Abstract
The Newport River Estuary (NPRE) is a high priority shellfish harvesting area in eastern North Carolina (NC) that is impaired due to fecal contamination, specifically exceeding recommended levels for fecal coliforms. A hydrologic-driven mean trend model was developed, as a function of antecedent rainfall, in the NPRE to predict levels of E. coli (EC, measured as a proxy for fecal coliforms). This mean trend model was integrated in a Bayesian Maximum Entropy (BME) framework to produce informative Space/Time (S/T) maps depicting fecal contamination across the NPRE during winter and summer months. These maps showed that during dry winter months, corresponding to the oyster harvesting season in NC (October 1st to March 30th), predicted EC concentrations were below the shellfish harvesting standard (14 MPN per 100 ml). However, after substantial rainfall 3.81 cm (1.5 inches), the NPRE did not appear to meet this requirement. Warmer months resulted in the predicted EC concentrations exceeding the threshold for the NPRE. Predicted ENT concentrations were generally below the recreational water quality threshold (104 MPN per 100 ml), except for warmer months after substantial rainfall. Once established, this combined approach produces near real-time visual information on which to base water quality management decisions.
INTRODUCTION
The growing impact of stormwater runoff with respect to land modifications and increased fecal indicator bacteria (FIB) concentrations has previously been documented in the State of North Carolina (NC) (1,2,3). The Newport River Estuary (NPRE), an estuarine system in the White River Oak Basin (approximately 35 km2 waterbody within a 453 km2 watershed), is one of 341 waterbodies in NC that is listed as impaired (i.e. on the EPA 303(d) list) for fecal coliforms (1; Figure 1). The NPRE is valuable to the local economy with respect to the shellfish and tourism industry, as well as being a vital environmental sanctuary. The population growth and development increases in the surrounding watersheds can reasonably be associated with stormwater runoff as a significant contributor to microbial contamination in the NPRE (3). The current water quality management program for the NPRE, as described in Coulliette and Noble (3), created a limited and biased water quality dataset that was not sufficient to assess the recent FIB impairment and subsequent listing of the NPRE on the United States EPA 303 (d) list of impaired waterbodies. One approach, that combines Space/Time Random Field (S/TRF) theory and Bayesian Maximum Entropy (BME), has proven successful in the statistical space/time estimation of chemicals and particulate for water and air quality, respectively (5,6).
Figure 1.
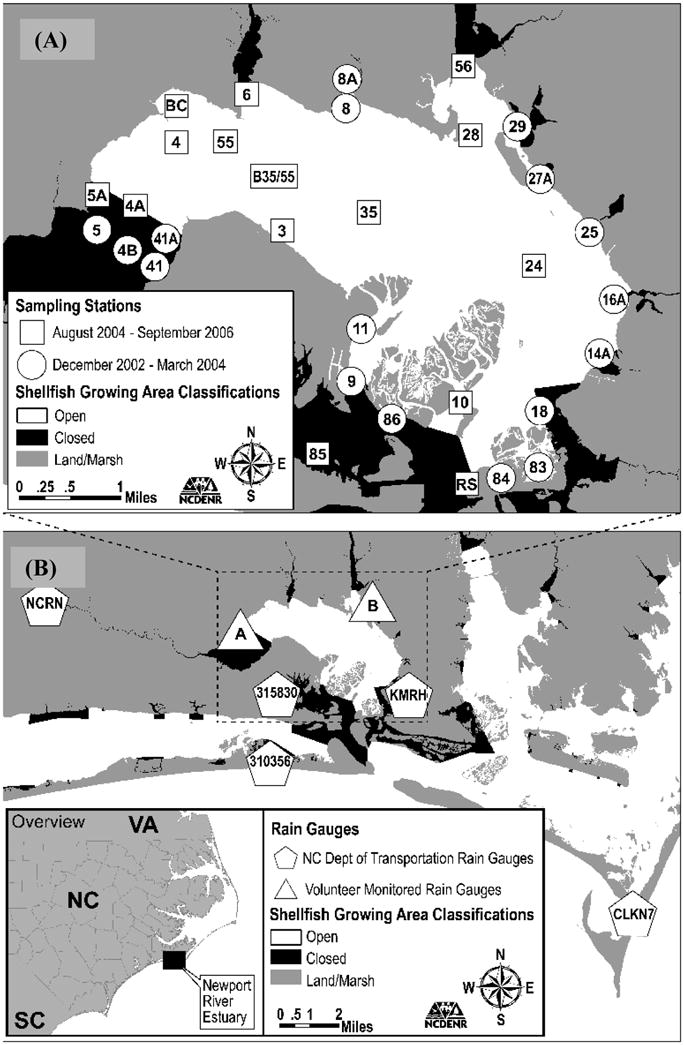
The Newport River Estuary (NPRE). The sampling stations (A) are identified by letters and numbers (sampling location identifiers), where circles relate to samples taken from August 2004 to September 2006, while squares relate to the historical samples taken by NCDENR-SSS from December 2002 to March 2004. Rain gauges (B) are illustrated in triangles for the rain data collected by NCDENR-SSS volunteers and the pentagon stations are those data reported from NCDOT.
The NPRE is a prime candidate for this modeling approach due to the complexity of the system (patchy coastal rainfall, various tributary inputs, diurnal tidal exchange), as no regular water quality sampling approach would capture the estuarine dynamics without a large budget and extensive manpower. Space/Time Random Field (S/TRF) theory provides a framework to model the uncertainty and variability of environmental parameters (i.e. microbial concentrations, rainfall volume) across space and time in terms of statistical moments such as the covariance, which models the magnitude at which the parameters are auto-correlated across space and time. Space/Time (S/T) BME is a modeling technique that allows for one to incorporate general (e.g., covariance) and site specific (e.g., measurements from multiple sources) knowledge about the environmental parameter to produce visual maps that represent the distribution of the parameter at any unsampled point of interest, resulting in informative maps of water quality. These maps allow microbiologists to further understand a waterbody, as traditional water quality research approaches produce limited information across space and time.
The presented research describes a combination of the S/T BME framework with an intensive water quality study regarding the fecal coliform impaired NPRE in NC, where a water quality dataset collected from September 2004 to August 2006, and supplemental historical data from December 2002 to March 2004, was evaluated and incorporated using the BME framework. This approach was used to thoroughly assess the impacts of stormwater runoff and associated factors on FC impairment of the NPRE. The presented work includes: (1) a hydrologic model for FIB across the NPRE that accounts for the effect of rainfall and distance to shore, as well as models the hydrologic-driven trends in microbial concentrations, (2) a covariance model that models space/time dependencies in residual microbial concentrations (i.e. detrended for the hydrological model), and (3) a BME integration of the developed hydrological model and the general and site-specific knowledge bases about concentration residuals that yield informative space/time maps depicting the distribution of microbial concentration in the NPRE during extreme seasonal conditions.
RESEARCH
Study Area
The NPRE (Figure 1) is located north of Morehead City and Beaufort in eastern NC and is in an area classified as Area E-4 by NC Department of Environment and Natural Resources (NCDENR)-Shellfish Sanitation Section (SSS; 7). This estuary has an average depth of 1 m and is a well-mixed system with an average residence time of 6 days or 12 tidal cycles, with flushing stemming from the Atlantic Ocean controlled through the Beaufort Inlet (8). The surrounding land-uses are described in more detail in Coulliette and Noble (3).
Sampling Locations and Collection
Sampling sites were chosen based on existing NCDENR-SSS stations and NCDENR-SSS sanitary surveys (7; Figure 1A). Our goal was to select sites that (1) were spatially distributed across the NPRE, (2) were in high priority shellfish harvest areas (i.e. areas where commercial and recreational shellfish harvesting is prevalent), and (3) were proximal to runoff from land.
Between September 2004 and August 2006, a total of 179 surface water samples were collected. Sampling occurred at least three times a season, where seasons were defined as winter: December 21st to March 20th, spring: March 21st to June 20th, summer: June 21st to September 20th, and fall: September 21st to December 20th. However, additional efforts were made to collect samples across varying weather conditions and a range of storm sizes to produce a robust dataset. Salinity (based upon the practical salinity scale) and temperature (°C) were measured at each site using a calibrated multi-probe instrument (YSI Inc., Yellow Springs, OH). One liter samples were collected within 3 hours of low tide in order to collect samples with minimal dilution from marine waters (NPRE is too shallow to navigate at peak low tide). The samples were collected in sterilized containers following sampling techniques outlined in standard methods (9). After collection, samples were placed on ice and transported immediately to the University of North Carolina at Chapel Hill, Institute of Marine Sciences in Morehead City, NC for processing.
To supplement the data, historical fecal coliform concentrations were obtained from the NC Department of Environment and Natural Resources, Shellfish Sanitation Section (NCDENR-SSS; Figure 1, A). A total of 79 samples that were collected from December 2002 to March 2004 (n = 79) during falling tide conditions were included.
Fecal Indicator Bacteria Analyses
All samples collected from September 2004 to August 2006 were tested for E. coli (EC) and Enterococcus (ENT) using the defined substrate technology test kits, Colilert®-18 and Enterolert® (IDEXX® Laboratories, Westbrook, ME). The kit procedure entailed diluting the sample volume (10 ml) into 90 ml of distilled water. The QuantiTray® then separated the volume into 49 wells of 1.86 ml and 48 wells of 0.186 ml. Conversion of positive wells from these tests to a most probable number (MPN) value was conducted following Hurley and Roscoe (10). Newport River Estuary samples collected by the NCDENR-SSS from December 2002 to March 2004 were processed by Multiple Tube Fermentation for the quantification of fecal coliform (FC) using A1-M, where a dilution series of samples were placed in A-1 broth, incubated overnight, and determined positive for fecal coliforms if gas production was present (12). Previous analyses of estuarine water samples taken throughout eastern NC have shown that 93% of the FC are EC (n=3020, Kirby-Smith and Noble, unpublished data). Thus, for the purposes of this study we consider the NCDENR-SSS FC measurements to be representative of EC concentrations.
For statistical analysis, the currently used geometric mean threshold of 14 FC MPN per 100 ml (13) for shellfish water quality management was applied for general comparison. The NPRE is also actively utilized for boating, sailing, and other forms of recreation. Thus, the “Tier 1” single sample threshold of 104 ENT MPN per 100 ml was applied as a means to compare this estuarine waterbody with other recreational waters.
Rainfall observations
Daily observed measurements were reported from seven rain gauges surrounding the NPRE to obtain full coverage and to account for the heterogeneous nature of NC coastal rainfall patterns (Figure 1B). Five (5) of the stations data were from the State Climate Office of North Carolina, NC Climate Retrieval and Observations Network Of the Southeast (NC CRONOS) Database: Morehead City 2 WNW (ID# 315830), Beaufort Smith Field (ID# KMRH), Atlantic Beach WP (ID# 310356), and Croatan (ID# NCRN). Two stations included daily rainfall observations from volunteers for the NCDENR-SSS: Newport/Mill Creek (“A”) and Beaufort/Ware Creek (“B”).
Hydrologic Model
Fecal contamination in estuaries is influenced by several hydrologic factors. Two factors that are particularly relevant in the NPRE are rainfall, which leads to increased microbial loading, and distance to shore, which addresses microbial fate and transport into the estuary. The dependency of FIB concentration B with these hydrologic factors may be expressed for sample i as
| (1) |
where Bi is the MPN estimate of FIB concentration for sample i, rain1i is the rainfall for that sample over an antecedent period of a few days, rain2i is the rainfall for an antecedent period preceding that of rain1i, di is the distance from the sample to the shore, εi is an error term, and β0, β1, β2 and β3 are linear regression coefficients. This model allows investigation of the effect of various levels of antecedent rainfall on fecal contamination, as well as the distance to shore, which tests whether overland flow into the estuary acts as a vector of fecal contamination (i.e. microbial concentration are expected to decrease with increasing distance to shore). Linear regression theory can be used to obtain the regression coefficients, which can then be used to model the hydrological trend in the (natural base) log-transform FIB concentration Y=log(B) as the space/time function
| (2) |
for any space/time point p=(s,t), where s=(s1,s2) is the spatial coordinate and t is time.
For each sample i, the straight-line distance between the sample site and its corresponding closet point on the shore of the NPRE was calculated (di in Eq. 1). Additionally, the daily rainfall recorded at seven rain gauges surrounding the NPRE (Fig. 1) was calculated using the space/time BMElib numerical implementation (14) of the classical ordinary kriging method (15,16). This leads to estimates of daily rainfall at the location of each sampling site and for the day of sampling as well as each of its preceding 14 days. The rain1i and rain2i variables (Eq. 1) were constructed from these kriging estimates using various non-overlapping antecedent periods of daily rainfall.
Bayesian Maximum Entropy Estimation Framework for Space/Time Mapping Analysis
The theory of space/time random field (S/TRF) was used to model the variability and uncertainty associated with the distribution of FIB concentrations across space and time. Let B(p) be the S/TRF describing the distribution of a FIB, and Y(p)=log(B(p)) be its log-transform. The log-transform residual S/TRF X(p) is defined as
| (3) |
This equation expresses that the S/TRF X(p) models the space/time variability and uncertainty associated with the difference between the S/TRF Y(p) and the hydrologic function hY(p).
BMElib, a powerful MATLAB numerical toolbox of Modern Spatiotemporal Geostatistics implementing the BME theory (17,18,19), was used to create space/time maps of FIB concentration across the NPRE. This framework has been successfully applied to model environmental contaminants (5,6). As demonstrated in these studies, BME presents the flexibility of providing the space/time simple, ordinary and universal kriging methods as its linear limiting case, while it can be expanded to a non-linear estimator if non-linear knowledge bases (e.g. soft data, non Gaussian distributions, etc.) need to be considered. Implementation of the BME method using the BMElib numerical package (14) requires specifying the general and site specific knowledge bases characterizing the Space/Time Random Function (S/TRF) X(p), and produces BME estimates at any space/time point of interest.
The general knowledge base describes systematic space/time trends and dependencies in the S/TRF X(p). In this work, the general knowledge base consists of the space/time mean trend function mX(p)=E[X(p)], and the covariance function cX(p, p′)=E[(X(p)−mX(p))(X(p′)−mX(p′))] of the S/TRF X(p), where E[.] is the expectation operator. The mean trend function characterizes the systematic space/time trends in X(p), while the covariance function describes the dependency of X between points p and p′. These mean trend and covariance functions are modeled from FIB data using the BMElib numerical package (14,19).
The site specific knowledge base describes information that is only relevant to specific points where measurements where performed. In this work the site specific knowledge base consists in the soft data provided by the results of the sample analysis at each of the space/time location pi where a sample i was collected. The FIB test kits (Colilert®-18 for EC and Enterolert® for ENT) provide for each sample i the number of wells found to be positive for the FIB of interest out of two sets of wells prepared at different volumes. From these two numbers of positive wells the Maximum Likelihood (ML) methodology provides for each sample the MPN estimate BMPN and the 95% Confidence Interval (CI) lower and upper bounds BL and BU, respectively, for the log-normally distributed concentration B(pi) expressed in FIB/100ml. It follows directly that, given site specific knowledge, the log-transform residual FIB concentration X(pi) is statistically characterized by a Gaussian probability density function (PDF) with mean log(BMPN)−hY(p) and standard deviation (log(BU)−log(BL))/(2*1.96). Hence the site specific knowledge base S consists of Gaussian soft data for X(p) at each space/time sampling points pi.
General knowledge and site specific knowledge was processed as described above using BMElib to obtain BME estimates of the log-transform residual S/TRF X(p) across the NPRE for each day of the period of study. The BME estimate for a given day is a function of data collected on that day, as well as data collected on days prior and following that day. The estimation error associated with a BME estimate of X(p) at an estimation point is fully characterized by the BME posterior PDF. The expected value and corresponding estimation error variance of the corresponding FIB concentration at that estimation point is then simply obtained by adding the hydrologic driven trend hY(p), and back log-transforming the BME posterior PDF for X(p). This results in BME maps showing the space/time distribution of FIB concentration across the NPRE.
The strength of our proposed approach is that it extracts information in two stages: In the first stage, we use the linear model (1) to obtain a mean trend from hydrologic variables. In the second stage, we use the BME method to finalize the estimate of FIB concentration at some estimation point using the additional information provided by the FIB concentrations measured in the neighborhood of that estimation point. Hence we expect a gain of information in each of these two stages. The gain of information in the first stage comes from pertinent hydrological variables. The gain of information in the second stage comes from the autocorrelation amongst residual FIB concentrations.
In order to investigate the gain of information at each stage of our approach, we calculate the Residual Mean Square Error (RMSE(k)) for some stage (k) of the analysis as
| (4) |
where n is the number of data points, Yi is the i-th measured log-transformed FIB concentration, and is its corresponding estimate at stage (k). At the first stage, is simply obtained from the hydrologic model (Eq. 2) as . At the second stage, corresponds to the BME cross-validation estimate, which is obtained from the FIB data in the neighborhood of Yi, but not Yi itself. The RMSE provides a measure of estimation error standard deviation. If our approach is successful, then we expect the RMSE to decrease from the first stage to the second stage of our analysis.
RESULTS AND DISCUSSION
Hydrologic Model
Ordinary kriging estimates of daily rainfall for the rain1i and rain2i variables were constructed from non-overlapping antecedent periods. The non-overlapping periods were selected that resulted in the most statistically significant fit for the linear regression model (Eq. 1), which corresponded for rain1i in the 4-day antecedent rainfall (day of sampling, 3 days prior) and for rain2i in the preceding non-overlapping a 10-day antecedent rainfall (day 13 to 4 prior to sampling). Antecedent rainfall (rain1i, rain2i) and distance shore (di), along with the associated error (εi), explain 61% and 4% of variability seen in measured EC (F=134.20, p<0.00001) and ENT concentrations (F=3.85, p=0.02), respectively, in the NPRE during the study period (Table 1). The coefficients determined for EC showed that for every centimeter of rainfall in the 4-day antecedent period (β1) and 10-day antecedent period (β2), there was an increase of 48.4 and 6.5 FIB per 100 ml, respectively (Table 1). The coefficients determined for ENT showed that for every centimeter of rainfall in the 4 day antecedent period (β1) and 10-day antecedent period (β2), there was an increase of 112.6 and a decrease of 33.3 FIB per 100 ml, respectively (Table 1). The distance coefficient (β3) revealed that for every kilometer away from the shore, EC decreased by 0.62 MPN/100 ml and ENT 0.11 MPN per 100 ml (Table 1).
Table 1.
Linear regression coefficients β1, β2 and β3 of the hydrologic driven model Bi=β0+β1rain1i+β2rain2i+β3di+εi (Eq. 1) obtained for E. coli and Enterococcus, where the coefficients represent fecal indicator bacteria (FIB) most probable number (MPN) per 100 ml per cm (β1, β2) or km (β3). The 90% confidence intervals are listed in parentheses under each regression coefficient.
| E. coli | Enterococcus | |
|---|---|---|
| R2 | 0.61 (F=134.20, p<0.00001) | 0.04 (F=3.85, p=0.02) |
| β1 | 48.4 (44.40 to 52.40) | 112.6 (36.10 to 189.20) |
| β2 | 6.5 (1.80 to 11.30) | −33.3 (−124.30 to 58.0) |
| β3 | −0.62 (−1.15 to −0.08) | −0.11 (−11.20 to 10.97) |
These findings suggest that a linear relationship between EC and rainfall is a suitable model, however, such a linear relationship is not a suitable model for ENT. Coulliette and Noble (3) reported similar findings in the NPRE with EC having a significant positive linear relationship with rainfall within the previous 48 hours (0.179, p=0.05), while ENT did not. This discrepancy between EC and ENT in their relative strengths with the hydrological model can be explained by the survival kinetics of each FIB. EC, which is recommended for use as a FIB primarily for fresh-water environments, entered the NPRE under favorable conditions of rainfall and sampled at low tide (i.e. fresh water conditions). The linear decrease in EC concentration as the distance from shore increased was due to the increase of salinity, thus the good fit in the hydrological model (R2=0.61, Table 1). ENT, being a FIB that persists for longer periods in more saline waters, does not fit the linear function of the hydrological model with distance to shore due to its ability to persist for longer periods of time.
The 4-day (day of sampling to 3 days prior) antecedent rainfall appears to contribute the greatest concentration of FIB, as compared to 10-day rainfall preceding the sampling date (day 4 to day 13 prior to sampling). The study period was noted to be a “wet year” as compared to historic rainfall averages (3), thus the 10-day antecedent rainfall may not have played a large of a role as would be expected in a “normal” or “dry” years (i.e. where long periods between storms might be expected to accumulate fecal contamination which can then be flushed into the estuarine system). The distance to shore illustrated a negligible decay for both FIB types. The negligible decay detected in measured FIB concentrations away from shore is mostly likely due to the NPRE being relatively shallow (few meters) and being impacted by wind and tide-induced mixing.
Covariance Model
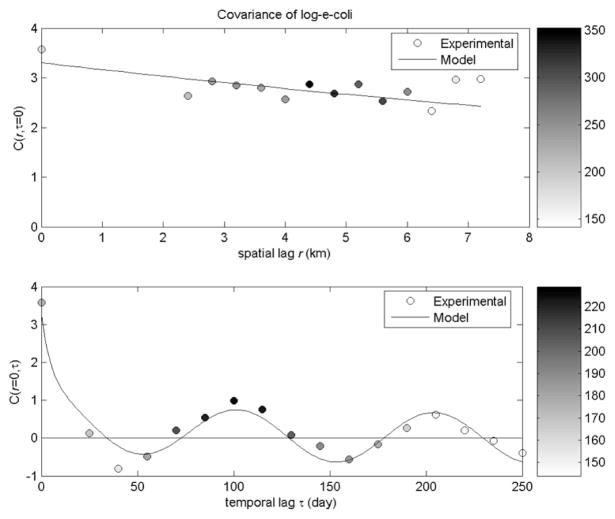
Based on the expected value of the soft X-data for EC, BMElib was used to calculate the covariance experimental values that are shown as circles in Fig. 2. These experimental covariance values were then used to fit the parameters of the covariance model shown in plain line in Fig. 2, which corresponds to the following equation:
where r and τ are the spatial distance and time difference, respectively, between two points p and p′. ENT resulted in a similar model (data not shown). A nugget effect (see supporting information) was removed for EC and ENT covariance models to account for measurement error, which was estimated from the average of the measurement error variance for each X-data points to be 0.27 and 0.67 (logFIB/100 ml)2, for EC and ENT, respectively. The variance σX2 and spatial range (ar)for EC were 3.30 (log-FIB/100 ml)2 and 70 km, respectively. The parameters of the three nested covariance functions modeling the temporal variability of EC were c1=0.6 and at1=15 days, c2=0.2 and at2=150 days, and c3=0.2 and at3=51 days, respectively. For ENT, the variance σ2X and spatial component (ar) were 8.68 (log-FIB/100 ml)2 and 70 km, respectively, and the parameters of the three nested temporal covariance functions were c1=0.5 and at1=15 days, c2=0.3 and at2=300 days, and c3=0.2 and at3=55 days, respectively.
Figure 2.
The spatial, cX(r,τ =0), and temporal, cX(r=0,τ), components of the covariance model for E. coli (EC), where a nugget effect was removed for EC (0.27 logFIB/100 ml)2 to account for measurement error. The color bars to the right shows the number of pairs of measurements used to calculate the corresponding experimental covariance value. Enterococcus results were similar (data not shown).
The variance σX2 for residual log-transformed EC and ENT correspond to a standard deviations of 1.8 (logFIB/100ml) and 2.95 (logFIB/100ml), respectively. The lower variability observed for the residual EC field than for the residual ENT field may be explained by a difference in the variability of the sources of bacterial contamination for these two FIBs. EC may persist or have a more constant source into the NPRE than ENT, which would lead to a lower residual variance, as suggested in the findings of Coulliette and Noble (3). The spatial range of the covariance model (70 km) for both residual EC and ENT is quite large, indicating that predicted FIB concentrations are being contributed from estuarine-wide sources rather than from one specific area of the NPRE. This valuable insight suggests that environmental influences may be providing uniform estuarine-wide fecal contamination (i.e. wildlife). An additional perspective includes that despite growing development and businesses, one specific upstream area around the estuary does not yet impact the NPRE more than other areas.
The temporal component of the covariance model consists of three structures of 15, 500, and 51 days for EC, and 15, 300, and 55 days for ENT. These different time scales may be due to interacting effects of seasons (which cycles over a one year period) and the lunar cycle of spring tides (lasting 14.76 days) where pronounced high and low tides are a result of when the Earth, Moon, and Sun are nearly aligned (20). The extreme high tides have the potential to wash FIB from neighboring banks (21), as well as allowing freshwater to flow into the NPRE during the ultra-low tides
Space/Time Bayesian Maximum Entropy Maps
The covariance model was used as general knowledge in BMElib to obtain the BME posterior PDF of FIB at any location and time of interest. The expected value and 2.5% and 97.5% quantiles of the BME posterior PDF provide a relevant BME estimate and BME 95% CI, respectively. These BME estimates and 95% CI can be used to construct plots showing how the FIB changes as a function of time at any particular site of interest (Fig. 3), or to construct maps showing how the spatial distribution of the FIB across the NPRE for any particular times or sampling events of interest (Fig. 4, Supplemental Figure 1). The figures shown illustrate predicted FIB concentrations across the NPRE on specific dates, however, EC and ENT were measured at the sampling sites on these dates.
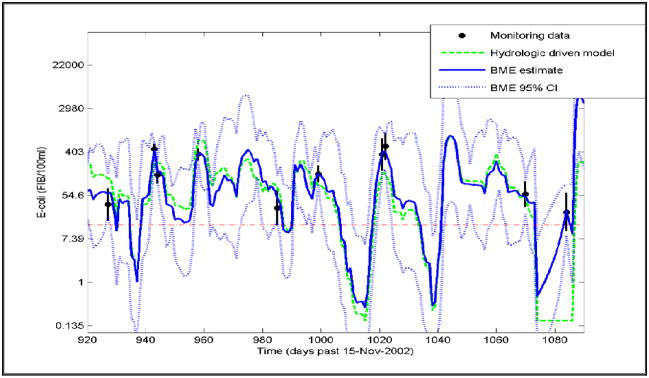
Figure 3.
Temporal plot of the monitoring data (dot centered at the MPN with error bars extending from the lower and upper bounds of the measurement error 95% CI), the hydrologic driven model (dashed line), the BME estimate (plain line) and the BME 95% CI (dotted lines) for E. coli (EC) at site 5A (the western-most sampling site shown in Fig. 1)
Figure 4.
Space/time maps of the NPRE illustrating the Bayesian Maximum Entropy (BME) estimate of E. coli (EC) for selected events where the color bar on the right of the maps represents EC Most Probable Number (MPN) per 100 ml. The x-axis and y-axis demonstrate the longitude and latitude coordinates, respectively.
Specific sampling events were chosen with regards to season (winter versus summer) and rainfall (0 cm versus >3.81 cm) to represent a range of BME estimations of predicted EC and ENT concentrations in the NPRE via space/time maps (EC: Figure 4, ENT: Supplemental Fig. S1). The winter examples had a temperature range of 10 to 14 (°C), salinity range of 11 to 36, and turbidity range of 1 to 11 (NTU), while the summer observations had a temperature range of 27 to 29 (°C), salinity range of 9 to 29, and turbidity range of 4 to 23 (NTU). The color scale to the right of S/T maps represent predicted FIB concentrations ranging from 2.2 to 1100.0 MPN per 100 ml for EC (Figure 4) and 7.4 to 2980.0 MPN per 100 ml for ENT (Supplemental Figure S1). The distribution of EC during the dry winter example indicates that predicted concentrations are near the standard for shellfish harvesting waters (14 MPN per 100 ml; Figure 4a). However, the predicted EC concentrations exceed the acceptable threshold for shellfish harvesting waters after substantial rainfall in the winter (>3.81 cm; Figure 4c). During warmer conditions, predicted EC concentrations exceed the standard regardless of rainfall (Figure 4b and d). For predicted ENT concentrations, the winter months illustrate that during dry conditions the estuary meets the recreational water quality water standard of 104 ENT MPN per 100 ml (Supplemental Figure 1a). The warmer dry conditions also show predicted ENT concentrations meeting the water quality standard with the exception of a western portion (Supplemental Figure 1b). However, after substantial rainfall (>3.81 cm), the northwest portion of the estuary exceeds the acceptable threshold in the winter and escalates far above the acceptable level during the summer (Supplemental Figure 1c and d). Overall, the model reveals that during cooler months when there is limited rainfall, such as during shellfish harvesting season (October to March), the waters meets the NSSP shellfish harvesting FC requirements. However, during the warmer months and/or after rainfall events exceeding 2.54 cm (3), ample time should be allowed for the estuary to return to acceptable water quality before areas are re-opened for shellfish harvesting and recreational contact.
While water quality has been monitored for decades to protect the public, using mathematics and graphics based on measured concentrations to represent water quality predictions for the entire watershed is a more recent integration of disciplines. Modeling and regression efforts to predict FIB has been successful in U.S. fresh water beaches (22, 23) and marine beaches (24). There is a need for such approaches to be published regarding U.S. shellfish harvesting waters, although one study in France utilized a hydrodynamic model to understand EC and virus inputs into shallow coastal waters containing shellfish (25). Riou and colleagues, unlike this study, found no significant differences with EC between seasons. However, both studies demonstrated rainfall having a profound impact on EC concentrations.
The BME framework presented provides a sound space/time geostatistical approach to integrate information from relevant hydrological variables, as well as the autocorrelation and errors amongst measurements. The RMSE (Eq. 4) provides an assessment of the standard deviation of logFIB estimation errors. In the case of logEC, the RMSE is found to be 1.96 and 1.33 (logFIB/100ml) at the first and second stage of the analysis, respectively. This means that when using the hydrologic variables alone (stage 1 of the analysis), there is a factor of about exp(1.96)≈7 in EC estimation errors, but this decreases to a factor of only about exp(1.33)≈3.8 when using the additional information provided by neighboring EC measurements. In the case of logENT, the RMSE is 3.65 and 1.28 (logFIB/100ml) at the first and second stage of the analysis, respectively. Hence, using only hydrologic variables, there is a factor of about exp(3.65)≈38 in ENT estimation errors, which is high, as expected since the hydrologic model for ENT has a low R2 of only 4%. However, this decreases to a factor of only about exp(1.28)≈3.6 in estimation errors when using neighboring ENT measurements, which demonstrates the usefulness of accounting for the autocorrelation amongst FIB measurements in the second stage of our analysis. Additional statistics and plots of the fit of the model at the first and second stages of the analysis are presented in the supporting information, which further demonstrate the advantage of using the BME method presented in this work over classical regression-based methods.
Once a hydrologic mean trend model and a covariance function have been established for a waterbody, such as the impaired NPRE, S/T maps can be produced in near-real time to provide visual information to user groups and stakeholders and provide critical information on which to base water quality management decisions. Over long periods of data collection these maps can become useful predictive tools for water quality management. Specifically, water quality managers would have the capability to identifying “hot spot” areas after heavy rainfall. Then, resources and efforts would be directed to re-open the least impacted harvesting areas, preserving the beneficial uses of at least part of the water body. Identification of these areas might also be useful for directing remediation efforts, such as specific types of stormwater filtration systems. Finally, the maps generated can quickly provide either a snapshot or an animated map, similar to existing precipitation maps or water quality forecasting animations (e.g., BeachCast for the Great Lakes Information Network), and posted online for commercial shellfish harvesters and the public alike to view. The established framework is promising, and has a variety of possible uses for future water quality monitoring and remediation.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute of Environmental Health Sciences (Grants no. 5 P42 ES05948 and P30ES10126), U.S. EPA NC Section 319 NPS Program NCDENR File#05-SC-DENR-1063, and NCDENR 319(h) Program.
References
- 1.Mallin MA, Willams KE, Esham EC, Lowe RP. Effect of Human Development on Bacteriological Water Quality in Coastal Watersheds. Ecological Applications. 2000;10(4):1047–1056. [Google Scholar]
- 2.Kirby-Smith WW, White N. Bacterial contamination associated with estuarine shoreline development. Journal of Applied Microbiology. 2006;100:648–657. doi: 10.1111/j.1365-2672.2005.02797.x. [DOI] [PubMed] [Google Scholar]
- 3.Coulliette AD, Noble RT. Impacts of Rainfall on the water quality of the Newport River Estuary (Eastern North Carolina, USA) Journal of Water and Health. 2008;6(4):473–482. doi: 10.2166/wh.2008.136. [DOI] [PubMed] [Google Scholar]
- 4.U.S. EPA. Final North Carolina Water Quality Assessment and Impaired waters List (2004 Integrated 305(b) and 303(d) Report) North Carolina Department of Environment and Natural Resources; 2004. [Google Scholar]
- 5.Akita Y, Carter G, Serre ML. Spatiotemporal non-attainment assessment of surface water tetrachloroethene in New Jersey. Journal of Environmental Quality. 2007;36(2) doi: 10.2134/jeq2005.0426. [DOI] [PubMed] [Google Scholar]
- 6.Puangthongthub S, Wangwongwatana S, Kamens RM, Serre ML. Modeling the Space/Time Distribution of Particulate Matter in Thailand and Optimizing Its Monitoring Network. Atmospheric Environment. 2007;41:7788–7805. [Google Scholar]
- 7.NCDENR-SSS 2005. E-4 Shoreline Survey. North Carolina Department of Environment and Natural Resources, Shellfish Sanitation Section.
- 8.Kirby-Smith WW, Costlow JD. The Newport River Estuarine System. UNC Sea Grant College Publication UNC-SG-89-04; 1989. [Google Scholar]
- 9.APHA (American Public Health Association) Standard Methods for the Examination of Water and Wastewater: Centennial Edition. 21. Baltimore, Maryland: 2005. [Google Scholar]
- 10.Hurley MA, Roscoe ME. Automated statistical analysis of microbial enumeration by dilution series. Journal of Applied Microbiology. 1983;55:26–164. [Google Scholar]
- 11.Pisciotta JM, Rath DF, Stanek PA, Flanery DM, Harwood VJ. Marine Bacteria Cause False-Positive Results in the Colilert-18 Rapid Identification Test for Escherichia coli in Florida Waters. Applied and Environmental Microbiology. 2002;68(2):539–544. doi: 10.1128/AEM.68.2.539-544.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.AOAC. AOAC International - Official Methods of Analysis. 15. Association of Official Analytical Chemists; Washington, D.C.: 1990. [A-1 Modified MPN test] [Google Scholar]
- 13.NSSP. Guide for the Control of Molluscan Shellfish. U.S. Department of Health and Human Services, Public Health Service, Food and Drug Administration; 2005. [Google Scholar]
- 14.Christakos G, Bogaert P, Serre ML. Temporal GIS: Advanced Functions for Field-Based Applications. Springer-Verlag; New York, N.Y.: 2002. p. 217. [Google Scholar]
- 15.Cressie NAC. Statistics for Spatial Data. John Wiley and Sons, Inc.; U.S.A: 1993. [Google Scholar]
- 16.Journel AG, Huijbregts ChJ. Mining Geostatistics. Academic Press; San Deigo, California: 1978. [Google Scholar]
- 17.Christakos G. A Bayesian/maximum-entropy view on the spatial estimation problem. Mathematical Geology. 1990;22(7):763–776. [Google Scholar]
- 18.Christakos G. Modern spatiotemporal geostatistics. 2 Oxford Univ. Press.; New York: 2001. [Google Scholar]
- 19.Serre ML, Christakos G. Modern geostatistics: computational BME in the light of uncertain physical knowledge--the equus beds study. Stochastic Environmental Research and Risk Assessment. 1999;13(1):1–26. [Google Scholar]
- 20.Kvale EP. The origin of neap-spring tidal cycles. Marine Geology. 2006;235:5–18. [Google Scholar]
- 21.Shibata T, Solo-Gabriele HM, Fleming LE, Elmir S. Monitoring marine recreational water quality using multiple microbial indicators in an urban tropical environment. Water Research. 2004;38:3119–3131. doi: 10.1016/j.watres.2004.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nevers MB, Whitman RL. Coastal strategies to predict Escherichia coli concentrations for beaches along a 35 km stretch of Southern Lake Michigan. Environmental Science and Technology. 2008;42(12):4454–4460. doi: 10.1021/es703038c. [DOI] [PubMed] [Google Scholar]
- 23.Nevers MB, Whitman RL. Nowcast modeling of Escherichia coli concentrations at multiple urban beaches of souther Lake Michigan. Water Research. 2007;39:5250–5260. doi: 10.1016/j.watres.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 24.He LM, He ZL. Water quality prediction of marine recreational beaches receiving watershed bsaeflow and stormwater runoff in southern California, USA. Water Research. 2008;42(10–11):2563–2573. doi: 10.1016/j.watres.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 25.Riou P, Le Saux JD, Dumas F, Caprais MP, Le Guyader SF, Pommepuy M. Microbial impact of small tributaries on water and shellfish quality in shallow coastal areas. Water Research. 2007;41:2774–2786. doi: 10.1016/j.watres.2007.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.