Abstract
Limited solubility and precipitation of amyloidogenic sequences such as the Alzheimer peptide (β-AP) are major obstacles to a molecular understanding of protein fibrillation and deposition processes. Here we have circumvented the solubility problem by stepwise engineering a β-AP homology into a soluble scaffold, the monomeric protein S6. The S6 construct with the highest β-AP homology crystallizes as a tetramer that is linked by the β-AP residues forming intermolecular antiparallel β-sheets. This construct also shows increased coil aggregation during refolding, and a 14-mer peptide encompassing the engineered sequence forms fibrils. Mutational analysis shows that intermolecular association is linked to the overall hydrophobicity of the sticky sequence and implies the existence of “structural gatekeepers” in the wild-type protein, that is, charged side chains that prevent aggregation by interrupting contiguous stretches of hydrophobic residues in the primary sequence.
Several neurodegenerative diseases are associated with the deposition of insoluble protein fibrils in the nervous tissue, so-called amyloid plaques (1). The most prevalent example is Alzheimer's disease, which is accompanied by the deposition of amyloid plaques in the brain (2). The major protein component of the Alzheimer plaque is the 39- to 43-aa β-peptide (β-AP), an aberrant proteolytic product of a 695- to 770-aa membrane-bound precursor protein (3). β-AP consists of a hydrophilic N-terminal part (amino acids 1–28) and a hydrophobic C-terminal sequence (amino acids 29–43), which is believed to be buried in the membrane in the precursor protein. Although β-AP readily forms fibrils in vitro, the fibrils' insolubility has been an obstacle to all attempts to solve their atomic structure. Furthermore, it has been difficult to isolate and characterize intermediates in the fibrillation process because it occurs by nucleation and growth (4). Nevertheless, evidence from low-angle x-ray diffraction (5) and infrared spectroscopy (6) suggests that β-AP forms a highly ordered and periodic arrangement of β-strands. The β-strands are widely believed to be organized in an antiparallel fashion (5–7), although some results favor a parallel arrangement (8).
Interestingly, no particular section of β-AP appears to be critical for fibrillation in vitro, and different sections of β-AP seem to form fibrils with different morphologies. Fragments of residues 18–28 assemble into ribbon-like structures (9), whereas fragments involving the hydrophobic C terminus may assume a fully extended filament structure (6). The assemblies are further sensitive to mutagenesis. For example, the Lys → Ala-16 mutation in the residues 1–28 fragment leads to amorphous aggregates, which, according to x-ray diffraction patterns, contain β-sheets (10). This spectrum of assemblies suggests that multiple modes of interaction may operate, so that β-AP aggregates specifically in several registers and by different quaternary contact patterns (2). Such a variability in the way the fibers may assemble has led to speculation that fibrillation is a generic property of the polypeptide chain (11). Consistently, proteins (12) and peptides (13) unconnected with deposition diseases can be induced to fibrillate, and amyloid fibrils from unrelated proteins appear to share a common β-sheet core as well as a similar fibril morphology (14). A model has been proposed in which a number of β-sheets run parallel to the filament axis, twisting to give a quaternary helical repeat of ca. 120 Å (14). Despite the ubiquity of fibrillation, some sequences are more disposed toward aggregation than others (15), but the basis for this bias is still unclear.
In this study we have solubilized various hydrophobic sequences homologous to the C-terminal part of β-AP by grafting them into a soluble protein scaffold, namely the monomeric 101-residue ribosomal protein S6 from Thermus thermophilus (cf. ref. 16). The procedure is aided by exploiting a preexisting homology between the β-AP sequence and the β-strand 2/loop region in wild-type S6. From the wild-type sequence, the β-AP homology was then increased in a stepwise manner while changes in the refolding kinetics and transient aggregation were analyzed. The quadruple mutation EA41/EI42/RM46/RV47 (S6-Alz) increases the identity of the two sequences to nine of 15 residues, and the remaining six residues are very similar (Fig. 1). In contrast to the wild-type protein, S6-Alz displays complex reversible aggregation in the refolding process and is disposed to form soluble aggregates in its folded state. The crystal structure of S6-Alz shows that the mutated protein assembles into tetramers joined by intermolecular β-sheets involving the β-AP sequence. Peptides encompassing the engineered sequence readily form fibrils, suggesting a link between transient aggregation, tetramerization, and fibrillation. The onset of aggregation is linked to the deletion of charged residues, which leaves contiguous hydrophobic stretches in the protein's primary sequence. The results are discussed in light of conformational gatekeepers, multiple aggregation registers, and amyloid funnels.
Figure 1.
Sequence alignment of S6 wild type, β-AP, and the three S6 mutants used in this study.
Materials and Methods
The mutants EA41/EI42, RM46/RV47, and EA41/EI42/RM46/RV47 (S6-Alz) were constructed as described (17). Expression and purification were essentially as described (17); however, the purification step on CM-Sepharose was followed by gel filtration on a Superdex 200 prep grade HiLoad 16/60 column.
Stopped-flow kinetics were performed with an Applied Photophysics SX18-MV instrument as described (17). All experiments were in 50 mM Mes (pH 6.3) at 25°C. Single-jump aggregation experiments were carried out by 1:10 dilution of the denaturant-unfolded protein into buffer. S6 wild-type and mutants were unfolded in 4.4 M guanidinium chloride.
S6-Alz and EA41/EI42 were crystallized in 0.2 M sodium citrate/0.1 M Tris (pH 8.5) and either 20% (S6-Alz) or 30% (EA41/EI42) polyethylene glycol 400 at a concentration of 6–8 mg/ml. We were unable to obtain suitable crystals of the mutant RM46/RV47 and other S6 mutants with higher homology to β-AP under these and other screened conditions. Crystal diffraction data were collected by using conventional radiation from a rotating anode x-ray source and the MAX-LAB synchrotron in Lund, Sweden, and analyzed as described (17). The space group for S6-Alz is P42212, the resolution is 2.2 Å, and the R factor is 20.5%. Two crystal forms of EA41/EI42 were analyzed: data from one crystal form, space group P6422, were collected to 2.1-Å resolution and refined to a current R factor of 22.0%. Data from the second crystal form, space group C2221, were recorded to 1.65-Å resolution and refined to an R factor of 24.1%. All crystal coordinates have been deposited in the Brookhaven Protein Data Bank, for S6-Alz with accession code 1QJH and for EA41/EI42 with accession codes 1CQN (space group P6422) and 1CQM (space group C2221).
Crosslinking of S6 wild type and S6-Alz by glutaraldehyde was carried out as described (18). Proteins at 0.3 mM were incubated in 100 mM borate (pH 8.9) and 0.013 M glutaraldehyde for 20 s, before the reaction was stopped by adding NaBH4 to 80 mM, and the protein mixture was run on a gel.
The peptides AcNH-RVEKVEELGLRRLA-CONH2, AcNH-RVEKVAILGLRRLA-CONH2, and AcNH-RVEKVAILGLMVLA-CONH2, corresponding to β-strand 2 in S6, were synthesized by Synt:em (Nîmes, France) and Neosystem (Strasbourg, France). They were dissolved in 50% acetonitrile/50% water (vol/vol). Fibrils were formed by incubating the peptides at 1 mg/ml in 25 mM sodium phosphate (pH 6)/50 mM NaCl. For electron microscopy studies, performed on a Philips CM120 BioTWIN cryo-transmission electron microscope, 5 μl of peptide solution was deposited on a glow-discharge copper grid, left for 1 min, blotted, and air-dried for 5 min before 5 μl of 1% phosphotungstic acid was applied in a similar manner.
CD spectra were recorded on a Jasco-720 spectrophotometer in 25 mM sodium phosphate (pH 6)/50 mM NaCl, using a path length of 1 mm and a peptide concentration of 0.5 mg/ml. Binding of thioflavin T was monitored in the same buffer with 0.1 mg/ml peptides in the presence of 10 μM thioflavin T, using excitation at 450 nm and emission at 482 nm (19).
Results
Increasing the Homology to β-AP Induces Transient Aggregation During Refolding.
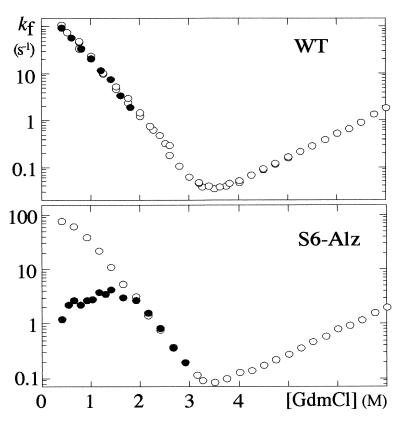
Wild-type S6 is a so-called two-state protein that folds in a highly concerted step directly from the denatured state D without intermediates (17). A kinetic hallmark of two-state behavior is a V-shaped “chevron plot,” i.e., a plot of the refolding and unfolding rate constants versus denaturant concentration (Fig. 2). The quadruple mutant S6-Alz also shows this typical two-state behavior at low concentrations of protein (<1 μM). At higher concentrations of protein, however, the refolding rate constant of S6-Alz decreases under poor solvent conditions (i.e., at low denaturant concentrations), causing a curvature in the refolding limb of the chevron plot. At 25 μM S6-Alz, the refolding rate constant is almost 100-fold less than at 1 μM (Fig. 2). A similar retardation is reported for the two-state proteins U1A and CI2 and is connected to transient aggregation of denatured protein (20–22). The protein may either fold rapidly in a two-state reaction or become trapped in aggregates that convert to native monomers more slowly. Aggregation and folding seem to occur in parallel, and the extent of aggregation is simply determined by protein concentration. Because the S6-Alz mutations only have a marginal effect on kf, the rate of direct folding (Table 1), the elevated level of transient aggregation can be linked to an increase in the association rate kagg (Scheme 1):
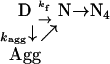 |
 |
That is, the mutations increase the protein's likelihood of getting stuck in an aggregate (Agg) upon collision with another molecule. Despite the high association rates, results from U1A suggest that the process is specific to a certain extent. U1A molecules, which also display a limited homology to β-AP, aggregate only with themselves and are unaffected by the presence of other denatured proteins (unpublished observations). Above 5 μM, S6-Alz displays a second additional aggregation step with a rate constant around 0.05 s−1. This step is absent at lower protein concentrations and hints that the protein also undergoes self-association in its native form. The end product, however, is well ordered and perfectly soluble and can be crystallized.
Figure 2.
Chevron plots for S6 wild type and the mutant S6-Alz at 1 μM (○) and 25 μM (●). GdmCl, guanidinium chloride. The refolding rate constant (kf) for S6-Alz becomes slower at high concentrations of protein, indicating that the denatured protein becomes temporarily trapped in transient aggregates.
Table 1.
Equilibrium and kinetic parameters for the S6 mutants
| Mutation | ΔΔGD−N, kcal⋅mol−1* | kfwater, s−1† | kf0.4M, s−1† | [P]30%, μM‡ | |
|---|---|---|---|---|---|
| WT | — | ≡0 | 384 ± 5 | 121 ± 1 | >25 |
| S6-A | EA41/EI42 | 0.37 ± 0.02 | 417 ± 5 | 123 ± 2 | 14.0 |
| S6-B | RM46/RV47 | −0.13 ± 0.01 | 367 ± 6 | 106 ± 1 | 7.2 |
| S6-Alz | EA41/EI42/RM46/RV47 | 0.07 ± 0.03 | 570 ± 7 | 119 ± 3 | 1.6 |
Calculated as in ref. 42, using an average m value of 1.91 ± 0.04 M−1.
Measured in water or 0.4 M guanidinium chloride at a concentration of 0.5 μM protein, where the effect of aggregation is small.
Protein concentration at which the slow phases constitute 30% of the total amplitude.
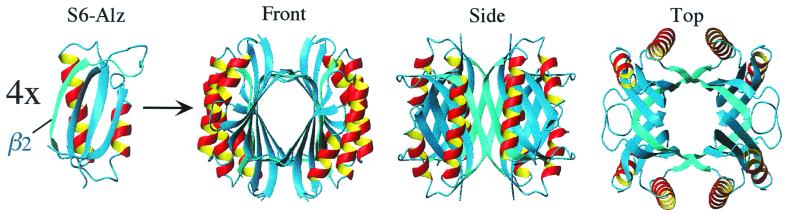
The Crystal Structure of S6-Alz Is a Tetramer.
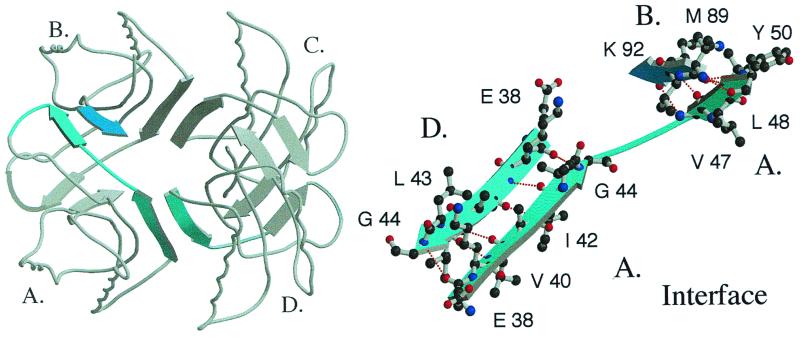
The crystal structure of folded S6-Alz reveals a beautifully assembled tetramer. Four S6-Alz molecules join to form a cylinder of contiguous antiparallel β-structure (Fig. 3). The interface between the molecules comprises the β-AP-like residues 38–53, adopting an extended β-strand that anneals the “central cylinder” in two directions. The major contact is along the cylinder axis, where residues 38–44 of molecule A form a perfect antiparallel β-sheet with the same residues in molecule D (A–D interface, Fig. 4). In wild-type S6, this fringe of β-strand 2 is solvent-exposed. The point of symmetry is at position 41, where the alanine side chains from the two strands are located side by side, pointing toward the central channel inside the tetramer (Fig. 4). The second and minor contact region completes the circumference of the cylinder. Here residues 47–50 in the far end of the β-AP homologous strand form a second antiparallel β-sheet with residues 89–92 in β-strand 4 of molecule B (A–B interface, Fig. 4). The gap between the strands joining molecules B and C occurs at positions 45 and 46 and is indicated in Fig. 4 Right, which emphasizes regions of intermolecular hydrogen bonds only.
Figure 3.
The build-up of the S6-Alz tetramer. The monomers are joined by the β-AP homology in strand 2 (β2), forming intermolecular β-sheets.
Figure 4.
Structure of the S6-Alz tetramer showing the linking β-sheets. The monomers are marked A, B, C, and D; a close-up of the interface is shown to the right. Residues 38–44 in β-strand 2 of molecule A form an antiparallel β-sheet with the same residues in molecule D, and residues 47–50 form a corresponding antiparallel β-sheet with residues 89–92 in molecule B.
Notably, residues 89–92 (MVVK) are not found in β-AP. The involvement of such odd stretches in the interface plausibly reflects the high degeneracy of β-sheet formation: if no direct obstacles interfere, any interface will do. It is most likely that this indiscriminate propensity for forming β-sheets enabled us to obtain the S6-Alz tetramer easily. The particular register we observe is simply selected because it is compatible with the intermolecular packing required for crystal formation. In solution, S6-Alz may therefore sample other intermolecular contact patterns and form larger oligomers. This is consistent with crosslinking experiments with glutaraldehyde, in which S6-Alz forms high-molecular-weight smears rather than distinct tetramers (data not shown). In contrast, S6 wild type forms a small amount of dimers, as expected from the nonspecific crosslinking effect of glutaraldehyde from random collisions of S6 monomers.
At the monomer level, the major difference between S6-Alz and wild-type S6 is in the large loop between β-strands 2 and 3 (residues 46–57), which is contorted in wild-type S6 and EA41/EI42 because of an extra loop involving residues 51–57. The kink arising from this extra loop is absent in S6-Alz, leading to a much smoother loop structure and the formation of an intermolecular β-sheet between residues 47–50 of one subunit and residues 89–92 of another. It is not clear whether this loop structure is a prerequisite for, or a consequence of, tetramerization.
To check whether the tetramerization of S6-Alz is induced by the crystallization conditions, we have also determined the structure of the S6 mutant EA41/EI42, which crystallizes under the same conditions as S6-Alz. The crystal structure of EA41/EI42, however, shows the same monomeric packing geometry as the wild-type protein.
Fibrillation of an S6-Alz Peptide.
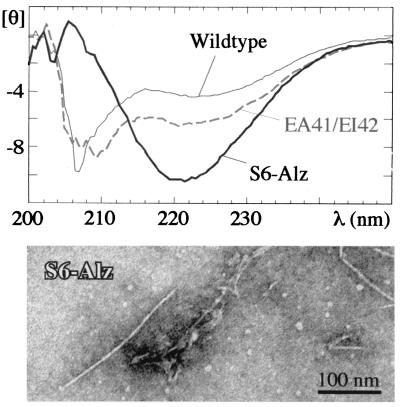
We have also investigated the conformational properties of peptides corresponding to residues 36–49 (β2) in S6 wild type and the two mutants EA41/EI42 and S6-Alz. The peptides from S6 wild type and the double mutant EA41/EI42 are soluble and largely unstructured by CD. In contrast, the CD spectrum of the S6-Alz peptide indicates a significant degree of β-sheet structure in water (Fig. 5). Only the S6-Alz peptide binds thioflavin T and Congo red, which are commonly used markers for amyloid structure (19, 23) (data not shown). This peptide also consistently forms visible precipitates, which according to electron microscopy contain amyloid-like fibrils (Fig. 5). Upon standing on ice for at least 12 h, the EA41/EI42 peptide forms a slightly translucent gel at high peptide concentrations (10 mg/ml), but at 1 mg/ml light-scattering experiments indicate no formation of high-molecular-weight structures. Because the EA41/EI42 peptide sequence in S6 allows the full-length protein to aggregate transiently in a fraction of a second, it is expected that the isolated peptide should be able to form very stable aggregates at equilibrium conditions. Yet electron microscopy reveals only a few “tattered” fibrils after 12 days of standing and none within the first few hours. Apparently the EA41/EI42 peptide forms only small soluble aggregates that are not fit to assemble into higher-order structures.
Figure 5.
CD spectra of the 14-mer peptides corresponding to residues 36–46 in S6 (units are in 10−3 deg⋅cm2⋅dmol−1). Below is an electron micrograph of the S6-Alz peptide after 7 days of incubation, showing fibrillar structures similar to those of β-AP.
Discussion
What Prevents the Wild-Type Protein from Aggregating?
Most amyloidogenic proteins seem to undergo significant conformational changes upon the formation of amyloid fibrils. For example, insulin, cystatin C, the amyloidogenic variants of lysozyme (24), and the prion protein (25) have extensive native α-helical structure in the monomeric state that is at least partially lost in the fibrillar state. This is clearly not the case with the S6-Alz tetramer, where the protein associates in its native state and without large conformational changes. Nevertheless, the S6-Alz tetramer encompasses features of real fibrils. First, the linking interface is the “sticky” hydrophobic edge of a β-strand with an amino acid composition similar to that of the amyloidogenic β-AP. Second, the interface in the S6-Alz tetramer is composed of antiparallel β-strands, which is the likely arrangement in amyloid fibers (5, 6, 8).
Is it then possible to identify particular amyloidogenic features of the β-AP interface? As already suggested from studies of β-AP fragments, the structural interpretation is complicated by the peptides' ability to form ordered aggregates in many different ways or registers (6, 9, 10). The antiparallel interface within the S6-Alz tetramer is probably only one of many possible interaction modes. Therefore, we have approached the question of amyloidicity from a somewhat alternative angle: instead of directly dealing with the forces driving the tetramerization, we start by focusing on the interactions that keep wild-type S6 monomeric. In the end, the polypeptides' ability to actively obstruct misfolded or aggregated states may be as important in the evolution of protein structures as their ability to adapt good packing geometry in the native state (11, 27).
A Link Between Tetramerization, Transient Aggregation, and Peptide Fibrillation.
Wild-type S6 contains two side chains, E41 and R46, that seem to be incompatible with the tetrameric assembly. Although these side chains would not cause any steric clashes in the tetramer packing, they would lead to unfavorable desolvation penalties (28): E41 from monomers A and D places two carboxylate groups in close contact in the center of the A–D interface, whereas R46 inserts a positive charge among nonpolar moieties at the A–B interface. Consistently, the mutant EA41/EI42, which retains R46, crystallizes as a monomer. We have not yet been able to obtain diffracting crystals of the “sibling” mutant RM46/RV47 to make the complementary test.
Interestingly, these side chains modulate not only the tetramerization of the native structure, but the transient aggregation of the coil as well. On their own, the mutants EA41/EI42 and RM46/RV47 show a small but significant degree of coil aggregation during refolding (Table 1). But when the mutations are combined in S6-Alz, extensive aggregation takes place. The same trend is seen for peptides of residues 36–49 in these mutants. The EA41/EI42 peptide is able to associate sufficiently at high concentrations to form a gel, but only the S6-Alz peptide is able to form fibrillar structures (Fig. 5). This coupling between tetramerization, transient aggregation, and fibrillation of peptides could mean that truncation of E41 and R46 results in a common type of intermolecular contact pattern, namely, intermolecular β-sheets.
Gatekeepers and Local Hydropathy.
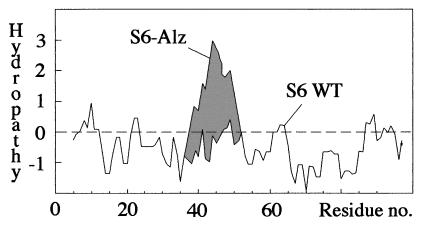
Because substitutions of the charged residues E41, E42, R46, and R77 have no marked effect on the folding or stability of the isolated protein (Table 1), it is tempting to believe that their role is to prevent S6 from aggregating. That is, they steer folding indirectly by obstructing certain misfolded states. In an earlier study we have referred to such residues as structural gatekeepers (27). It was demonstrated that S6 accumulates a misfolded refolding intermediate in the presence of salt and that certain mutations stabilize the misfolded species by allowing the coil to collapse in the wrong way. Although the mechanism by which the “gatekeeping” side chains obstruct nonnative interactions is still unclear, it is noteworthy that these side chains were found to be both hydrophilic and hydrophobic (27). In the case of S6 aggregation we discern a more distinct pattern: aggregation is triggered by the presentation of contiguous hydrophobic stretches in the primary sequence. The charge deletions in EA41/EI42, RM46/RV47, and S6-Alz produce new stretches of 6, 11, and 14 hydrophobic residues, respectively (Fig. 1). Correspondingly, the protein concentrations at which the mutants undergo 30% transient aggregation are 14 μM, 7 μM, and 1.6 μM ([P]30%, Table 1). The correlation is even clearer in a hydropathy plot, where the wild-type sequence is balanced around and below zero, whereas the mutants produce large overshoots (Fig. 6). Because folding and stability remain unaffected by these mutations, it is possible that the bias toward short hydrophobic stretches found in natural sequences (29, 30) is dictated to some extent by the need to minimize aggregation. The key feature of the C-terminal part of β-AP may thus be its overall hydrophobicity rather than local specificity.
Figure 6.
Hydropathy plot (41) showing how the charge truncations in S6-Alz produce a large hydrophobic overshoot.
Given that gatekeeper residues work indirectly, they are not necessarily conserved as strictly as residues that are critical for protein function and stability. For example, other charged residues might substitute for Glu-41 to induce electrostatic repulsion upon aggregation. However, sequence analysis of 17 genes reveals that position 41 is Glu in eight cases, Asp in two, Lys in two, polar in three, and hydrophobic in two, indicating some degree of conservation. When position 41 is polar or hydrophobic, there is a charged residue in position 42 or 43 that could act as a substitute gatekeeper. In the case of Arg-46, however, a strong functional conservation obscures the picture, because this residue is also involved in RNA binding (31).
Specific Aggregation in Multiple Registers.
There are several indications that protein fibrillation involves sampling of multiple packing and interaction modes. For example, β-AP sometimes forms protofibril precursors that are morphologically distinct from the final fibrils (32, 33), and protofibril-like assemblies, in turn, may spring from large granular or amorphous aggregates, according to electron microscopy studies on acylphosphatase (12). Gradual interconversions between intermediate species and mature fibrils are also observed for certain transthyretin mutants (E. Lundgren and A. Olofsson, personal communication). The process thus resembles proteins that precipitate transiently en route to forming crystals, and experimental support for a link between fibrillation and protein crystallization was recently reported by atomic force microscopy studies (34). Together with the evidence of multiple interaction registers from β-AP fragments (6, 9, 35), it seems reasonable to assume that variations in aggregate morphologies stem from alternative registers within a common β-packing. In the case of S6, however, the aggregation process is terminated at an early stage: the drive to fold rips the primary aggregates apart. Notably, the observation that the protein dissociates and folds into a monomeric native state after the initial burst of aggregation provides further evidence that the transient aggregates involve multiple registers. If they were all joined as the tetramer, S6-Alz could fold without having to separate. In contrast, most of the burst registers seem incompatible with the tetramer and need to be broken.
A corollary of multiple aggregation registers is that the effect of deleting obstructing gatekeepers will be synergistic. For example, during refolding S6-Alz may sample not only the contact registers of the two double mutants EA41/EI42 and RM46/RV47 but also registers that rely on interactions in both sites. Accordingly, S6-Alz is 5–10 times more prone to transient aggregation than either of the double mutants (Table 1). Conversely, the introduction of even a single charged mutant into β-AP is expected to dramatically reduce its aggregation propensity.
The “Amyloid Funnel,” a Competing Force in Protein Folding.
Assuming that the sampling of different aggregated states involves shuffling of registers within β-sheets, it is tempting to describe the aggregation process in terms of an “amyloid funnel,” i.e., a reaction scheme that shows how a broad ensemble of early aggregates in multiple registers progressively narrows into a highly ordered fibril with only one allowed register. Similar funnels are used to describe the relation between conformational entropy (the width of the funnel) and contact energy (the depth of the funnel) in protein folding (26). For simplicity, we approximate here the aggregate entropy with the number of accessible β-sheet registers; the depth of the amyloid funnel is a reaction coordinate based on contact energy. (Low-dimensional funnel projections of the aggregation process do not explicitly capture the complexity of mixed aggregate sizes and hence give an incomplete picture of the system entropy. Nevertheless, the amyloid funnel provides a minimalistic platform for discussing protein aggregation.)
Transient aggregation of denatured protein is then likely to occur at the very entrance of the funnel, where the peptide is free to form productive contacts in many different registers. The large number of accessible contact registers at this stage would explain how this type of aggregation can be selective and still very fast. However, the structural heterogeneity of these primary aggregates makes them difficult to organize into larger assemblies. Further progression down the funnel relies on the reduction of contact registers. At the bottom, there may be only one or a few registers that are compatible with the well-ordered amyloid fibril.
It is easy to imagine that interconversion between different aggregated states could seize up and come to a halt before the amyloid is reached. In particular, under conditions of high contact energies, where the shuffling of backbone hydrogen bonds is accompanied by high local barriers; the aggregate becomes kinetically trapped in a rugged energy surface (cf. ref. 26). The amorphous aggregates observed in the presence of stabilizing salts such as ammonium sulfate could be the result of such trapping. If, on the other hand, the contacts are weak, only the most well-packed assemblies may overcome the entropic cost of association. This can lead to situations where just monomeric precursors and the final fibrils populate at equilibrium, and the species in between form a kinetic barrier of high-energy states (4). To overcome the barrier a certain number of precursors need to assemble into a critical nucleus. This assembly may be slow. Once the fibril is formed, however, its leading edge will rapidly catalyze its own growth by providing a “sticky” template for the monomeric precursors. Such nucleated growth is indicated by a lag phase in the aggregation kinetics and is commonly encountered in amyloid formation (36). Accordingly, protein fibrillation resembles a macroscopic phase transition between monomeric polypeptides and a generic form of linear crystals, the helical periodicity of which stems from the chirality of the peptide bond; the linearity is simply dictated by the polypeptide's deviation from radial symmetry.
The existence of a generic fibrillar phase of the polypeptide has been inferred recently from observations that the addition of methanol and other organic cosolvents induces fibrillation of proteins that are not normally associated with amyloidosis (11, 12). In the light of the S6 data, the induction may be due in part to the suppression of desolvation penalties of polar and charged gatekeeping residues. Another effect of the cosolvents could be to increase the occupancy of protein conformations that are structurally fit to aggregate, that is, to lower the stability of the native state relative to that of the coil and certain “sticky” intermediates (11).
Avoiding Traps by Smoothing the Folding Energy Landscape?
In the crowded and dynamic interior of the cell, it is possible that part of the aggregation problem is solved generically by the proteins' disposition against nonnative contact patterns, i.e., the “principle of minimal frustration” (26). The features that prevent nonnative contacts will also prevent nonspecific aggregation. Part of this conformational bias may be governed by gatekeepers that smooth out the folding energy landscape and level competing free-energy minima of aggregated species. Similar smoothing, but at the level of reducing local barriers, also is observed to arise from symmetry effects (P. Wolynes and S. Plotkin, personal communication). However, there are cases where complications may still arise. For instance, proteins may assemble by native-like contacts between complementary segments of identical molecules, forming so-called domain-swapped units (37, 38). Domain swapping occurs naturally in many protein dimers (39) and is suggested to be the basis for the polymerization of α1-antitrypsin (40). Another Achilles' heel is provided by contacts that are not normally encountered in the cell and hence constitute “dead angles” from an evolutionary perspective. Examples include interactions that cannot be established within a single chain because of backbone restrictions and interactions between polypeptides outside their normal biological context. The local stretches of hydrophobic residues within the normally membrane-bound β-AP, the U1A protein (20), and the aggregation-prone S6 mutants would fall in the latter category.
Although fibrillation of the C-terminal part of β-AP may thus be driven by rather promiscuous hydrophobic interactions, it is clear that fibrillation of other proteins relies on specific pairing of complementary charges. As in native proteins, the β-sheets of the fibrils can be built up in many different ways; the only requirement may be that the interaction pattern complies with a constructive periodicity. In a general perspective, this leaves two determinants for protein fibrillation: the quality of the “sticky” sequence and its exposure. If the sequence is hidden in a native protein structure, fibrillation must be preceded by unfolding and is correlated with the stability of the hosting protein (11). If the sequence, like β-AP, cannot fold on its own and therefore remains constantly exposed in the monomer, aggregation is determined by the side-chain properties and concentration only.
Acknowledgments
We thank Peter Wolynes and Håkan Wennerström for valuable discussions and the Swedish Natural Science Council for financial support. D.E.O. gratefully acknowledges a postdoctoral stipend from the European Molecular Biology Organization.
Abbreviation
- β-AP
Alzheimer β-peptide
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1QJH, 1CQN, and 1CQM).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.160086297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.160086297
References
- 1.Kelly J W. Structure (London) 1997;5:595–600. doi: 10.1016/s0969-2126(97)00215-3. [DOI] [PubMed] [Google Scholar]
- 2.Iversen L L, Mortishire-Smith R J, Pollack S J, Shearman M S. Biochem J. 1995;311:1–16. doi: 10.1042/bj3110001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang J, Lemaire H-G, Unterbeck A, Salbaum J M, Masters C L, Grzeschik K H, Multhaup G, Beyreuther K, Muller-Hill B. Nature (London) 1987;325:1124–1126. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 4.Harper J D, Lansbury P T J. Annu Rev Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- 5.Kirschner D A, Abraham C, Selkoe D J. Proc Natl Acad Sci USA. 1986;83:503–507. doi: 10.1073/pnas.83.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halverson K, Fraser P E, Kirschner D A, Lansbury P T., Jr Biochemistry. 1990;29:2639–2644. doi: 10.1021/bi00463a003. [DOI] [PubMed] [Google Scholar]
- 7.Lansbury P T, Jr, Costa P R, Griffiths J M, Simon E J, Auger M, Halverson K J, Kocisko D A, Hendsch Z S, Ashburn T T, Spencer R G, et al. Nat Struct Biol. 1995;2:990–998. doi: 10.1038/nsb1195-990. [DOI] [PubMed] [Google Scholar]
- 8.Benzinger T L S, Gregory D M, Burkoth T S, Miller-Auer H, Lynn D G, Botto R E, Meredith S C. Proc Natl Acad Sci USA. 1998;95:13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorevic P D, Castano E M, Sarma R, Frangione B. Biochem Biophys Res Commun. 1987;147:854–862. doi: 10.1016/0006-291x(87)91008-4. [DOI] [PubMed] [Google Scholar]
- 10.Kirschner D A, Inouye H, Duffy L K, Sinclair A, Lind M, Selkoe D J. Proc Natl Acad Sci USA. 1987;84:6953–6957. doi: 10.1073/pnas.84.19.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobson C M. Trends Biochem Sci. 1999;24:329–332. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- 12.Chiti F, Webster P, Taddei N, Clark A, Stefani M, Ramponi G, Dobson C M. Proc Natl Acad Sci USA. 1999;96:3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross M, Wilkins D K, Pitkeathly M C, Chung E W, Higham C, Clark A, Dobson C M. Protein Sci. 1999;8:1350–1357. doi: 10.1110/ps.8.6.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sunde M, Serpell L C, Bartlam M, Fraser P E, Pepys M B, Blake C C F. J Mol Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 15.Helms L R, Wetzel R. J Mol Biol. 1996;257:77–86. doi: 10.1006/jmbi.1996.0148. [DOI] [PubMed] [Google Scholar]
- 16.Stott K, Blackburn J M, Butler P J, Perutz M. Proc Natl Acad Sci USA. 1995;92:6509–6513. doi: 10.1073/pnas.92.14.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otzen D E, Kristensen O, Proctor M, Oliveberg M. Biochemistry. 1999;38:6499–6511. doi: 10.1021/bi982819j. [DOI] [PubMed] [Google Scholar]
- 18.Eisenstein E, Schachman H K. In: Protein Function: A Practical Approach. Creighton T E, editor. Oxford: IRL; 1989. pp. 135–176. [Google Scholar]
- 19.Levine H I. Protein Sci. 1993;2:404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silow M, Oliveberg M. Proc Natl Acad Sci USA. 1997;94:6084–6086. doi: 10.1073/pnas.94.12.6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oliveberg M. Acc Chem Res. 1998;31:765–772. [Google Scholar]
- 22.Silow M, Tan Y-J, Fersht A R, Oliveberg M. Biochemistry. 1999;38:13006–13012. doi: 10.1021/bi9909997. [DOI] [PubMed] [Google Scholar]
- 23.Klunk W E, Jacob R F, Mason R P. Anal Biochem. 1999;266:66–70. doi: 10.1006/abio.1998.2933. [DOI] [PubMed] [Google Scholar]
- 24.Booth D R, Sunde M, Bellotti V, Robinson C V, Hutchinson W L, Fraser P E, Hawkins P N, Dobson C M, Radford S E, Blake C C, Pepys M B. Nature (London) 1997;385:787–793. doi: 10.1038/385787a0. [DOI] [PubMed] [Google Scholar]
- 25.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. Nature (London) 1996;362:553–557. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 26.Bryngelson J, Onuchic J N, Socci N D, Wolynes P. Proteins Struct Funct Genet. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 27.Otzen D E, Oliveberg M. Proc Natl Acad Sci USA. 1999;96:11746–11751. doi: 10.1073/pnas.96.21.11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fersht A R, Shi J P, Knill-Jones J, Lowe D M, Wilkinson A J, Blow D M, Brick P, Carter P, Waye M M, Winter G. Nature (London) 1985;314:235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- 29.Irbäck A, Peterson C, Potthast F. Proc Natl Acad Sci USA. 1996;93:9533–9538. doi: 10.1073/pnas.93.18.9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pande V S, Grosberg A Y, Tanaka T. Proc Natl Acad Sci USA. 1994;91:12972–12975. doi: 10.1073/pnas.91.26.12972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindahl M, Svensson L A, Liljas A, Sedelnikova S E, Eliseikina I A, Fomenkova N P, Nevskaya N, Nikonov S V, Garber M B, Muranova T A, et al. EMBO J. 1994;13:1249–1254. doi: 10.2210/pdb1ris/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harper J D, Wong S S, Lieber C M, Lansbury P T. Chem Biol. 1997;4:119–125. doi: 10.1016/s1074-5521(97)90255-6. [DOI] [PubMed] [Google Scholar]
- 33.Walsh D M, Lomakin A, Benedek G B, Condron M M, Teplow D B. J Biol Chem. 1997;272:22364–22372. doi: 10.1074/jbc.272.35.22364. [DOI] [PubMed] [Google Scholar]
- 34.Kowalewski T, Holtzman D M. Proc Natl Acad Sci USA. 1999;96:3688–3693. doi: 10.1073/pnas.96.7.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hilbich C, Kisters-Woike B, Reed J, Masters C, Beyreuther K. J Mol Biol. 1991;218:149–163. doi: 10.1016/0022-2836(91)90881-6. [DOI] [PubMed] [Google Scholar]
- 36.Jarrett J T, Lansbury P T., Jr Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 37.Taniuchi H, Anfinsen C B. J Biol Chem. 1971;246:2291–2297. [PubMed] [Google Scholar]
- 38.Taniuchi H, Parr G R, Juillerat M A. Methods Enzymol. 1986;131:185–217. doi: 10.1016/0076-6879(86)31042-5. [DOI] [PubMed] [Google Scholar]
- 39.Schlunegger M P, Bennettt M J, Eisenberg D. Adv Protein Sci. 1997;50:61–122. doi: 10.1016/s0065-3233(08)60319-8. [DOI] [PubMed] [Google Scholar]
- 40.Elliott P R, Lomas D A, Carrell R W, Abrahams J P. Nat Struct Biol. 1996;3:678–681. doi: 10.1038/nsb0896-676. [DOI] [PubMed] [Google Scholar]
- 41.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 42.Clarke J, Fersht A R. Biochemistry. 1993;32:4322–4329. doi: 10.1021/bi00067a022. [DOI] [PubMed] [Google Scholar]