3 insertion into Cox1 (Smith et al., 2005; Khalimonchuk et al., 2007; Bundschuh et al., 2008; Pierrel et al., 2008). Interestingly, respiratory growth of a yeast shy1Δ mutant is improved by dominant suppressor mutations in MSS51 and by overexpression of wild-type MSS51 (Barrientos et al., 2002).
Finally, Cox1-containing assembly intermediates that retain Cox14 and Shy1 but not Mss51 proceed toward further assembly by insertion of metal ions and heme a moieties into Cox1, and association of additional enzyme subunits (Mick et al., 2007; Pierrel et al., 2007; Khalimonchuk and Winge, 2008; Barrientos et al., 2009). At this step, Mss51 is released from the assembly pathway and available to activate additional rounds of COX1 mRNA translation (Figure 6, steps 5 and 6).
Sequestration of Mss51 in early complexes formed during the assembly process could cause decreased COX1 mRNA translation if the early complexes over accumulate due to downstream blocks in assembly, as proposed previously (Barrientos et al., 2004). Our data support this model by showing that Mss51 levels can limit COX1 mRNA translation and that a significant fraction of cellular Mss51 is associated with early assembly complexes containing Cox1. Furthermore, our data explain why mutations eliminating Cox14 do not prevent Cox1 synthesis, despite preventing cytochrome c oxidase assembly: Mss51 is not stably associated with newly synthesized Cox1 in the absence of Cox14 and therefore remains available to activate futile synthesis of Cox1. However, the Cox1 produced in the absence of Cox14 is highly unstable and present at lower steady-state levels than thos% observed in mutants blocked further downstream in the assembly process (Barrientos et al., 2004). This feedback system, observed in mutants, may reflect a mechanism that normally coordinates the level of newly synthesized Cox1 with the levels of assembly factors in wild-type cells and thereby helps to protect against oxidative damage (Khalimonchuk et al., 2007).
In contrast to expectation, we did not observe a significant increase in the fraction of total Mss51-3xMyc coprecipitated with Cox1-3xHA when cytochrome c oxidase assembly was blocked by a nuclear mss2Δ (Broadley et al., 2001) mutation. However, the available methodology for quantifying these solubilized coprecipitated complexes is not highly accurate, and the physiologically relevant differences may not be great. Furthermore, we cannot determine whether the presence of epitope tags exerts subtle effects on the behavior of these proteins. Thus, these data do not argue strongly against the model. When we compared wild type with an assembly defective mitochondrial cox2 mutant (Saracco, 2003) for the fraction of total unmodified Cox1 coprecipitated with Mss51-3xHA, we observed a dramatic difference. In contrast to wild type, virtually all of the Cox1 present in the mutant was associated with Mss51, consistent with the idea that early assembly intermediates containing Mss51 and Cox1 accumulate when assembly is blocked.
Mutants lacking the assembly factor Coa1 seem to resemble cox14Δ mutants insofar as they exhibit normal pulse labeling of Cox1 (Mick et al., 2007; Pierrel et al., 2007, 2008). However, in contrast to the tight respiratory negative phenotype of cox14Δ mutants, coa1Δ mutants exhibit low levels of respiration (Pierrel et al., 2007) and weak growth on nonfermentable carbon sources (Mick et al., 2007). In these respects, and their suppressibility by over expressed MSS51, the coa1Δ and shy1Δ mutations are similar (Barrientos et al., 2002; Pierrel et al., 2007). Furthermore, coa1Δ does not disrupt the interaction between Mss51 and Cox14 (Pierrel et al., 2007). Thus, Coa1 is probably not required for the interaction between Mss51 and newly synthesized Cox1 but could reinforce it. Robust Cox1 synthesis in the coa1Δ mutant may depend on the low level of cytochrome c oxidase assembly that takes place in its absence.
Photosynthetic complexes are composed of protein subunits encoded by genes in both chloroplast and nuclear DNA, analogously to mitochondrial respiratory complexes. In Chlamydomonas reinhardtii, translation of chloroplast encoded mRNAs specifying certain key subunits of photosystem I, photosystem II, and the cytochrome b6f complex has been shown to be coupled to the assembly of those complexes (Choquet and Wollman, 2002). In each case, regulation of translation by assembly was dependent upon the 5′-UTRs of the chloroplast encoded mRNAs (Choquet et al., 1998; Wostrikoff et al., 2004; Minai et al., 2006). In the case of cytochrome f synthesis, assembly feedback regulation depends upon C-terminal residues of cytochrome f itself, which is hypothesized to interact with an effector protein that directly regulates translation (Choquet et al., 2003). These findings suggest that mRNA-specific coupling of organellar translation to the assembly of energy transducing complexes may be widespread in eukaryotes, and raise interesting questions about how such regulation could be achieved in mammalian mitochondria whose mRNAs lack 5′-UTRs.
ACKNOWLEDGMENTS
We thank Adam C. Martin for isolation of the cox2-N15I mutation, David Mick for helpful discussions, and Soledad Funes for critically reading the manuscript. This work was supported by a Pew Charitable Trusts Fellowship, research grants Consejo Nacional de Ciencia y Tecnologia (82505) and Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica, Universidad Nacional Autónoma de México (IN201805) (to X.P.-M.), and by National Institutes of Health grant GM-29362 (to T.D.F.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-06-0522) on August 26, 2009.
REFERENCES
- Banting G. S., Glerum D. M. Mutational analysis of the Saccharomyces cerevisiae cytochrome c oxidase assembly protein Cox11p. Eukaryot. Cell. 2006;5:568–578. doi: 10.1128/EC.5.3.568-578.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A., Gouget K., Horn D., Soto I. C., Fontanesi F. Suppression mechanisms of COX assembly defects in yeast and human: insights into the COX assembly process. Biochim. Biophys. Acta. 2009;1793:97–107. doi: 10.1016/j.bbamcr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A., Korr D., Tzagoloff A. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh's syndrome. EMBO J. 2002;21:43–52. doi: 10.1093/emboj/21.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A., Zambrano A., Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004;23:3472–3482. doi: 10.1038/sj.emboj.7600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefoy N., Fox T. D. In vivo analysis of mutated initiation codons in the mitochondrial COX2 gene of Saccharomyces cerevisiae fused to the reporter gene ARG8m reveals lack of downstream reinitiation. Mol. Gen. Genet. 2000;262:1036–1046. doi: 10.1007/pl00008646. [DOI] [PubMed] [Google Scholar]
- Bonnefoy N., Remacle C., Fox T. D. Genetic transformation of Saccharomyces cerevisiae and Chlamydomonas reinhardtii mitochondria. Methods Cell Biol. 2007;80:525–548. doi: 10.1016/S0091-679X(06)80026-9. [DOI] [PubMed] [Google Scholar]
- Broadley S. A., Demlow C. M., Fox T. D. A peripheral mitochondrial inner membrane protein, Mss2p, required for export of the mitochondrially coded Cox2p C-tail in Saccharomyces cerevisiae. Mol. Cell Biol. 2001;21:7663–7672. doi: 10.1128/MCB.21.22.7663-7672.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundschuh F. A., Hoffmeier K., Ludwig B. Two variants of the assembly factor Surf1 target specific terminal oxidases in Paracoccus denitrificans. Biochim. Biophys. Acta. 2008;1777:1336–1343. doi: 10.1016/j.bbabio.2008.05.448. [DOI] [PubMed] [Google Scholar]
- Calder K. M., McEwen J. E. Deletion of the COX7 gene in Saccharomyces cerevisiae reveals a role for cytochrome c oxidase subunit VII in assembly of remaining subunits. Mol. Microbiol. 1991;5:1769–1777. doi: 10.1111/j.1365-2958.1991.tb01926.x. [DOI] [PubMed] [Google Scholar]
- Choquet Y., Stern D. B., Wostrikoff K., Kuras R., Girard-Bascou J., Wollman F. A. Translation of cytochrome f is autoregulated through the 5′ untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc. Natl. Acad. Sci. USA. 1998;95:4380–4385. doi: 10.1073/pnas.95.8.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y., Wollman F. A. Translational regulations as specific traits of chloroplast gene expression. FEBS Lett. 2002;529:39–42. doi: 10.1016/s0014-5793(02)03260-x. [DOI] [PubMed] [Google Scholar]
- Choquet Y., Zito F., Wostrikoff K., Wollman F. A. Cytochrome f translation in Chlamydomonas chloroplast is autoregulated by its carboxyl-terminal domain. Plant Cell. 2003;15:1443–1454. doi: 10.1105/tpc.011692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobine P. A., Pierrel F., Winge D. R. Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochim. Biophys. Acta. 2006;1763:759–772. doi: 10.1016/j.bbamcr.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Cohen J. S., Fox T. D. Expression of green fluorescent protein from a recoded gene inserted into Saccharomyces cerevisiae mitochondrial DNA. Mitochondrion. 2001;1:181–189. doi: 10.1016/s1567-7249(01)00012-5. [DOI] [PubMed] [Google Scholar]
- Costanzo M. C., Fox T. D. Specific translational activation by nuclear gene products occurs in the 5′ untranslated leader of a yeast mitochondrial mRNA. Proc. Natl. Acad. Sci. USA. 1988;85:2677–2681. doi: 10.1073/pnas.85.8.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decoster E., Simon M., Hatat D., Faye G. The MSS51 gene product is required for the translation of the COX1 mRNA in yeast mitochondria. Mol. Gen. Genet. 1990;224:111–118. doi: 10.1007/BF00259457. [DOI] [PubMed] [Google Scholar]
- Demlow C. M., Fox T. D. Activity of mitochondrially synthesized reporter proteins is lower than imported proteins, and is increased by lowering cAMP in glucose-grown Saccharomyces cerevisiae cells. Genetics. 2003;165:961–974. doi: 10.1093/genetics/165.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekert K., de Kroon A. I., Kispal G., Lill R. Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 2001;65:37–51. doi: 10.1016/s0091-679x(01)65003-9. [DOI] [PubMed] [Google Scholar]
- Dunstan H. M., Green-Willms N. S., Fox T. D. In vivo analysis of Saccharomyces cerevisiae COX2 mRNA 5′-untranslated leader functions in mitochondrial translation initiation and translational activation. Genetics. 1997;147:87–100. doi: 10.1093/genetics/147.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi F., Soto I. C., Horn D., Barrientos A. Assembly of mitochondrial cytochrome c oxidase, a complicated and highly regulated cellular process. Am. J. Physiol. Cell Physiol. 2006;291:C1129–C1147. doi: 10.1152/ajpcell.00233.2006. [DOI] [PubMed] [Google Scholar]
- Fox T. D. Genetics of mitochondrial translation. In: Hershey J.W.B., Matthews M. B., Sonenberg N., editors. Translational Control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 733–758. [Google Scholar]
- Fox T. D., Folley L. S., Mulero J. J., McMullin T. W., Thorsness P. E., Hedin L. O., Costanzo M. C. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gray M. W., Lang B. F., Burger G. Mitochondria of protists. Annu. Rev. Genet. 2004;38:477–524. doi: 10.1146/annurev.genet.37.110801.142526. [DOI] [PubMed] [Google Scholar]
- Green-Willms N. S., Butler C. A., Dunstan H. M., Fox T. D. Pet111p, an inner membrane-bound translational activator that limits expression of the Saccharomyces cerevisiae mitochondrial gene COX2. J. Biol. Chem. 2001;276:6392–6397. doi: 10.1074/jbc.M009856200. [DOI] [PubMed] [Google Scholar]
- Herrmann J. M., Funes S. Biogenesis of cytochrome oxidase-sophisticated assembly lines in the mitochondrial inner membrane. Gene. 2005;354:43–52. doi: 10.1016/j.gene.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Herrmann J. M., Westermann B., Neupert W. Analysis of protein-protein interactions in mitochondria by coimmunoprecipitation and chemical cross-linking. Methods Cell Biol. 2001;65:217–230. doi: 10.1016/s0091-679x(01)65013-1. [DOI] [PubMed] [Google Scholar]
- Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Horan S., Bourges I., Taanman J. W., Meunier B. Analysis of COX2 mutants reveals cytochrome oxidase subassemblies in yeast. Biochem. J. 2005;390:703–708. doi: 10.1042/BJ20050598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalimonchuk O., Bird A., Winge D. R. Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J. Biol. Chem. 2007;282:17442–17449. doi: 10.1074/jbc.M702379200. [DOI] [PubMed] [Google Scholar]
- Khalimonchuk O., Rodel G. Biogenesis of cytochrome c oxidase. Mitochondrion. 2005;5:363–388. doi: 10.1016/j.mito.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Khalimonchuk O., Winge D. R. Function and redox state of mitochondrial localized cysteine-rich proteins important in the assembly of cytochrome c oxidase. Biochim. Biophys. Acta. 2008;1783:618–628. doi: 10.1016/j.bbamcr.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar A. A., Hatzoglou M. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J. Biol. Chem. 2005;280:23425–23428. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- Kuersten S., Goodwin E. B. The power of the 3′ UTR: translational control and development. Nat. Rev. Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- Labouesse M. The yeast mitochondrial leucyl-tRNA synthetase is a splicing factor for the excision of several group I introns. Mol. Gen. Genet. 1990;224:209–221. doi: 10.1007/BF00271554. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Manthey G. M., McEwen J. E. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 1995;14:4031–4043. doi: 10.1002/j.1460-2075.1995.tb00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashkevich G., Repetto B., Glerum D. M., Jin C., Tzagoloff A. SHY1, the yeast homolog of the mammalian SURF-1 gene, encodes a mitochondrial protein required for respiration. J. Biol. Chem. 1997;272:14356–14364. doi: 10.1074/jbc.272.22.14356. [DOI] [PubMed] [Google Scholar]
- Meunier B., Lemarre P., Colson A.-M. Genetic screening in Saccharomyces cerevisiae for large numbers of mitochondrial point mutations which affect structure and function of catalytic subunits of cytochrome-c oxidase. Eur. J. Biochem. 1993;213:129–135. doi: 10.1111/j.1432-1033.1993.tb17742.x. [DOI] [PubMed] [Google Scholar]
- Mick D. U., Wagner K., van der Laan M., Frazier A. E., Perschil I., Pawlas M., Meyer H. E., Warscheid B., Rehling P. Shy1 couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 2007;26:4347–4358. doi: 10.1038/sj.emboj.7601862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minai L., Wostrikoff K., Wollman F. A., Choquet Y. Chloroplast biogenesis of photosystem II cores involves a series of assembly-controlled steps that regulate translation. Plant Cell. 2006;18:159–175. doi: 10.1105/tpc.105.037705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naithani S., Saracco S. A., Butler C. A., Fox T. D. Interactions among COX1, COX2 and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:324–333. doi: 10.1091/mbc.E02-08-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijtmans L. G., Taanman J. W., Muijsers A. O., Speijer D., Van den Bogert C. Assembly of cytochrome-c oxidase in cultured human cells. Eur. J. Biochem. 1998;254:389–394. doi: 10.1046/j.1432-1327.1998.2540389.x. [DOI] [PubMed] [Google Scholar]
- Perez-Martinez X., Broadley S. A., Fox T. D. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 2003;22:5951–5961. doi: 10.1093/emboj/cdg566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrel F., Bestwick M. L., Cobine P. A., Khalimonchuk O., Cricco J. A., Winge D. R. Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 2007;26:4335–4346. doi: 10.1038/sj.emboj.7601861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrel F., Khalimonchuk O., Cobine P. A., Bestwick M., Winge D. R. Coa2 is an assembly factor for yeast cytochrome c oxidase biogenesis facilitating the maturation of Cox1. Mol. Cell Biol. 2008;28:4927–4939. doi: 10.1128/MCB.00057-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poutre C. G., Fox T. D. PET111, a Saccharomyces cerevisiae nuclear gene required for translation of the mitochondrial mRNA encoding cytochrome c oxidase subunit II. Genetics. 1987;115:637–647. doi: 10.1093/genetics/115.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pungartnik C., Kern M. F., Brendel M., Henriques J. A. Mutant allele pso7–1, that sensitizes Saccharomyces cerevisiae to photoactivated psoralen, is allelic with COX11, encoding a protein indispensable for a functional cytochrome c oxidase. Curr. Genet. 1999;36:124–129. doi: 10.1007/s002940050481. [DOI] [PubMed] [Google Scholar]
- Rödel G. Two yeast nuclear genes, CBS1 and CBS2, are required for translation of mitochondrial transcripts bearing the 5′-untranslated COB leader. Curr. Genet. 1986;11:41–45. doi: 10.1007/BF00389424. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Sanchirico M. E., Fox T. D., Mason T. L. Accumulation of mitochondrially synthesized Saccharomyces cerevisiae Cox2p and Cox3p depends on targeting information in untranslated portions of their mRNAs. EMBO J. 1998;17:5796–5804. doi: 10.1093/emboj/17.19.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco S. A. Ithaca, NY: Cornell University; 2003. Analysis of the export of Saccharomyces cerevisiae Cox2p from the mitochondrial matrix. Ph.D. Thesis. [Google Scholar]
- Schneider B. L., Seufert W., Steiner B., Yang Q. H., Futcher A. B. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- Siep M., van Oosterum K., Neufeglise H., van der Spek H., Grivell L. A. Mss51p, a putative translational activator of cytochrome c oxidase subunit-1 (COX1) mRNA, is required for synthesis of Cox1p in Saccharomyces cerevisiae. Curr. Genet. 2000;37:213–220. doi: 10.1007/s002940050522. [DOI] [PubMed] [Google Scholar]
- Smith D., Gray J., Mitchell L., Antholine W. E., Hosler J. P. Assembly of cytochrome c oxidase in the absence of the assembly protein surf1p leads to loss of the active site heme. J. Biol. Chem. 2005;280:17652–17656. doi: 10.1074/jbc.C500061200. [DOI] [PubMed] [Google Scholar]
- Steele D. F., Butler C. A., Fox T. D. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc. Natl. Acad. Sci. USA. 1996;93:5253–5257. doi: 10.1073/pnas.93.11.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares-Carreon F., Camacho-Villasana Y., Zamudio-Ochoa A., Shingu-Vazquez M., Torres-Larios A., Perez-Martinez X. The pentatricopeptide repeats present in Pet309 are necessary for translation but not for stability of the mitochondrial COX1 mRNA in yeast. J. Biol. Chem. 2008;283:1472–1479. doi: 10.1074/jbc.M708437200. [DOI] [PubMed] [Google Scholar]
- Tiranti V., et al. Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 1998;63:1609–1621. doi: 10.1086/302150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towpik J. Regulation of mitochondrial translation in yeast. Cell Mol. Biol. Lett. 2005;10:571–594. [PubMed] [Google Scholar]
- Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- Westermann B., Herrmann J. M., Neupert W. Analysis of mitochondrial translation products in vivo and in organello in yeast. Methods Cell Biol. 2001;65:429–438. doi: 10.1016/s0091-679x(01)65025-8. [DOI] [PubMed] [Google Scholar]
- Williams J. C., Sue C., Banting G. S., Yang H., Glerum D. M., Hendrickson W. A., Schon E. A. Crystal structure of human SCO 1, implications for redox signaling by a mitochondrial cytochrome c oxidase “assembly” protein. J. Biol. Chem. 2005;280:15202–15211. doi: 10.1074/jbc.M410705200. [DOI] [PubMed] [Google Scholar]
- Williams S. L., Valnot I., Rustin P., Taanman J. W. Cytochrome c oxidase subassemblies in fibroblast cultures from patients carrying mutations in COX10, SCO1, or SURF1. J. Biol. Chem. 2004;279:7462–7469. doi: 10.1074/jbc.M309232200. [DOI] [PubMed] [Google Scholar]
- Woolhead C. A., Johnson A. E., Bernstein H. D. Translation arrest requires two-way communication between a nascent polypeptide and the ribosome. Mol. Cell. 2006;22:587–598. doi: 10.1016/j.molcel.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Wostrikoff K., Girard-Bascou J., Wollman F. A., Choquet Y. Biogenesis of PSI involves a cascade of translational autoregulation in the chloroplast of Chlamydomonas. EMBO J. 2004;23:2696–2705. doi: 10.1038/sj.emboj.7600266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano A., Fontanesi F., Solans A., de Oliveira R. L., Fox T. D., Tzagoloff A., Barrientos A. Aberrant translation of cytochrome c oxidase subunit 1 mRNA species in the absence of Mss51p in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:523–535. doi: 10.1091/mbc.E06-09-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., et al. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 1998;20:337–343. doi: 10.1038/3804. [DOI] [PubMed] [Google Scholar]
- Ziegler J., Vogt T., Miersch O., Strack D. Concentration of dilute protein solutions prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Anal. Biochem. 1997;250:257–260. doi: 10.1006/abio.1997.2248. [DOI] [PubMed] [Google Scholar]