Abstract
The pannexin family of mammalian proteins, composed of Panx1, Panx2, and Panx3, has been postulated to be a new class of single-membrane channels with functional similarities to connexin gap junction proteins. In this study, immunolabeling and coimmunoprecipitation assays revealed that Panx1 can interact with Panx2 and to a lesser extent, with Panx3 in a glycosylation-dependent manner. Panx2 strongly interacts with the core and high-mannose species of Panx1 but not with Panx3. Biotinylation and dye uptake assays indicated that all three pannexins, as well as the N-glycosylation-defective mutants of Panx1 and Panx3, can traffic to the cell surface and form functional single-membrane channels. Interestingly, Panx2, which is also a glycoprotein and seems to only be glycosylated to a high-mannose form, is more abundant in intracellular compartments, except when coexpressed with Panx1, when its cell surface distribution increases by twofold. Functional assays indicated that the combination of Panx1 and Panx2 results in compromised channel function, whereas coexpressing Panx1 and Panx3 does not affect the incidence of dye uptake in 293T cells. Collectively, these results reveal that the functional state and cellular distribution of mouse pannexins are regulated by their glycosylation status and interactions among pannexin family members.
INTRODUCTION
The pannexin family of proteins is composed of three members, Panx1, Panx2, and Panx3 in mouse and PANX1, PANX2, and PANX3 in human. They were discovered based on their sequence similarity to innexins, the invertebrate gap junction proteins (Panchin et al., 2000). Recent studies have started to elucidate their characteristics and potential function. Panx1 has been characterized more extensively and has been postulated to be a conduit for release of ATP from erythrocytes and taste receptor cells (Locovei et al., 2006; Huang et al., 2007b) and to contribute to ATP release when expressed apically in airway epithelial cells (Ransford et al., 2009). Panx1 single-membrane channels are mechanosensitive (Bao et al., 2004; Pelegrin and Surprenant, 2007) and can also be activated by high extracellular potassium (Silverman et al., 2009). Interestingly, Panx1 channels have been implicated in cell death after brain ischemia and epileptiform seizure activity (Thompson et al., 2006, 2008). In other studies, Panx1 and Panx2 expression patterns in the rodent brain have been shown to be highly regulated during development (Ray et al., 2005). For example, Panx1 is more abundant in the embryonic and postnatal brain, with a subsequent decrease in adult rats, whereas Panx2 transcripts are low in prenatal brains but increase significantly during postnatal development (Vogt et al., 2005).
Panx1 and Panx3 have been reported to be glycoproteins, capable of assembling into functional single-membrane channels at the plasma membrane (Boassa et al., 2007; Penuela et al., 2007). In vivo, Panx1 and Panx3 have been reported to be coexpressed in mouse skin, osteoblasts, and specialized cartilage (Baranova et al., 2004; Penuela et al., 2007). Panx1 and Panx2, in contrast, have been found in retina, lens, cochlea, cerebellum, and brain (Bruzzone and Dermietzel, 2006; Ray et al., 2006; Dvoriantchikova et al., 2006a,b; Tang et al., 2008). Very little is known about the posttranslational modifications or functional properties of Panx2. Even though two or more Pannexin (Panx) family members are coexpressed within many cell types, it is not known whether they intermix or whether intermixing would regulate channel function. Previous evidence that such intermixing might exist for any combination of Panxs was deduced from Xenopus oocyte studies where coexpression of Panx2 attenuated Panx1 channels (Bruzzone et al., 2003, 2005).
Connexins, in contrast, the vertebrate family of gap junction proteins, have been extensively reported to form mixed channels of co-oligomerized connexins (Harris, 2007). The connexin family of gap junction proteins has 20 members in rodent genomes and based on their sequence homologies have been divided into several subgroups (Willecke et al., 2002). Connexins of the same subgroup often have the capacity to hetero-oligomerize and form mixed channels that confer unique properties to the channels that select for transjunctional permeable molecules (Koval, 2006; Harris, 2007). However, although connexins provide proof-of-principle that family members can intermix, these frequently phosphorylated molecules (Solan and Lampe, 2005) are quite distinct from the pannexin family of glycoproteins. Thus, in the present study, we further assessed the glycosylation and functional status of all mouse pannexin family members and examined how glycosylation regulates the channel function of pannexins and their ability to cointeract.
MATERIALS AND METHODS
Sequence Analysis and Generation of Pannexin-specific Antibodies
Protein sequences corresponding to the current National Center for Biotechnology Information reference sequences (RefSeq) for mouse Panx1 (NP_062355.2), Panx2 (NP_001002005.2), and Panx3 (NP_766042.2) were analyzed using Blast2seq (Tatusova and Madden, 1999) for pairwise sequence alignment, accounting for differences in protein length. Toppred algorithm (Claros and Heijne, 1994) was used to predict their topology and identify the predicted location of their transmembrane domains. The NetN-Glyc1.0 server (Blom et al., 2004) was used to identify predicted consensus sites for N-glycosylation in all three family members.
Peptide sequences from each of the main domains of the pannexin proteins were tested for potential antigenicity and used to generate site-directed rabbit polyclonal antibodies by Genemed Synthesis (San Antonio, TX). Specific peptides (Table 1) were synthesized, tagged with keyhole limpet hemocyanin through an extra C-terminal cysteine, and injected into rabbits for generation of antisera. Immunoblotting and immunofluorescent analysis were used to test preimmune sera and antisera before the selection of the antibodies that would be affinity purified against the corresponding peptides by Genemed Synthesis (Table 1). Selected antibodies were denoted by their corresponding domain (EL1, first extracellular loop; EL2, second extracellular loop; IL, intracellular loop; CT, carboxy terminus) and the first amino acid in the peptide sequence used to generate the antibody. All of the affinity-purified antibodies were used in immunoblotting and immunofluorescent assays for characterizing the pannexins. For most studies, the Panx1 CT-395 and Panx3 CT-379 antibodies (reported previously by Penuela et al., 2007) were used. For Panx2 immunolabeling, the CT-494 sera was used, whereas the rabbit anti-mouse Panx2 antibody from Invitrogen (Burlington, ON, Canada) was used for all Panx2 immunoblots shown. Thus, to simplify the description of these antibodies in the text and figures, they are referred to as anti-Panx1, Panx2, and Panx3 antibodies.
Table 1.
Comparison of mouse pannexins and peptides used for antibodies
| Pannexin | Coding sequence (aa) | Protein size (kDa) | Glycosylation sites | Protein domain | Peptides |
|---|---|---|---|---|---|
| Panx1 | 426 | ∼41–48 | N254 (EL2) | EL2 (247–265) | SIKSGVLKNDSTIPDRFQC |
| CT (395–409) | QRVEFKDLDLSSEAR | ||||
| Panx2 | 677 | ∼80 | N86 (EL1)a | CT (494–508) | ASEKKHTRHFSLDVH |
| Panx3 | 392 | ∼43 | N71 (EL1) | EL1 (84–100) | CWDSLAHHTQDKAGQYK |
| IL (169–185) | SDPHVFWDELEKARKER | ||||
| CT (379–392) | KPKHLTQHTYDEHA |
a Predicted N-glycosylation consensus site.
Pannexin Expression Constructs and N-Glycosylation-deficient Mutants
Untagged or green fluorescent protein (GFP)-tagged mouse Panx1 and Panx3 expression constructs in pEGFP-N1 (Clontech, Mountain View, CA), were generated as described previously (Penuela et al., 2007). Site-directed mutagenesis was used to generate Panx1N254Q and Panx3N71Q mutants as described previously (Penuela et al., 2007). Mouse Panx2 was originally cloned from total RNA from mouse brain collected using Isogen (Nippon Gene, Tokyo, Japan) and was reverse transcribed using SuperScript III First-Strand Synthesis kit as per the manufacturer's protocol (Invitrogen). Mouse Panx2 open reading frame was amplified from the brain cDNA by using specific primers (sense, ATCGAATTCACC-ATGCACCACCTCCTGGAGCAGT; antisense, ATCTCGAGTCAAAACTCCA-CAGTACTCACAACT) and KOD Plus DNA polymerase (Toyobo, Osaka, Japan). EcoRI and XhoI restriction digestion sites were introduced into the polymerase chain reaction (PCR) product through sense and antisense primers, respectively. The purified PCR product was inserted into the respective site of pcDNA3 vector (Invitrogen) after digestion with restriction enzyme. The clone was confirmed by restriction digestion and DNA sequencing. To express mouse Panx2 in the same expression vector as generated Panx1 and Panx3 cDNA constructs, we excised Panx2 from pcDNA3 vector and inserted into the EcoR1–Sal1 site of pEGFP-N1 vector (Clonetech), rendering an untagged version of Panx2 in this expression vector. All clones were confirmed by sequencing to contain the complete coding sequence of each pannexin gene, according to RefSeq and encoded proteins of 426 (Panx1), 677 (Panx2), and 392 (Panx3) amino acids.
Cell Lines and Culture Conditions
Media and reagents were obtained from Invitrogen, Sigma-Aldrich (St. Louis, MO), and BD Biosciences (Mississauga, ON, Canada). Normal rat kidney (NRK), mouse osteoblastic cells (MC3T3-E1), and human embryonic kidney cells (293T) were obtained from American Type Culture Collection (Manassas, VA). Cells were grown in high-glucose DMEM, or α-minimum essential medium (MEM) for MC3T3-E1, supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine.
Transient Expression
For transient transfections, mammalian cells grown to 50–75% confluence in 35- or 100-mm (for biotinylation and coimmunoprecipitation [co-IP]) culture dishes were transfected in Opti-MEM I media with Lipofectamine2000 (Invitrogen) and 1 or 5 μg of plasmid DNA, respectively, for 4 h at 37°C as described previously (Penuela et al., 2007). For coexpression assays, equal amounts of each construct were transfected at a 1:1 ratio. After 48 h, transfections were evaluated by immunofluorescence.
Immunocytochemistry
Coverslips with cultured cells were immunolabeled as described previously for Panx1 CT-395 and Panx3 CT-379 (Penuela et al., 2007), by using a final concentration of 2 μg/ml for each affinity-purified antibody. Preimmune sera and antisera for the new anti-pannexin antibodies were tested at 1:500 dilutions, and the affinity-purified antibodies were used at 2 μg/ml. Peptide preadsorption assays were performed for all antibodies at 20:1 M excess of the peptide with respect to the antibody, as described previously (Penuela et al., 2007). Because all antibodies were generated in rabbits, for double labeling of Panx1 with Panx2 or Panx3, we generated a Panx1 CT-395 antibody directly labeled with Fluor 488, by using the DyLight 488-microscale antibody labeling kit (Pierce Chemical, Rockford, IL) according to manufacturer's instruction. This fluorochrome-labeled antibody was used for direct labeling of Panx1 after immunolabeling of coexpressed pannexins in cultured cells. The labeled antibody was used at 2 μg/ml and incubated for 1 h at room temperature, followed by washes with phosphate-buffered saline (PBS) and distilled deionized water, before mounting. Analysis was performed on an LSM 510 inverted confocal microscope (Carl Zeiss, Thornwood, NY) by using a 63× oil immersion objective.
Western Blotting and Deglycosylation Assays
Cell lysates from control 293T cells or 293T cells expressing pannexins were collected from cultures by using a Triton-based extraction buffer as described previously (Penuela et al., 2007). N-Glycosidase F assays were performed according to the manufacturer's instructions (Roche Applied Science, Laval, QC, Canada). Using 35 μg of total protein with or without 10 U of the glycosidase and incubated for either 60 or 15 min at 37°C (the different incubation times allow for the estimation of the number of glycan chains bound to the glycoprotein) (Tarentino and Plummer, 1987). Endoglycosidase H (EndoH) digestion of 35 μg of total protein per sample was done at 37°C for 1 h, by using 5000 U of the enzyme, in parallel with control samples that did not contain EndoH. Digestion with EndoH was used to detect the presence of high-mannose modification, whereas more complex forms of glycosylation are known to be resistant to digestion. Protein samples were separated on an 8% SDS-polyacrylamide gel electrophoresis gel (PAGE) and transferred to nitrocellulose membranes. Membranes were processed for immunoblotting as described previously (Penuela et al., 2007) by using the selected antibodies for Panx1, Panx2 (Zymed Laboratories, South San Francisco, CA/Invitrogen), and Panx3, as well as for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a loading control. All protein standards are depicted in kilodaltons.
Cell Surface Biotinylation Assays
293T cells expressing Panx1 and/or Panx2, Panx3, or the N-glycosylation–deficient mutants of Panx1 and Panx3 described by Penuela et al. (2007) were subjected to cell surface biotinylation 48 h after transfection. All reagents and cultures were kept on ice during the biotinylation assay. In brief, cultures were washed three times with ice-cold PBS. Control plates were incubated in PBS, whereas test plates were incubated with the same solution containing EZ-link NHS-LC-biotin or EZ-link Sulfo NHS-LC biotin (1 mg/ml; Pierce Chemical) for 20 min at 4°C. Cultures were washed once with PBS containing 100 mM glycine and then incubated in glycine buffer for 15 min at 4°C to quench the biotin. Cells were then lysed with SDS lysis buffer (1% Triton X-100 and 0.1% SDS in PBS). Lysates were rocked at 4°C for 1 h, and supernatants were subjected to bicinchoninic acid test (Pierce Chemical) for quantification. Equal amounts of protein lysate (750–1000 μg) from control and biotin samples were incubated overnight with 50 μl of NeutrAvidin-agarose beads (Pierce Chemical). Beads were washed three times with IP lysis buffer (150 mM NaCl, 10 mM Tris-HCl, pH 7.4, 1 mM EDTA, 0.5% NP-40, and 1% Triton X-100) containing 1 mM NaF and 1 mM Na3VO4, and once with PBS, and then dried by aspiration. The beads were resuspended in 2× Laemmli buffer (under reducing conditions), boiled for 5 min, resolved by SDS-PAGE, and transferred to nitrocellulose membranes to be probed with the corresponding anti-Panx1, -Panx2, or -Panx3 antibodies. In parallel, an aliquot of 20 μg of total protein lysate was subject to immunoblot analysis. GAPDH (an intracellular protein) was used as a control to detect any unexpected biotin internalization. Quantification of biotinylated proteins in immunoblots of Panx2-expressing cell lysates or Panx1- and Panx2-coexpressing cells was processed using the Odyssey infrared-imaging system (LI-COR, Lincoln, NE). Densitometry readings of three independent experiments were analyzed using a Student's t test (p < 0.05).
Coimmunoprecipitation
CoIP assays were performed as described by Langlois et al. (2008). In brief, 1 mg of protein from each lysate was incubated in IP buffer (see above) overnight at 4°C in the presence of 10 μg/ml anti-Panx1, -Panx2 (Zymed/Invitrogen) or -Panx3 affinity purified antibodies. Complexes were collected by incubating the mixtures for 2 h at 4°C with 30 μl (50% slurry) of protein A-Sepharose beads. Several washes with 500 μl of IP buffer removed unspecific binding proteins. Beads were dried by aspiration and resuspended in 2× Laemmli buffer (reducing conditions), boiled for 5 min, run in 8% SDS-PAGE gels and transferred to nitrocellulose membranes to be probed with the specific anti-pannexin antibodies.
Because all antibodies used were generated in rabbits, it was necessary to use the Clean-Blot IP Detection Reagent (horse radish peroxidase; Pierce Chemical) as a secondary antibody. This reagent binds preferentially to native immunoglobulin G (IgG) and not denatured IgG, minimizing the appearance of IgG bands in the blots. It was used in a 1:4000 dilution and after washes with 0.05% Tween 20 in PBS, the signal was detected with a SuperSignal West Femto chemiluminescent kit (Pierce Chemical). When more than one probing was necessary for the same blot, the membrane was not stripped (to avoid reduction in signal) and was subsequently probed with the second rabbit-derived antibody, followed by a goat anti-rabbit Alexa Fluor 680 secondary antibody for detection on an Odyssey infrared imaging system (LI-COR). In this case, IgG bands were visible after using the anti-rabbit fluorescent secondary antibody.
Immunoprecipitation controls for all three antibodies were performed by IP of singly transfected 293T cells expressing Panx1 or Panx2 and pulled down with the opposite antibody (Panx2 or Panx3 antibodies) to rule out any cross-talk between antibody specificities (Supplemental Figure 1).
Dye Uptake Experiments and Statistics
One hundred thousand 293T cells per dish were plated in 35-mm culture dishes coated with 0.5 mg/ml collagen I in 60% ethanol. The cells were transfected the next day with the various pannexin constructs or cotransfected with two constructs as described above. Control 293T cells were transfected with free GFP to monitor transfection efficiency and background dye uptake. After 48–72 h posttransfection, all cultures were kept on ice and subjected to sulforhodamine B (mol. wt. 558.7; Invitrogen) dye uptake assays under conditions of physiological Ca++ and Mg++ and mechanical stimulation as we described in detail previously (Penuela et al., 2007). ImageJ (National Institutes of Health, Bethesda, MD) was used for quantification of the dye uptake incidence per field (three random 20× fields near the stimulation target were averaged per dish). Parallel control experiments were done using dextran rhodamine dye. Experiments included four independent transfections and were assessed by one-way analysis of variance (ANOVA) followed by a Tukey test.
RESULTS
Sequence Analysis and Cloning of Mouse Panx2
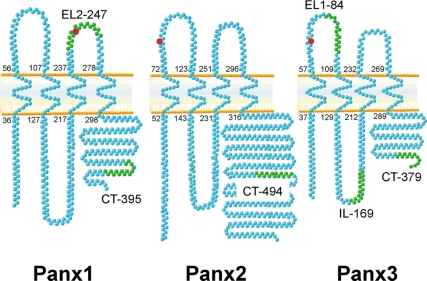
The three National Center for Biotechnology Information protein reference sequences (RefSeq) annotated for mouse Panx1, Panx2, and Panx3 were analyzed with a Toppred algorithm (Claros and Heijne, 1994) to predict their topology and identify the location of their transmembrane domains. As shown in Figure 1, all three murine pannexins are predicted to have intracellular amino tails and carboxy tails, as well as four transmembrane domains, each of 20 amino acids. As we reported previously, Panx1 and Panx3 have a consensus site for N-linked glycosylation on amino acids N254 and N71, respectively (Penuela et al., 2007). Using the NetN-Glyc1.0 server (Blom et al., 2004), we found that Panx2 also encodes a potential N-linked glycosylation site at position N86 (Table 1) in its first extracellular loop domain, similar to Panx3. Based on the Toppred-predicted topology, Panx2 also exhibits a substantially larger carboxy tail compared with the other two pannexin family members.
Figure 1.
Transmembrane topology of pannexin family members and peptides used for antibody generation. Toppred algorithms predicted a similar membrane tetra spanning topology for all three mouse pannexin proteins. Numbers indicate the amino acid position of transmembrane domains. N-Glycosylation consensus sites are depicted in red. Peptides from different domains of the three pannexins (green residues) were used to generate rabbit polyclonal antibodies (CT, carboxy terminus, EL, extracellular loop; and IL, intracellular loop, followed by the first amino acid position of the peptide).
We cloned mouse Panx2 by reverse transcription-PCR from mouse brain, and the complete coding sequence, represented by a band of ∼2 kb, was cloned into expression vectors, validated by sequencing, and found to be identical to the annotated RefSeq with a predicted protein product of 677 amino acids (Table 1).
Site-directed Antibodies and Pannexin Characterization
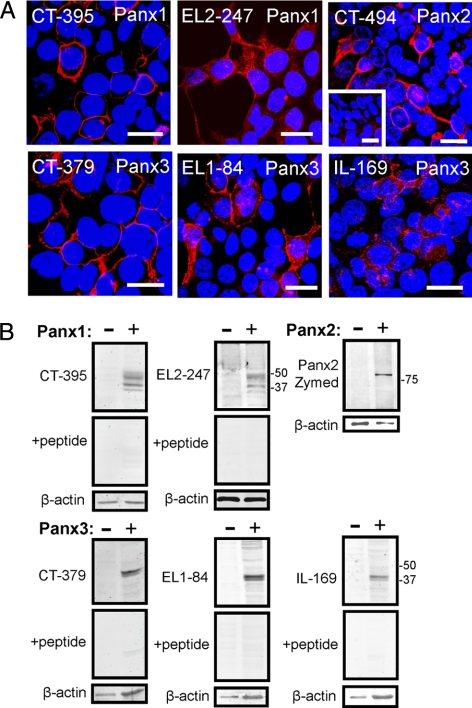
Peptides for the generation of site-directed rabbit polyclonal antibodies (Table 1) were designed for each of the extracellular and intracellular domains of the pannexins (Figure 1). Out of those, six peptides and their corresponding antibodies were affinity purified and used in the characterization of pannexins in permeabilized 293T cells expressing Panx1, Panx2, or Panx3 (Figure 2). The Panx1 antibody, targeting the second extracellular loop (EL2-247), showed specific labeling at both cell surface and intracellular compartments (Figure 2A). In Western blots, EL2-247, revealed a similar banding profile to our previously reported anti-Panx1 CT-395 antibody (Penuela et al., 2007), and the corresponding peptide competition indicated that it is specific (Figure 2B). The Panx2 antibody (CT-494) exhibited specificity for Panx2, whereas the control preimmune serum did not show any significant labeling (Figure 2A, inset). Western blots of Panx2-expressing cells by using the CT-494 antibody highlighted a band of ∼80 kDa, but because the antibody presented higher than desirable background signal (data not shown), in further immunoblotting experiments we chose to use the commercially available rabbit anti-Panx2 antibody C terminus (Zymed Laboratories/Invitrogen). The Zymed anti-Panx2 antibody also recognized a band of ∼80 kDa (Figure 2B) that occasionally presented as a doublet in Western blots of 293T cells expressing mouse Panx2. All Panx3 antibodies derived from three distinct protein epitopes, recognized a doublet band of ∼43 kDa as was reported earlier for CT-379 (Penuela et al., 2007), and the labeling was effectively eliminated by preadsorption with the cognate peptides (Figure 2B). Both, Panx3 EL1-84 and IL-169 antibodies, when used for immunolabeling of Panx3-expressing 293T cells, were particularly effective in identifying both intracellular and cell surface Panx3. From this library of novel anti-pannexin antibodies, we used the CT-395 anti-Panx1 antibody, the CT-379 anti-Panx3 antibody, and the CT-494 anti-Panx2 antibody for all further immunolabeling studies reported here. The Zymed Laboratories/Invitrogen rabbit anti-mouse Panx2 antibody was used for all immunoblotting assays.
Figure 2.
Characterization of pannexin-specific antibodies. Panx1, Panx2, and Panx3 were localized in transfected 293T cells by using various anti-pannexin affinity-purified antibodies denoted by CT, carboxy terminus; EL, extracellular loop; and IL, intracellular loop, followed by the first amino acid position of the peptide (A). As a control, preimmune Panx2 sera revealed no significant labeling (inset). Bars, 20 μm. Immunoblotting of cell lysates from pannexin-expressing 293T cells (B) revealed similar banding profiles for the two Panx1 antibodies (∼41–47 kDa), as well as the three Panx3 antibodies (doublet ∼43 kDa). The anti-Panx2 rabbit antibody (Zymed Laboratories/Invitrogen) identified an ∼80-kDa band (sometimes resolved as a doublet), and this antibody was chosen along with CT-395 and CT-379 for further immunoblotting assays. Peptide preadsorption assays revealed that all antibodies were specific for pannexins. β-Actin was used as loading control.
Pannexin Family Members Are Distinctly Different Glycoproteins
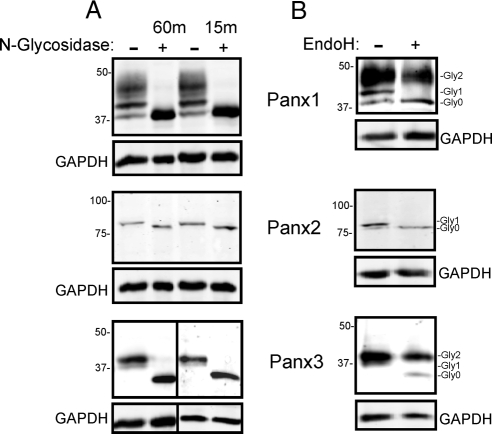
As was reported previously (Boassa et al., 2007; Penuela et al., 2007), N-glycosidase assays and site-directed mutagenesis revealed that both rat and mouse Panx1 are N-linked glycoproteins, whereas at least mouse Panx3 is a glycoprotein. To expand our understanding of the carbohydrate entities associated with each pannexin family member, we evaluated the number of glycan chains that are linked to each protein when expressed in 293T cells by using limited enzymatic digestions (Tarentino and Plummer, 1987). In only 15 min, enzymatic N-glycosidase F digestions removed all the carbohydrate from all pannexins, indicating that only one glycan chain is probably present in Panx1, Panx2, and Panx3 (Figure 3A). These results are consistent with the sequence-based prediction of a single consensus site for N-glycosylation in the extracellular domains of the pannexin proteins (Figure 1). Notably, this is the first reporting that Panx2 is also a glycoprotein, but the level of posttranslational modification seems to be reduced compared with the other two pannexins, as evidenced by the minor shift in band mobility (Figure 3A). To further characterize the carbohydrate moieties of these proteins, digestion with EndoH, an enzyme specific for the high-mannose form of N-linked glycosylation was used. As reported by Boassa et al. (2007), the intermediate band of rat Panx1, namely, Gly1, was selectively reduced upon digestion with EndoH. Interestingly, the minor band shift upon glycosidase treatment for Panx2 was also sensitive to EndoH, indicating that Panx2 is only subjected to high-mannose glycosylation in 293T cells. The lower band of the Panx3 doublet (Gly1) was also reduced to the level of Gly0, identifying this band as the high-mannose component, whereas the Gly2 band corresponds to the more complex glycoprotein (Figure 3B). Collectively, these results suggest that both Panx1 and Panx3 are subject to both high mannose and complex carbohydrate processing, whereas Panx1 can also exist in the cell as a core protein escaping any glycosylation. Panx2 seems to exist only as a core protein or as a high mannose species.
Figure 3.
Pannexin family members are diversely glycosylated. Cell lysates from 293T cells expressing Panx1, Panx2, and Panx3 were treated with N-glycosidase F (A). Gel banding patterns revealed band shifts for each of the pannexins at both the 15- and 60-min time points, suggesting that each pannexin has a single glycan chain. N-Glycosidase F (A) and Endo H (B) enzymatic digestion revealed that Panx2 only exists as core (Gly0) and high mannose (Gly1) species, whereas Panx1 and Panx3 also exist as complex glycoprotein species (Gly2). Panx1 also exists in an unglycosylated state (Gly0). GAPDH was used as loading control.
Cell Surface Localization of Pannexin Species
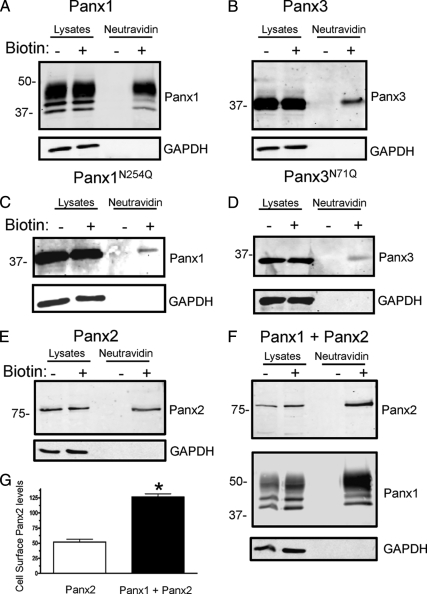
To determine whether all three pannexin family members and their intermediate glycosylation species were capable of localizing to the plasma membrane, we conducted biotinylation assays by using EZ-link NHS-LC-Biotin and EZ-link Sulfo NHS-LC biotin (Pierce Chemical) in a manner to label proteins at the cell surface of live 293T cells. As assessed by Panx1 immunoblotting of cell lysates and the NeutrAvidin-isolated fraction, all species of Panx1 were found to be able to traffic to the cell surface, but there was a preference for the Gly2 species (Figure 4A). Confirmation that the unglycosylated species of Panx1 can traffic and reside at the cell surface was provided by the fact that the Panx1N254Q species could also be biotinylated at the cell surface (Figure 4C). A subpopulation of glycosylated Panx3 was also identified at the cell surface as was its corresponding glycosylation-deficient mutant (Panx3N71Q) (Figure 4, B and D). When 1 mg of the Panx2 biotinylated lysate was pulled down with the NeutrAvidin beads, a subpopulation of Panx2 was evident at the cell surface (Figure 4E). Moreover, cell surface Panx2 increased over twofold when coexpressed with Panx1 (Figure 4, F and G). In all cell surface biotin labeling experiments, GAPDH was used as a control to verify that biotin had not entered the cell (Figure 4).
Figure 4.
Glycosylated and unglycosylated pannexin species reach the cell surface. Cell surface biotinylation of 293T cells expressing Panx1 (A), Panx3 (B), Panx1N254Q (C), Panx3N71Q (D), Panx2 (E), or cells expressing both Panx1 and Panx2 (F) revealed that all pannexin species and their glycosylation-defective mutants have at least some capacity to traffic to the cell surface. Coexpression of Panx1 and Panx2 caused over a twofold increase in the cell surface biotinylation of Panx2 (G) (p < 0.05, n =3 independent transfections). Lysates with (+) or without (−) biotin were precipitated with NeutrAvidin beads and run in parallel with control lysates. GAPDH was used as a control for biotin internalization.
Panx1 Facilitates the Cell Surface Localization of Panx2
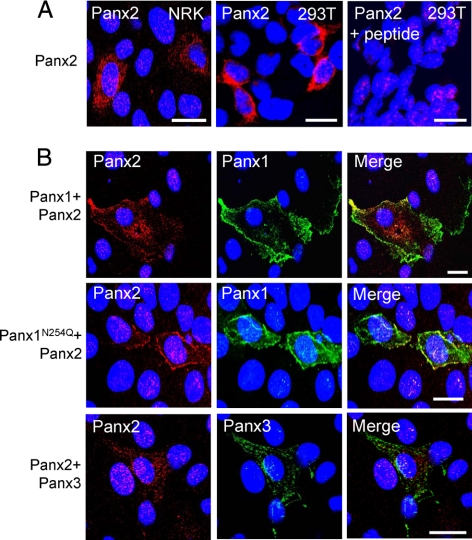
When Panx2 was ectopically expressed in 293T or NRK cells and detected with the CT-494 antibody, it was found to be predominantly localized to intracellular compartments with some, but limited, evidence for cell surface localization (Figure 5A). A peptide preadsorption assay revealed some residual nuclear staining that was nonspecific (Figure 5A). Given their shape, large nuclei, and little cytoplasmic volume, 293T cells are not ideal for immunolocalization studies; therefore, we typically used NRK cells for localization studies. When Panx2 was coexpressed with Panx1 there was an increase in the cell surface localization of Panx2 (Figure 5B). Even when coexpressed with the mutant Panx1N254Q, there was an increase in the Panx2 signal detectable at the plasma membrane, indicating a potential interaction between Panx1 and Panx2. Conversely, when Panx3 was coexpressed with Panx2, there was no change in the Panx2 distribution pattern (Figure 5B).
Figure 5.
Coexpression of Panx1, but not Panx3, increased the cell surface localization of Panx2. Ectopic Panx2 was localized to intracellular compartments in NRK and 293T cells with some, but limited, evidence for its localization at the cell surface (A). Peptide preadsorption competed the antibody binding for Panx2, whereas some nonspecific nuclear labeling remained (A). In NRK cells, the coexpression of Panx1 or Panx1N254Q with Panx2 induced an increase in the cell surface population of Panx2. The subcellular localization of Panx2 did not change when coexpressed with Panx3 (B). Bars, 20 μm.
Panx1 and Panx3 Colocalize
Using Blast2seq (Tatusova and Madden, 1999) for pairwise sequence alignment we observed that the protein sequence identities and amino acid conservations among the three mouse pannexins ranged from 48 to 59% (Table 2), whereas the human pannexins were even more conserved (51–62%). It is also noteworthy that there is 93–94% conservation between the mouse and human pannexin proteins (Table 2) when comparing the current RefSeq protein accessions for all six genes. The sequence homology is highest between mouse Panx1 and Panx3 as well as between human PANX1 and PANX3. The genes encoding Panx1 and Panx3 are located on the same chromosome in both the mouse and human genomes (Baranova et al., 2004), and these pannexins are also coexpressed in many tissues (Penuela et al., 2007). This observation led us to explore the potential colocalization and interaction between mouse Panx1 and Panx3.
Table 2.
Protein conservation among pannexin family members
| Mouse | Sequence homologyaidentical (%)/conserved (%) | Human | Sequence homologyaidentical (%)/conserved (%) | Human tomouse | Sequence homologyaidentical (%)/conserved (%) |
|---|---|---|---|---|---|
| Panx1 and Panx2 | 28/50 | PANX1 and PANX2 | 33/56 | PANX1 and Panx1 | 86/94 |
| Panx1 and Panx3 | 41/59 | PANX1 and PANX3 | 42/62 | PANX2 and Panx2 | 91/93 |
| Panx2 and Panx3 | 29/48 | PANX2 and PANX3 | 30/51 | PANX3 and Panx3 | 89/93 |
a Blast2Seq pairwise alignment.
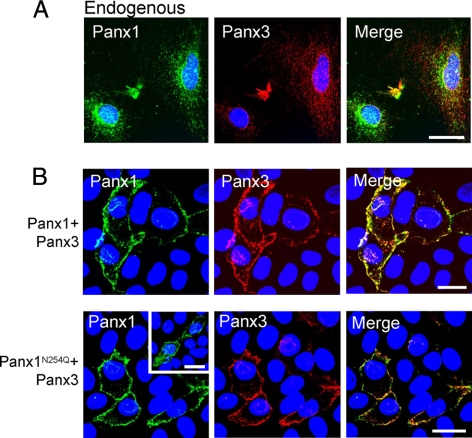
We had reported in previous studies that Panx1 and Panx3 are coexpressed in several mouse tissues such as skin, spleen, and cartilage (Penuela et al., 2007) and in cultures of osteoblastic cells, namely, MC3T3-E1 (Penuela et al., 2008). Double labeling of MC3T3-E1 cells with antibodies to Panx1 and Panx3 confirmed their endogenous colocalization and revealed a predominantly intracellular profile (Figure 6A), with isolated instances of cell surface distribution. Immunofluorescent labeling revealed colocalized Panx1 and Panx3 when coexpressed in NRK cells (Figure 6B). Interestingly, when the N-glycosylation mutant Panx1N254Q was coexpressed with Panx3, there was also an increase in cell surface localization compared with the predominantly intracellular profile of the mutant when expressed alone (Figure 6B, inset).
Figure 6.
Colocalization of Panx1 and Panx3. Double immunofluorescent labeling revealed that endogenous Panx1 and Panx3 partially colocalized in MC3T3-E1 cells (A). Coexpression of Panx1 or Panx1N254Q with Panx3 in NRK cells revealed colocalization of these pannexins and an increase in the cell surface distribution of the N-glycosylation mutant Panx1N254Q (B), compared with the more intracellular profile observed for Panx1N254Q when expressed alone (B; inset). Bars, 20 μm.
Pannexin Interactions Are Dependent on Glycosylation Status
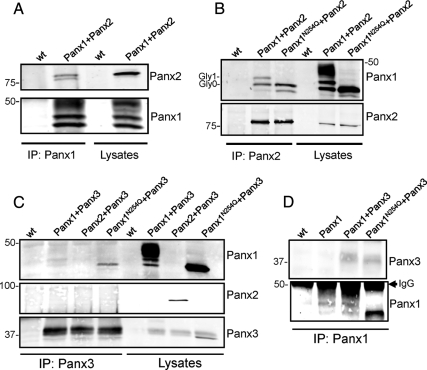
When Panx1 and Panx2 were coexpressed in 293T cells and immunoprecipitated with the Panx1 antibody, both the core and high mannose species of Panx2 coimmunoprecipitated with Panx1, indicating a strong interaction between the two proteins (Figure 7A). Reciprocally, the Panx2 antibody was able to pull down the Gly0 and Gly1 forms of Panx1, as well as the Panx1 glycosylation-defective mutant (Figure 7B). These findings indicate an interaction between the lower glycosylated forms of Panx1 and Panx2 but not with the Gly2 (complex glycoprotein form) of Panx1. In 293T cells coexpressing Panx1 and Panx3, the immunoprecipitation of Panx3 revealed the pull-down of only trace amounts of Panx1, but a substantial amount of Panx1N254Q (Figure 7C) was coimmunoprecipitated. In reciprocal experiments, immunoprecipitation of Panx1 or Panx1N254Q revealed the pull-down of small amounts of Panx3 (Figure 7D). These findings indicate that the weak interaction between Panx1 and Panx3 favors the core, unglycosylated species of Panx1 but that the interaction is not as robust as that observed between Panx1 and Panx2 (Figure 7A). The potential interaction of Panx2 and Panx3 was also assessed, but there was no significant coimmunoprecipitation of these pannexins when antibodies to either Panx3 (Figure 7C) or Panx2 (Supplemental Figure 2A) were used.
Figure 7.
Interactions among pannexin family members are regulated by glycosylation. An interaction between the Gly0 and Gly1 species of Panx1 with Panx2 was revealed when lysates from 293T cells expressing both pannexins were immunoprecipitated (IP) with Panx1 (A) or Panx2 (B) antibodies and immunoblotted for Panx1 and Panx2. (An occasional band seen below Gly0 seems to be a proteolytic product). Coimmunoprecipitation assays revealed an interaction between Panx1 and Panx3 that was more pronounced when the Panx1 glycosylation-deficient mutant was coexpressed with Panx3 (C). Panx2 and Panx3 did not interact when coexpressed in 293T cells (C). The reciprocal IP for Panx1 and Panx3 was performed with equivalent results (D). Twenty micrograms of the corresponding protein lysates were run in parallel to assess the expression levels of each pannexin.
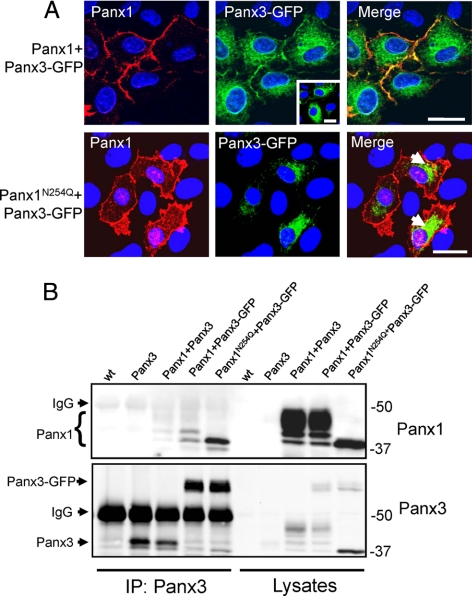
Interestingly, when the trafficking mutant Panx3-GFP (Penuela et al., 2007) was cotransfected with Panx1, there was an increase in the Panx3-GFP population at the cell surface, in contrast to its more typical intracellular profile observed when expressed alone (Figure 8A and inset). However, when the glycosylation-deficient mutant Panx1N254Q was coexpressed with Panx3-GFP, it partially colocalized with Panx3-GFP at intracellular sites but was essentially ineffective at rescuing the trafficking of Panx3-GFP to the cell surface (Figure 8A). Similar findings to those obtained in NRK cells were observed when Panx1, Panx3, Panx3-GFP, and Panx1N254Q were coexpressed in 293T cells, with the additional finding that Panx3 was quite effective at rescuing the trafficking of Panx3-GFP, and Panx1N254Q colocalized with Panx3-GFP (Supplemental Figure 3). The diverse localization profiles of Panx1N254Q in different cell lines have been previously reported (Penuela et al., 2008). Surprisingly, in coimmunoprecipitation studies using an antibody to Panx3, Panx3-GFP had increased ability to interact with the lower glycosylated species of Panx1 in comparison with untagged Panx3 (Figure 8B), perhaps due to the increased presence of Panx3-GFP in the endoplasmic reticulum. A strong interaction between Panx1 and Panx3-GFP was also observed in a reciprocal coimmunoprecipitation where an antibody to Panx1 was used to pull down Panx3-GFP (Supplemental Figure 2B). Collectively, these pannexin coexpressing and coIP experiments would suggest that Panx1 interacts strongly with Panx2 and can moderately interact with Panx3, whereas Panx2 and Panx3 do not interact under the conditions tested.
Figure 8.
Cell surface localization of Panx3-GFP was evident when coexpressed with Panx1 in NRK cells. When Panx3-GFP was coexpressed with Panx1N254Q it exhibited a similar intracellular localization profile (A; arrows) as when it was expressed alone (A; inset). Bars, 20 μm (A). Antibodies to Panx3 coimmunoprecipitated the Gly0 and Gly1 species of Panx1 and Panx3-GFP (B) in 293T cells. The same blot was probed sequentially with Panx3 after Panx1, by using different secondary antibodies (see Materials and Methods). Twenty micrograms of the corresponding protein lysates were run in parallel to assess pannexin expression levels.
Selective Pannexin Intermixing Can Alter Dye Uptake
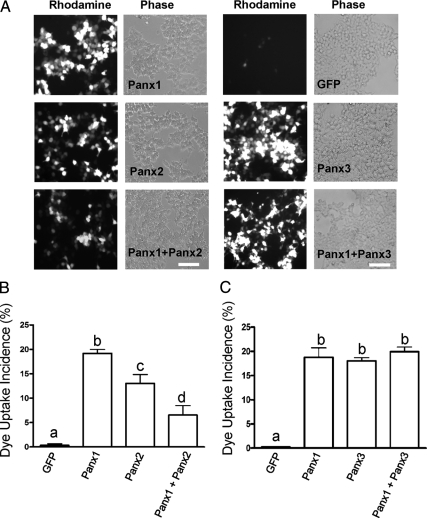
Pannexin-expressing 293T cells were mechanically stimulated to uptake sulforhodamine B dye under physiological Ca++ and Mg++ conditions. Cells expressing Panx1 or Panx3 were active in dye uptake whereas controls expressing free GFP did not show significant dye uptake (Figure 9A). Intriguingly, Panx2 expressing cells were also capable of dye uptake, but they showed lower uptake incidence than Panx1-expressing cells. Furthermore, when Panx1 and Panx2 were coexpressed in 293T cells the incidence of dye uptake was significantly reduced (p < 0.01) in comparison with cells expressing only one pannexin (Figure 9B). In contrast, when Panx1 and Panx3 were coexpressed in the same cells, there was no apparent change in dye uptake incidence (Figure 9C).
Figure 9.
Panx2, but not Panx3, intermixing with Panx1 affects dye uptake. 293T cells expressing Panx1, Panx2, or Panx3 were active in sulforhodamine dye uptake, whereas cells expressing GFP exhibited negligible dye uptake (A). The expression of Panx2 together with Panx1 reduced the incidence of dye uptake compared with Panx1 or Panx2 alone (B), whereas mixing Panx1 and Panx3 had no functional effect (C). Parallel experiments using dextran rhodamine dye revealed no uptake (data not shown). Experiments are composites of four independent transfections and assessed by one-way ANOVA followed by a Tukey test, p < 0.01. Bars indicate SEM, and letters depict statistical significance among groups. Bars, 20 μm.
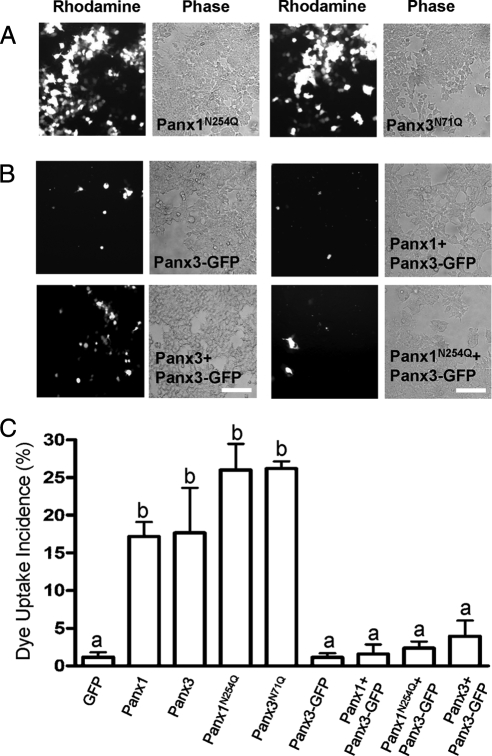
N-Glycosylation-defective Pannexin Mutants Are Capable of Dye Uptake
Interestingly, glycosylation-defective mutants Panx1N254Q and Panx3N71Q were also capable of taking up dye, indicating that a subpopulation of these mutants reach the plasma membrane and are capable of forming functional single-membrane channels (Figure 10A). Surprisingly, cells expressing Panx3-GFP or any combination of this GFP-tagged pannexin with Panx1, Panx3, or Panx1N254Q were incapable of significant dye uptake (Figure 10B). Although no significant differences were observed in dye uptake between untagged Panx1 and Panx3 and their glycosylation mutants, they were all significantly different (p < 0.001) from the GFP control and to all the pannexin coexpression combinations that included Panx3-GFP. These findings indicate that Panx3-GFP dominantly inhibits pannexin channel function (Figure 10C).
Figure 10.
Glycosylation-defective mutants of Panx1 and Panx3 can form functional single-membrane channels. Uptake of sulforhodamine B dye by 293T cells expressing pannexins, N-glycosylation–defective mutants, or combinations of pannexins was assessed (A). Dextran-rhodamine dye uptake was used as a negative control to assess damaged cells (data not shown). Significant dye uptake was observed in cells expressing Panx1, Panx3, and their corresponding N-glycosylation–deficient mutants but not in any cells coexpressing pannexins or pannexin mutants together with Panx3-GFP (B). n =3 (one-way ANOVA, p < 0.001). Bars indicate SEM, and letters depict statistical significance among groups (C). Bars, 20 μm.
DISCUSSION
Since the discovery of the pannexin family of proteins encoded within the mammalian genome (Panchin et al., 2000), there has been an increasing interest in their potential function as intercellular and/or single-membrane channels. All three family members are predicted to have a tetraspanning transmembrane topology with intracellular N and C termini, two conserved cysteines in each extracellular loop (Baranova et al., 2004), plus Panx1 and Panx3 have been characterized as N-linked glycoproteins (Boassa et al., 2007; Penuela et al., 2007). Panx1 has been shown to oligomerize into a hexamer to form a pannexon (Boassa et al., 2007) or a single-membrane channel-forming protein (Bao et al., 2004; Locovei et al., 2006; Pelegrin and Surprenant, 2006). This functional channel has been proposed recently to activate caspase-1 and the inflammasome in neural cells in response to elevated extracellular K+ (Silverman et al., 2009). However, our understanding of the other two pannexins and the potential interactions among family members is far more limited (Bruzzone et al., 2003). Therefore, in the present study, we set out to further characterize all three members of the pannexin family in terms of posttranslational glycosylation, cell surface distribution, single-membrane channel function, and the potential interactions among family members.
To aide in the characterization of this diverse family of proteins, we generated a new set of site-specific antibodies that could target not only the C termini of each protein as reported previously (Penuela et al., 2007) but also antibodies specific for their extracellular domains and intracellular loops. Having antibodies to two or three pannexin epitopes that exhibited similar patterns of immunolabeling and immunoblotting further validated the specificity of these antibodies and the identity of all expressed pannexin species. This is particularly important for pannexins because they are encoded on four to five exons (Baranova et al., 2004). This gene arrangement allows for the possibility of splice variants that can result in proteins of different sizes and, possibly, alternative functions. For example, the human PANX1 and PANX2 genes have recently been reported to be differentially spliced (Baranova et al., 2004; Ma et al., 2008), generating two splice variants of each protein, and PANX1a and b seem to have unique expression patterns (Ma et al., 2008).
Even though all three pannexins have a single predicted glycosylation site in their first (Panx2 and Panx3) or second (Panx1) extracellular loops, their level of glycosylation is highly diverse. Although Panx1 and Panx3 present core, high-mannose and complex glycoprotein forms, Panx2 is only glycosylated to a high-mannose form that manifests as a minor shift in gel mobility. Interestingly, the ability of all these forms to traffic to the cell surface is also varied. Complex glycoprotein species of Panx1 and Panx3 readily traffic to the cell surface, but although less efficient, the core and intermediate forms of Panx1 and Panx3 also reach the cell surface and function as channels in dye uptake. Previous reports revealed that the Gly0 form of mouse Panx1 did in fact reach the cell surface in HeLa cells (Huang et al., 2007a), and to a lesser extent, so did the glycosylation-deficient form of rat Panx1 (Boassa et al., 2007). The capacity of unglycosylated Panx1 to reach the cell surface may be governed by the cell type, other coexpressed pannexins or even other unknown pannexin-binding partners. A recent report by Prochnow et al. (2009) showed that in zebrafish, the glycosylation-deficient mutant zfPanx1-N246K can traffic to the cell surface, but its ability to take up ethidium bromide was impaired compared with wild-type zfPanx1. Thus, in vivo, unglycosylated species of Panx1 and Panx3 may have the potential to reach and function at the cell surface as single-membrane channels or possibly, without the steric interference of their large glycan chains, even dock with opposed pannexons to form intercellular channels. The concept of unglycosylated Panx1 forming an intercellular channel was postulated in a report that saw increased junctional conductance of frog oocytes expressing rat Panx1 upon removal of carbohydrate moieties with an N-glycosidase F treatment (Boassa et al., 2008). Given that immunofluorescent studies revealed the bulk of Panx2 within intracellular compartments, it was somewhat surprising to detect a subpopulation of Panx2 at the plasma membrane, and furthermore, to find that cell surface Panx2 could function in dye uptake, albeit less well than Panx1 or Panx3. In one other study, rat Panx2 was reported to localize to the cell surface when expressed in HeLa cells (Zappala et al., 2006). Collectively, these studies strongly suggest that a population of all three pannexin family members can reside and function at the cell surface as single-membrane channels. In the future, it will be interesting to determine whether both glycosylated and unglycosylated pannexin species are in fact present at the cell surface in vivo.
To begin to understand the functional characteristics of pannexin family members, it is critical to determine which pannexin family members interact. In the present study, we investigate all possible pannexin interactions through a series of colocalization, coimmunoprecipitation and rescue studies. The strongest and most robust interactions were found between the Gly0 and Gly1 species of Panx1 and Panx2. These data support an interaction between Panx1 and Panx2 that is initiated, while these pannexins are still residents of the endoplasmic reticulum. Although it is not possible to confirm a direct interaction between these two family members, these findings are consistent with the potential formation of mixed Panx1/2 channels that result in decreased channel function as suggested by our dye uptake assays. These new findings are in keeping with previous data in which the coinjection of rat Panx1 and Panx2 RNA in Xenopus laevis oocytes resulted in “hemichannels” or intercellular channels with attenuated functional properties from channels formed by Panx1 alone (Bruzzone et al., 2003). Although these authors never demonstrated the interaction of untagged Panx1 and Panx2 in this study, they later showed that Myc tagged rat Panx1 did coimmunoprecipitate with GFP-tagged Panx2 (Bruzzone et al., 2005). Together with our findings, it is tempting to postulate that the function of Panx2 is mainly to modulate Panx1 channels.
We had shown previously that many tissues and some cultured cell types coexpress Panx1 and Panx3 (Penuela et al., 2007, 2008), and because they share the highest homology levels among the family members, we hypothesized that they could also interact. Panx1 and Panx3 partially colocalize in NRK and 293T cells, and coimmunoprecipitation assays revealed that they physically interact. However, Panx1 and Panx3 interaction is limited and favors situations when Panx1 is not glycosylated or glycosylated to only the Gly1 species. These studies would predict that when Panx1 and Panx3 are fully glycosylated they are less likely to interact and more likely exist in homomeric arrangements as single-membrane channels at the cell surface. It is important to note that the results obtained with the Panx1 glycosylation mutant, could be attributable to misfolding of the protein, because glycosylation is important for quality control in the cell. However, the naturally occurring Gly0 form of Panx1 displayed similar properties, indicating that it is not merely an aberrant form of Panx1 but an entity that can traffic and function. Not surprisingly, because Panx2 and Panx3 are rarely if ever coexpressed in the same cell all of our evidence would suggest that they do not interact.
Further support of selective interpannexin interactions can be obtained using the trafficking and functional defective Panx3-GFP variant of Panx3. For example, we found that the Panx1 and Panx3 glycosylation-deficient mutants could uptake sulforhodamine B dye, yet when coexpressed with any combination of Panx3-GFP, no significant dye uptake was observed. Clearly, this finding provides supportive evidence that Panx1 and Panx3 can in fact interact, although intermixing of the untagged versions of Panx1 and Panx3 did not seem to alter channel function. Furthermore, the GFP tag not only affects normal Panx3 trafficking but also transforms the Panx3-GFP chimera into a dominant-negative mutant that cross-talks with Panx1 in addition to blocking the channel function of coexpressed Panx3.
In summary, the diverse subcellular distribution profiles observed for the glycoprotein family of channel forming pannexins in vitro and in vivo can be partially explained by their interactions with other members of the family. Furthermore, these interactions seem to be highly regulated by the glycosylation state of each pannexin and their intermixing can significantly affect the functional outcome. Further studies on the behavior of homomeric and heteromeric pannexin channels will be crucial to facilitate the complete understanding of the scope of pannexin functions in the cell.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Stephanie Langlois, Isabelle Plante, and Qing Shao for assistance in the optimization of coimmunoprecipitation and biotinylation assays and for helpful discussions on the manuscript. This study was supported by grants from the Canadian Institute of Health Research (to D.W.L.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-01-0067) on August 19, 2009.
REFERENCES
- Bao L., Locovei S., Dahl G. Pannexin membrane channels are mechanosensitive conduits for ATP. FEBS Lett. 2004;572:65–68. doi: 10.1016/j.febslet.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Baranova A., et al. The mammalian pannexin family is homologous to the invertebrate innexin gap junction proteins. Genomics. 2004;83:706–716. doi: 10.1016/j.ygeno.2003.09.025. [DOI] [PubMed] [Google Scholar]
- Blom N., Sicheritz-Ponten T., Gupta R., Gammeltoft S., Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Boassa D., Ambrosi C., Qiu F., Dahl G., Gaietta G., Sosinsky G. Pannexin1 channels contain a glycosylation site that targets the hexamer to the plasma membrane. J. Biol. Chem. 2007;282:31733–31743. doi: 10.1074/jbc.M702422200. [DOI] [PubMed] [Google Scholar]
- Boassa D., Qiu F., Dahl G., Sosinsky G. Trafficking dynamics of glycosylated Pannexin1 proteins. Cell. Commun. Adhes. 2008;15:119–132. doi: 10.1080/15419060802013885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzone R., Barbe M. T., Jakob N. J., Monver H. Pharmacological properties of homomeric and heteromeric pannexin hemichannels expressed in Xenopus oocytes. J. Neurochem. 2005;92:1033–1043. doi: 10.1111/j.1471-4159.2004.02947.x. [DOI] [PubMed] [Google Scholar]
- Bruzzone R., Dermietzel R. Structure and function of gap junctions in the developing brain. Cell Tissue Res. 2006;26:239–248. doi: 10.1007/s00441-006-0287-0. [DOI] [PubMed] [Google Scholar]
- Bruzzone R., Hormuzdi S. G., Barbe M. T., Herb A., Monyer H. Pannexins, a family of gap junction proteins expressed in brain. Proc. Natl. Acad. Sci. USA. 2003;100:13644–13649. doi: 10.1073/pnas.2233464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros M. G., Heijne G. V. TopPred II: an improved software for membrane protein structure predictions. CABIOS. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- Dvoriantchikova G., Ivanov D., Panchin Y., Shestopalov V. I. Expression of pannexin family of proteins in the retina. FEBS Lett. 2006a;580:2178–2182. doi: 10.1016/j.febslet.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Dvoriantchikova G., Ivanov D., Pestova A., Shestopalov V. Molecular characterization of pannexins in the lens. Mol. Vis. 2006b;12:1417–1426. [PubMed] [Google Scholar]
- Harris A. L. Connexin channel permeability to cytoplasmic molecules. Prog. Biophys. Mol. Biol. 2007;94:120–143. doi: 10.1016/j.pbiomolbio.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Grinspan J. B., Abrams C. K., Scherer S. S. Pannexin1 is expressed by neurons and glia but does not form functional gap junctions. Glia. 2007a;55:46–56. doi: 10.1002/glia.20435. [DOI] [PubMed] [Google Scholar]
- Huang Y.-J., Maruyama Y., Dvoryanchikov G., Pereira E., Chaudhari N., Roper S. D. The role of pannexin 1 hemichannels in ATP release and cell-cell communication in mouse taste buds. Proc. Natl. Acad. Sci. USA. 2007b;104:6436–6441. doi: 10.1073/pnas.0611280104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koval M. Pathways and control of connexin oligomerization. Trends Cell Biol. 2006;16:159–166. doi: 10.1016/j.tcb.2006.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois S., Cowan K. N., Shao Q., Cowan B. J., Laird D. W. Caveolin-1 and -2 interact with connexin43 and regulate gap junctional intercellular communication in keratinocytes. Mol. Biol. Cell. 2008;19:912–928. doi: 10.1091/mbc.E07-06-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locovei S., Bao L., Dahl G. Pannexin 1 in erythrocytes: function without a gap. Proc. Natl. Acad. Sci. USA. 2006;103:7655–7659. doi: 10.1073/pnas.0601037103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Hui H., Pelegrin P., Surprenant A. Pharmacological characterization of pannexin-1 currents expressed in mammalian cells. J. Pharmacol. Exp. Ther. 2008;328:409–418. doi: 10.1124/jpet.108.146365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchin Y., Kelmanson I., Matz M., Lukyanov K., Usman N., Lukyanov S. A ubiquitous family of putative gap junction molecules. Curr. Biol. 2000;10:R473–R474. doi: 10.1016/s0960-9822(00)00576-5. [DOI] [PubMed] [Google Scholar]
- Pelegrin P., Surprenant A. Pannexin-1 mediates large pore formation and interleukin-1beta release by the ATP-gated P2X7 receptor. EMBO J. 2006;25:5071–5082. doi: 10.1038/sj.emboj.7601378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelegrin P., Surprenant A. Pannexin-1 couples to maitotoxin- and nigericin-induced interleukin-1beta release through a dye uptake-independent pathway. J. Biol. Chem. 2007;282:2386–2394. doi: 10.1074/jbc.M610351200. [DOI] [PubMed] [Google Scholar]
- Penuela S., Bhalla R., Gong X.-Q., Cowan K. N., Celetti S. J., Cowan B. J., Bai D., Shao Q., Laird D. W. Pannexin 1 and pannexin 3 are glycoproteins that exhibit many distinct characteristics from the connexin family of gap junction proteins. J. Cell Sci. 2007;120:3772–3783. doi: 10.1242/jcs.009514. [DOI] [PubMed] [Google Scholar]
- Penuela S., Celetti S. J., Bhalla R., Shao Q., Laird D. W. Diverse subcellular distribution profiles of pannexin1 and pannexin3. Cell. Commun. Adhes. 2008;15:133–142. doi: 10.1080/15419060802014115. [DOI] [PubMed] [Google Scholar]
- Prochnow N., Hoffmann S., Vroman R., Klooster J., Bunse S., Kamermans M., Dermietzel R., Zoidl G. Pannexin1 in the outer retina of the zebrafish, Danio rerio. Neuroscience. 2009;162:1039–1054. doi: 10.1016/j.neuroscience.2009.04.064. [DOI] [PubMed] [Google Scholar]
- Ransford G. A., Fregien N., Qiu F., Dahl G., Conner G. E., Salathe M. Pannexin 1 contributes to ATP release in airway epithelia. Am. J. Respir. Cell Mol. Biol. 2009 doi: 10.1165/rcmb.2008-0367OC. DOI: 10.1165/rcmb.2008-0367OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., Zoidl G., Wahle P., Dermietzel R. Pannexin expression in the cerebellum. Cerebellum. 2006;5:189–192. doi: 10.1080/14734220500530082. [DOI] [PubMed] [Google Scholar]
- Ray A., Zoidl G., Weickert S., Wahle P., Dermietzel R. Site-specific and developmental expression of pannexin1 in the mouse nervous system. Eur. J. Neurosci. 2005;21:3277–3290. doi: 10.1111/j.1460-9568.2005.04139.x. [DOI] [PubMed] [Google Scholar]
- Silverman W. R., De Rivero Vaccari J. P., Locovei S., Qiu F., Carlsson S. K., Scemes E., Keane R. W., Dahl G. The pannexin 1 channel activates the inflammasome in neurons and astrocytes. J. Biol. Chem. 2009;284:18143–18151. doi: 10.1074/jbc.M109.004804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solan J. L., Lampe P. D. Connexin phosphorylation as a regulatory event linked to gap junction channel assembly. Biochim. Biophys. Acta. 2005;1711:154–163. doi: 10.1016/j.bbamem.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Tang W., Ahmad S., Shestopalov V. I., Lin X. Pannexins are new molecular candidates for assembling gap junctions in the cochlea. Neuroreport. 2008;19:1253–1257. doi: 10.1097/WNR.0b013e32830891f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarentino A. L., Plummer T.H.J. Peptide-N4-(N-acetyl-beta-glucosaminyl) asparagine amidase and endo-beta-N-acetylglucosaminidase from Flavobacterium meningosepticum. Methods Enzymol. 1987;138:770–778. doi: 10.1016/0076-6879(87)38065-6. [DOI] [PubMed] [Google Scholar]
- Tatusova T. A., Madden T. L. Blast 2 sequences—a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Thompson R. J., Jackson M. F., Olah M. E., Rungta R. L., Hines D. J., Beazely M. A., MacDonald J. F., MacVicar B. A. Activation of pannexin-1 hemichannels augments aberrant bursting in the hippocampus. Science. 2008;322:1555–1559. doi: 10.1126/science.1165209. [DOI] [PubMed] [Google Scholar]
- Thompson R. J., Zhou N., MacVicar B. A. Ischemia opens neuronal gap junction hemichannels. Science. 2006;312:924–927. doi: 10.1126/science.1126241. [DOI] [PubMed] [Google Scholar]
- Vogt A., Hormuzdi S. G., Monyer H. Pannexin1 and pannexin2 expression in the developing and mature rat brain. Mol. Brain Res. 2005;141:113–120. doi: 10.1016/j.molbrainres.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Willecke K., Eiberger J., Degen J., Eckardt D., Romualdi A., Guldenagel M., Deutsch U., Sohl G. Structural and functional diversity of connexin genes in the mouse and human genome. Biol. Chem. 2002;383:725–737. doi: 10.1515/BC.2002.076. [DOI] [PubMed] [Google Scholar]
- Zappala A., Cicero D., Serapide M. F., Paz C., Catania M. V., Falchi M., Parenti R., Panto M. R., La Delia F., Cicirata F. Expression of pannexin1 in the CNS of adult mouse: cellular localization and effect of 4-aminopyridine-induced seizures. Neuroscience. 2006;141:167–178. doi: 10.1016/j.neuroscience.2006.03.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.