Abstract
TAS loci in plant genomes encode transacting small interfering RNAs (ta-siRNAs) that regulate expression of a number of genes. The function of TAS3 precursor in Arabidopsis thaliana is controlled by two miR390 target sites flanking two ta-siARF sequences targeting mRNAs of ARF transcription factors. Cleavage of the 3′-miR390-site initiates ta-siRNAs biogenesis. Here we describe the new method for identification of plant ta-siRNA precursors based on PCR with oligodeoxyribonucleotide primers mimicking miR390. The method was found to be efficient for dicotiledonous plants, cycads, and mosses. Based on sequences of amplified loci and a database analysis, a novel type of miR390-dependent TAS sequences was identified in dicots. These TAS loci are characterized by a smaller distance between miR390 sites compared to TAS3, a single copy of ta-siARF, and a sequence conservation pattern pointing to the possibility that processing of novel TAS-like locus is initiated by cleavage of the 5′-terminal miR390 target site.
1. Introduction
Small RNA-mediated gene silencing plays important roles in many cellular processes, including development, genome maintenance and integrity, and adaptive responses to biotic and abiotic stress in most eukaryotes. Small RNAs, usually 20–25 nucleotides (nt) in length, guide heterochromatin formation, mRNA degradation, translational inhibition, and DNA elimination. In plants, small RNAs are highly diverse and significant progress has been achieved in unraveling the components and mechanisms involved in their biogenesis and function [1–8]. MicroRNAs (miRNAs) are a class of small noncoding RNAs (21-22-nt long) transcribed from the genomes of all multicellular organisms and some viruses [5, 9–13]. Plant miRNA biogenesis starts with the transcription of miRNA precursors by RNA polymerase II. These precursors contain the mature miRNA sequence within the stem of a long imperfect RNA hairpin which is processed in the nucleus by RNaseIII-like protein DCL1 to give an imperfect RNA duplex with 2 nt 3′ overhangs on each strand [5, 14, 15]. In total, several dozens of plant miRNA families (hundreds of individual miRNA species) are currently identified and found to target mainly protein-coding mRNAs [7, 12, 13, 15, 16]. However, some microRNAs guide the cleavage of the non-protein-coding primary transcripts of TAS genes directing the formation of trans-acting siRNAs (ta-siRNAs). In this case, miRNAs represent a cleavage guider for ARGONAUTE (AGO) proteins with RNaseH-like activity that cleaves TAS single-stranded RNA transcript in the region complementary to small RNA [4]. TAS cleavage product(s) is converted by cooperative action of RNA-binding protein SGS3 and RNA-polymerase RDR6 to double-stranded form and subsequently processed by DCL4 to produce a cluster of ta-siRNAs that are phased in 21 nt increment relative to the original cleavage site on both strands [4, 5, 13, 17, 18]. The generated 21 nt ta-siRNAs further work as components of RISC complex to guide AGO-dependent cleavage of their target mRNAs. Recent studies have reported that some ta-siRNAs also work as cleavage guiders to cut other TAS RNA precursors similarly to microRNA and thus generate secondary ta-siRNAs with new specificity [19]. Most TAS RNA precursors have only single miRNA target motif (e.g., miR173) positioned 5′ to ta-siRNA-producing site, and the target motif is cleaved by AGO1 guided by the respective miRNA. However, two miR390 target sites (5′ and 3′ to ta-siRNA site) were shown to be necessary for TAS3 precursor RNA cleavage that appeared to be dependent on specific interaction between AGO7 and miR390 [4, 18, 20].
In this study, we analyzed TAS3-related sequences encoded by genomes of different land plants. Using multiple alignments of TAS3-like RNAs in dicotyledonous plants [17, 18], we synthesized oligonucleotide primers complementary to miR390 target sites positioned 5′ and 3′ to ta-siRNA site. In a control PCR reaction with Arabidopsis thaliana chromosomal DNA, we obtained a PCR product of expected size. However, in Nicotiana benthamiana and tobacco, in addition to the PCR products corresponding to TAS3-like RNAs, we found smaller PCR fragments. Sequencing of these fragments and further database analysis revealed that they represent a new, previously undescribed, type of miR390-mediated TAS genes producing potential precursors of ta-siARF RNA in different dicotyledonous plants.
2. Materials and Methods
2.1. Plant Material
Plants were taken from collections of Main Botanic Garden of the Russian Academy of Sciences and Department of Virology, Moscow State University. Solanaceous plants were germinated in standard growth chamber conditions for several weeks until stage of 6–8 leaves.
2.2. Analysis of Nucleic Acids
Genomic plant DNA was isolated from 200 mg of plant material by DNA extraction kit (Macherey-Nagel) according to the protocol of the manufacturer. Total RNA was isolated from young tobacco leaves with the Trizol reagent according to the manufacturer's instructions (Invitrogen). Digestion of any contaminating DNA was achieved by treatment of samples with RQI RNase-free DNase (Promega). Reverse transcription was performed with 1 μg of total RNA and oligo (dT)-primer using the RT system (Invitrogen) according to the protocol of the manufacturer. Primers for dicotiledonous plants and Cycas revoluta were: forward primer: TAS-P 5′-GGTGCTATCCTATCTGAGCTT-3′ and reverse primers TAS-Mcaa 5′-AGCTCAGGAGGGATAGCAA-3′ and TAS-Maca 5′-AGCTCAGGAGGGATAGACA-3′.
The primers for mosses (Physcomitrella-specific) were: forward primers: Bryo TAS-P1 5′-GGCGCTATCCCTCCTGAGCT-3′ and Bryo TAS-P3 5′-GACGCTACCCTTCCTGAGCT-3′, reverse primer: Bryo TAS-M 5′-TAGCTCAGGAGTGATA(G/T)A(C/A)AA-3′. For PCR, 25–35 cycles were used for amplification with a melting temperature of 95°C, an annealing temperature of 58°C (for seed plants) or 60°C (for mosses), and an extending temperature of 72°C, each for 30 seconds, followed by a final extension at 72°C for 3 minutes. PCR products were visualized by electrophoresis of samples in 1% agarose gel. For cloning, the PCR-amplified DNA bands were isolated from gel and ligated into pGEM-T (Promega). In each case, 7–18 independent clones were sequenced. Sequences of 20–50% of clones depending on particular plant species showed no presence of tasi-ARF RNA sequences and these clones were regarded as false ones. In some cases (less than 10% of clones), nucleotide sequences of tasi-ARF precursor genes have the ends (primer sites) which are different by single nucleotide substitutions or deletions from the primer sequences. We propose that this discrepancy may result from rare mutations after cloning of PCR products in Escherichia coli.
2.3. GeneBank Accession Numbers
DNA sequences were deposited at NCBI databank under the following accession numbers: Nicotiana benthamiana—FJ804742; Nicotiana tabacum—FJ804743 (“short” TAS) and FJ804751 (classical TAS); Datura stramonium—FJ804744; Solanum demissum—FJ804745; Physalis longifolia—FJ804746; Brachythecium latifolium (clone 50 Br)—FJ804747; Brachythecium latifolium (clone 47 Br)—FJ804748; Hookeria lucens—FJ804749; Cycas revolute—FJ804750.
3. Results
To design primers for PCR and cloning of TAS3-like genes from Nicotiana benthamiana and Nicotiana tabacum, we used a multiple alignment of TAS3-like RNA precursors from flowering plants published by Axtell and others [18]. PCR analysis was performed with two pairs of primers: P-Tas3 corresponded to the 5′ miR390 target site and either M-Tas3/caa or M-Tas3/aca, both complementary to the 3′ miR390 target/cleavage site [18, 20]. Control PCR reaction using chromosomal Arabidopsis thaliana DNA as template and both paires of primers resulted in efficient synthesis of a single PCR-fragments with expected size of 260 bp (Figure 1(a)) that was in agreement with calculated distance between 5′ and 3′ miR390 target sites in Arabidopsis ta-siARF precursor RNAs [17, 18]. Sequencing of these PCR-amplified and cloned DNA fragments confirmed specific amplification of at least two of three known A. thaliana TAS3 chromosomal loci (data not shown) [17, 21].
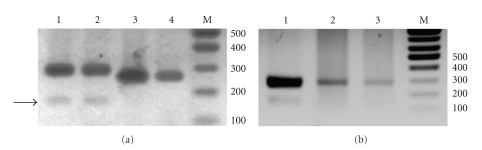
Figure 1.
Agarose gel analysis of PCR products. (a) Amplification of genomic DNA sequences flanked by miR390 sites. PCR products were obtained on Nicotiana tabacum genomic DNA with primers P-Tas3 and M-Tas3/caa (Lane 1), on N. tabacum genomic DNA with primers P-Tas3 and M-Tas3/aca (Lane 2), on Arabidopsis thaliana genomic DNA with primers P-Tas3 and M-Tas3/caa (Lane 3), and on A. thaliana genomic DNA with primers P-Tas3 and M-Tas3/aca (Lane 4). Arrow points to the 170-nt PCR product. M is DNA size markers. Marker fragment sizes in base pairs are indicated on the left. (b) Amplification of N. tabacum genomic DNA and cDNA preparation. PCR products were obtained on N. tabacum genomic DNA with primers P-Tas3 and M-Tas3/caa (Lane 1), on N. tabacum cDNA with primers P-Tas3 and M-Tas3/caa (Lane 2), and on N. tabacum cDNA with primers P-Tas3 and M-Tas3/aca (Lane 3). M is DNA size markers. Marker fragment sizes in base pairs are indicated on the left.
PCR amplification of chromosomal DNA from Nicotiana benthamiana and Nicotiana tabacum (cv. Samsun) also resulted in synthesis of one major band of 250 bp and 280 bp, respectively (Figure 1(a) and data not shown). Cloning and sequencing of the obtained DNA bands revealed that the amplified sequences contained putative ta-siARF site composed of two tandem copies of ARF-specific ta-siRNAs and located between miR390 target sites corresponding to PCR primers. Moreover, amplified sequences showed obvious similarity to TAS3-like genes from other dicotyledonous plants (Figure 2) [17, 18]. Bioinformatic analysis of the putative TAS3-like sequences from Nicotiana benthamiana and Nicotiana tabacum using NCBI Blast revealed closely related sequences in representatives of the family Solanaceae. For example, Nicotiana tabacum (cv. Samsun) TAS3-like sequence showed almost 100% identity to the Nicotiana tabacum (cv. Hicks Broadleaf) genomic sequence obtained in the frame of Tobacco Genome Initiative (GenBank accession no. FH434354) and 78% identity to the Solanum lycopersicum (cv. Heinz 1706) chromosome 11 BAC clone (GenBank accession no. DU917444) (Figure 2).
Figure 2.
Nucleotide sequence alignment of “280-nt-long” ta-siARF loci of dicot plants. Sequences of Nicotiana tabacum and Nicotiana benthamiana (undelined) were determined in this study, the others were from GeneBank. Boxes indicate miR390 complementary sites and ta-siARF-encoding sequence regions. The length of nonconserved sequence between the 5′-miR390 complementary site and ta-siARF sequence not included in the alignment is indicated. Shading shows residues identical for the given position in most sequences. Sequences of N. tabacum and N. benthamiana miR390 complementary sites originate from primers used for the amplification of ta-siARF loci. GeneBank accession numbers of the sequences included in the alignment are as follows: Nicotiana tabacum, FJ804751; Nicotiana benthamiana, FJ804742; Arabidopsis thaliana, BX838290; Glycine max, BE330988; Manihot esculenta, CK652751; Malus domestica, CN490861; Vitis vinifera, DT025007; Populus trichocarpa, DT498974; Lycopersicon esculentum, DV105041.
Unlike Arabidopsis thaliana DNA showing only one visible DNA band after PCR amplification, Nicotiana tabacum DNA consistently produced a second minor PCR product of about 170 bp in length (Figure 1(a)). Since no indications on existence of miR390-dependent TAS3-like genes with so closely arranged miRNA target sites were found in the literature and sequence databases, cloning and sequencing of the minor PCR fragment was carried out. The 170 bp DNA sequence was found to contain the putative ta-siARF site composed of one copy of ARF-specific ta-siRNAs (Figure 3). To rule out the possibility of PCR/cloning artifact we compared this sequence to the available nucleotide sequence databases. As it was found for the 280-nucleotide-long Nicotiana tabacum (cv. Samsun) PCR product, 170 bp fragment contained sequence closely related to some yet uncharacterized database entries (Figure 3). Particularly, it showed 96% identity to Nicotiana tabacum (cv. Hicks Broadleaf) genomic survey sequences (GSS) obtained in the frame of Tobacco Genome Initiative (GenBank accessions nos. FH734100, FH203124 and FH011695), 91% identity to EST sequence of Nicotiana tabacum (cv. SNN) (GenBank accession nos. AM791738), 79% identity to the Solanum tuberosum EST sequence (GenBank accession no. FG548921), 78% identity to the Solanum lycopersicum chromosome 12 clone LE_HBa-26C13 (GenBank accession nos. AC209585) and the Solanum lycopersicum EST sequence (GenBank accession nos. BE459870). Somewhat lesser identity (73-74%) was also revealed for EST sequences from Solanum phureja (GenBank accession nos. FG647537) and Solanum tuberosum (GenBank accessions nos. BQ514736 and BI431636) (Figure 3 and data not shown). These data indicated that 170 bp-long PCR-amplified fragments corresponded to the genuine Nicotiana tabacum (cv. Samsun) genome fragment potentially encoding a new type of ta-siARF RNA precursors. Sequence analysis revealed that, except their length, the 170-nt-long ta-siARF RNA differ from well-characterized 280-nt-long precursors in the structure of ta-siARF-encoding region: while the longer precursors contain two tandem copies of ta-siARF arranged as the conserved 42-nt region, the smaller precursors described in this paper have only one monomeric 21-nt ta-siARF copy (Figures 2 and 3).
Figure 3.
Nucleotide sequence alignment of “170-nt-long” ta-siARF loci of dicot plants. Sequences of Nicotiana tabacum, Datura stramonium, Solanum demissum, and Physalis longifolia (undelined) were determined in this study the others were from GeneBank. Boxes indicate miR390 complementary sites and ta-siARF-encoding sequence regions. The length of nonconserved sequence between the ta-siARF sequence and 3′-miR390 complementary site not included in the alignment is indicated. Dashes in the sequences of Cichorium endivia, Citrus aurantium, Citrus sinensis, and Populus tremula indicate the sequence region lacking the 3′-miR390 complementary site that could not be aligned with other sequences. Shading shows residues identical for the given position in most sequences. Sequences of N. tabacum, D. stramonium, S. demissum, and P. longifolia miR390 complementary sites originate from primers used for the amplification of ta-siARF loci. GeneBank accession numbers of the sequences included in the alignment are as follows: Nicotiana tabacum, FJ804743; Datura stramonium, FJ804744; Solanum demissum, FJ804745; Physalis longifolia, FJ804746; Nicotiana tabacum, AM791738; Solanum lycopersicum, BE459870; Vitis vinifera, CU775162; Beta vulgaris, CV301446; Manihot esculenta, DV447689; Gossypium hirsutum, DW502659; Helianthus exilis 1, EE630512; Helianthus exilis 2, EE657417; Cyamopsis tetragonoloba, EG977206; Vigna unguiculata, FG922881; Cynara scolymus, GE588140; Cichorium endivia, EL348656; Citrus aurantium, EY848024; Citrus sinensis, CK935773; Populus tremula, DN500355.
We further experimentally analyzed the occurrence of such putative TAS precursor RNAs in other Solanaceae representatives. Genomic DNAs of Datura stramonium, Solanum demissum and Physalis longifolia were taken as templates for PCR with primers P-Tas3 and either M-Tas3/caa or M-Tas3/aca. In all cases, major 280-nucleotide-long and minor 170 bp amplified fragments were easily detected similarly to Nicotiana tabacum (data not shown). Sequencing of the smaller DNA band confirmed the occurrence of novel putative monomeric ta-siARF precursor sequences in different Solanaceae representatives (order Solanales) (Figure 3).
The TAS3 loci and miR390 are known to be well conserved among diverse flowering plants [18, 21, 22]. To explore the possibility that the novel monomeric ta-siARF precursor sequences might have emerged in plant evolution earlier than the appearance of family Solanaceae, we searched EST database at NCBI. We found that many dicot plants code for putative monomeric ta-siARF loci surrounded in most cases by complementary sites for miR390 positioned 160–190 bp from each other, thus confirming that targeting of ta-siRNA precursors is an evolutionarily conserved function of miR390. These plants include representatives of eurosids—orders Malpighiales, Fabales, Malvales, and Sapindales, asterids—orders Asterales and Solanales, as well as orders Caryophyllales and Vitales (Figure 3). Analysis of all identified monomeric ta-siARF precursor sequences revealed, in addition to the only copy of ta-siARF sequence, one more characteristic feature. The region between the 5′-miR390 target site and the ta-siARF sequence appeared to be well conserved in length and, to some extent, in sequence, whereas the region between the ta-siARF sequence and the 3′-miR390 target site was found to be variable both in length and sequence (Figure 3). This arrangement of the novel 170 bp-TAS loci differed considerably from that of the well characterized 280 bp-TAS loci, where the sequence and distance conservation was observed for the region between the tandem ta-siARF sequence and the 3′-miR390 target site and not for the sequence between the 5′-miR390 target site and the ta-siARF sequence (Figure 2).
To verify expression of novel putative monomeric ta-siARF precursor RNAs in plant leaves, total RNA was isolated from tobacco plants at 6-8-leaf stage and used as a template for cDNA synthesis with oligo(dT)-primer. It was found that PCR with primers P-Tas3, and either M-Tas3/caa or M-Tas3/aca carried out on this cDNA preparation gave rise only to 280-nucleotide-long DNA fragment but not detectable 170 bp-fragment (Figure 1(b)).
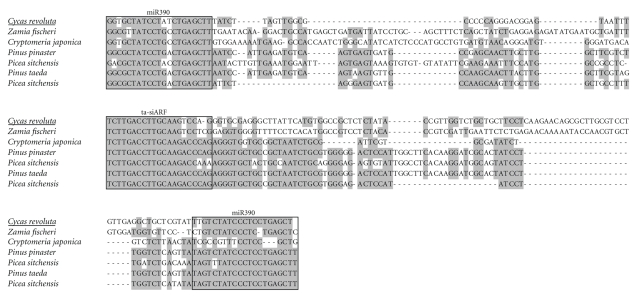
Conifers (Pinus taeda) TAS-like locus, like that of higher plants, was shown to contain the 42-nt ta-siARF region flanked by two miR390 complementary sites. However, unlike TAS3 of flowering plants, only the 5′ miR390-target site is cleaved in conifer TAS3-like RNA precursor [18]. To reveal the possible conservation of TAS3-like loci in lower seed plants, we performed PCR amplification of total DNA from Cycads (Cycas revoluta) using dicot-specific primers P-Tas3, M-Tas3/caa and M-Tas3/aca. As it was found for tobacco, PCR on Cycas revoluta DNA gave rise to one major DNA fragment of 250 bp and, in addition, several larger and smaller minor bands including those of 170–190 bp in length (data not shown). Sequencing and BLAST analysis of the latter sort of bands revealed that they corresponded to the monomeric ta-siARF precursors of flowering plants (Figure 4). Analysis of databases revealed conifers-encoded sequences showing similarity to the sequenced Cycas revoluta putative ta-siARF precursor (Figure 4), showing therefore that this type of regulatory element might have emerged early in seed plant evolution.
Figure 4.
Nucleotide sequence alignment of “short” ta-siARF loci of cycad and conifer plants. The sequence of Cycas revoluta (undelined) was determined in this study, the others were taken from GeneBank. Boxes indicate miR390 complementary sites and ta-siARF sequence regions. Shading shows residues identical for the given position in most sequences. Sequences of C. revoluta miR390 complementary sites originate from primers used for the amplification of ta-siARF loci. GeneBank accession numbers of the sequences included in the alignment are as follows: Cycas revoluta, FJ804750; Picea sitchensis, EF086492; Pinus pinaster, BX682439; Pinus taeda, CV034496; Picea sitchensis, CO220369; Zamia fischeri, DY034932; Cryptomeria japonica, DC432705.
Previously, four ta-siRNA loci targeted by miR390 were found in moss Physcomitrella patens and referred to as PpTAS3a-d. All four loci contain 5′ and 3′ miR390-target sites, and ta-siRNAs derived from these moss loci regulate, similarly to those encoded by the angiosperm TAS3 loci, ARF genes and, additionally, AP2 transcription factor genes [18, 23, 24]. To confirm applicability of the approach based on PCR amplification of TAS3-like loci to lower plants, we designed new primers BryoTAS-P1, Bryo TAS-P3, and Bryo TAS-M complementary to miR390-target sites of PpTAS3a-d. These primers were used for PCR on total DNA isolated from Brachythecium latifolium and Hookeria lucens. Sequencing of major amplified DNA bands showed that Brachythecium latifolium encodes at least two types of miR390-dependent AP2/ARF-targeting TAS loci demonstrating sequence similarity to different elements of this kind characterized for Physcomitrella patens, namely, PpTAS3d (Figure 5(a)) and PpTAS3b/3c (Figure 5(b)), whereas Hookeria lucens, despite its distant taxonomic relation to Physcomitrella patens, code for a rather close homolog of PpTAS3b/3c (Figure 5(b)).
Figure 5.
Nucleotide sequence alignment of ta-siARF loci of mosses. (a) Alignment of Brachythecium latifolium clone 50 Br sequence and Physcomitrella patens TAS3d. (b) Alignment of B. latifolium clone 47 Br sequence and Hookeria lucens with P. patens TAS3b and TAS3c. Sequences of B. latifolium and H. lucens (undelined) were determined in this study, the others were from GeneBank. Boxes indicate miR390 complementary sites, ta-siARF sequence regions and ta-siAP2 regions. Shading shows residues identical for the given position in most sequences. Sequences of B. latifolium and H. lucens miR390 complementary sites originate from primers used for the amplification of ta-siARF loci. GeneBank accession numbers of the sequences included in the alignment are as follows: Brachythecium latifolium clone 50 Br, FJ804747; Brachythecium latifolium clone 47 Br, FJ804748; Hookeria lucens, FJ804749; Physcomitrella patens TAS3d, BK005828; P. patens TAS3b, BK005826; P. patens TAS3c, BK005827.
4. Discussion
MicroRNAs and ta-siRNAs are small RNAs around 21 nt in length and have widespread roles as posttranscriptional regulators of plant physiology and development [25]. They can affect target genes through a variety of mechanisms, including transcript cleavage and translational repression. The mature miRNAs and ta-siRNAs are derived from larger precursor transcripts that are a few hundred base pairs in size. These precursor RNAs are capped and polyadenylated and can be spliced, in agreement with them being transcribed by RNA polymerase II [12, 26, 27]. Many plant miRNAs and ta-siRNAs are encoded by gene families. For example, four ta-siRNA loci targeted by miR390 were found in P. patens and three miR390-dependent TAS loci were identified in A. thaliana [17, 18, 21, 23]. Given their similar complementarity to miR390, it was reasoned that multiple TAS3-like loci might be paralogs derived from duplication and divergence of an ancestral locus, and perhaps the areas producing biologically relevant ta-siRNAs may have been preferentially conserved [18, 23]. However, despite obvious progress in studies of miRNA precursor evolution in plants [12, 28, 29], little is known about TAS gene molecular evolution.
Comparing sequences of ta-siRNA precursors within plant genera and between distantly related species should help to determine patterns of molecular evolution that is subject to different evolutionary constraints in comparison with patterns of protein-coding gene evolution. However, the discovery of ta-siRNA genes in plants is still an ongoing process. Although much effort has been directed toward ta-siRNA identification in Arabidopsis, poplar and moss Physcomitrella patens, many economically or evolutionarily important species have yet to be examined, and more ta-siRNAs likely remain to be discovered even in the species that have been extensively studied. One limiting factor in the ta-siRNA discovery process is the availability of a sequenced genome, without which a comprehensive analysis of the potential precursor structures of cloned small RNAs is not possible. In Arabidopsis, three major methods have been used for ta-siRNA discovery: forward genetics, direct cloning and sequencing, and bioinformatic prediction. Although forward genetic studies have only resulted in the identification of a few ta-siRNAs, this method provides hints to the functions of these miRNAs in addition to their isolation [3]. A second approach to miRNA discovery was bioinformatic prediction. While a number of published studies employed several different algorithms to predict ta-siRNAs, the features that the algorithms search for in the genomic sequences are based on our current knowledge of plant miRNAs and are largely similar among the studies [19]. A third, and perhaps the most effective, method for ta-siRNA discovery was direct cloning and sequencing. In particular, deep sequencing of cloned small RNA libraries using massively parallel signature sequencing (MPSS) or pyrosequencing allowed the identification of numerous small RNAs from Arabidopsis and provided a picture of the genomic landscape of small RNAs [18, 23]. One added advantage of this approach is that most small RNA species from a particular genomic loci are exhibited, which helps discern whether the locus gives rise to a single or multiple siRNAs. If multiple small RNAs mapping to both strands of a locus are present, these small RNAs are most likely siRNAs.
Here, we describe a new method for identification of plant miR390-flanked ta-siRNAs precursors based on PCR on the template of genomic DNA or cDNA using oligodeoxyribonucleotide primers mimicking in their specificity miR390. General idea of this experimental approach was suggested by the recently published paper on cloning of animal miRNA-regulated target genes by reverse transcription with direct use of miRNA as a primer [30]. Importantly, the recognition of ta-siRNA precursor by miR390 occurs in most cases by forming imperfect base pairs with 5′ and 3′ target sites [18, 23]. Assuming that imperfect base-pairing is tolerated by Taq polymerase, there is a high chance that designed miR390-related oligodeoxyribonucleotides will recognize both target sites in phylogenetically distant species. To investigate functional constraints on sequence evolution of ta-siARF precursor genes, we have studied sequence variation in the putative precursors of miR390-dependent TAS-like genetic elements between species from the Solanaceae. The application of the novel PCR-based method to Nicotiana tabacum showed obvious difference from A. thaliana: N. tabacum DNA directed a major 280-nucleotide-long PCR product and the second minor PCR product of about 170 bp in length (Figure 1). When the method was also applied to Datura stramonium, Solanum demissum, and Physalis longifolia, the minor PCR short product was again identified. Sequencing of 280-nucleotide-long PCR product in the Solanaceae and Arabidopsis as a control showed that we amplified and cloned the chromosomal region closely related to typical TAS3-related precursor sequences found in diverse dicot plants [17, 18]. This region included 42-nt region from AtTAS3 tasiRNAs that target ARF3 and ARF4 (tasiARFs) and 5′- and 3′- terminal 21-nt regions corresponding to the miR390 complementary sites (PCR primer sites) (Figure 2). These results demonstrate that the PCR-based method is a useful way to clone and identify targets on the basis of base pairing with individual miRNAs. Nucleotide sequence of the minor 170-nucleotide-long product of four selected solanaceous plants revealed the putative 21-nt ta-siARF site composed of one copy of ARF-specific ta-siRNAs and positioned between the miR390 complementary sites (PCR primer sites) (Figure 3). We searched publicly available nucleotide databases of genome survey sequences (GSSs), high-throughput genomics sequences (HTGSs), expressed sequenced tags (ESTs), and nonredundant (NR) nucleotides and identified 16 putative “short” miR390-dependent TAS sequences in 15 diverse plant species (Figure 3 and data not shown).
Using this PCR approach, ta-siARF precursors characterized by the monomeric ta-siARF were found in the cycad Cycas revolute. Database search confirmed this experimental finding and revealed novel miR390-dependent TAS sequences in one more cycad (Zamia fischeri) and four conifer species (Figure 4). The experimental approach used in this paper was further verified by the analysis of mosses with primers based on Physcomitrella patens miR390 complementary; the PCR products obtained on Brachythecium latifolium and Hookeria lucens DNA represented ta-siRNA precursor sequences that were not known before for these species and related to the previously characterized P. patens ta-siRNA loci (Figures 5(a) and 5(b)).
To analyze expression of the novel putative ta-siARF precursor RNAs in plants, total RNA was isolated from tobacco plants at 6-8-leaf stage and used as template for cDNA synthesis with oligo(dT)-primer. PCR analysis of leaf cDNA revealed only 280-nucleotide-long DNA fragment which sequence corresponded to classical tobacco TAS3-like element (Figure 1(b)). On the other hand, RNA template for the EST sequence of Nicotiana tabacum (cv. SNN) (GenBank accession no. AM791738) with 91% identity to our “short” TAS sequence from Nicotiana tabacum (cv. Samsun) (Figure 3) was isolated from 5-day seedlings. These data suggest that the novel type of miR390-dependent TAS elements might be expressed in tobacco only at early stages of ontogenesis.
Importantly, alignment of the putative novel TAS elements revealed that the length of a region between ta-siARF site and 5′ miR390-target site is highly preserved in contrast to classical TAS3-like genes in which perfect size conservation was revealed between ta-siARF site and 3′ miR390-target site (Figures 2 and 3). In addition, “core consensus” in classical ta-siARF site was found to represent sequence: TCTTGACCTTGTAAGACCPy, whereas consensus in “short” TAS is different: TCTTGACCTTGCAAGPyCCPu. Significance of this variation remains obscure.
Strikingly, in four cases we failed to reveal the 3′ miR390-target site in “short” miR390-dependent TAS sequences (Figure 3). This type of organization of ta-siRNA precursors is common for the other, except TAS3, TAS genes in Arabidopsis, and other plants [4, 12] in which miRNA-dependent cleavage occurs 5′ to ta-siRNA sites. Therefore, we speculate that in “short” miR390-dependent TAS sequences miRNA-dependent cleavage also can occur 5′ to ta-siRNA sites. This hypothesis requires experimental verification. In addition to identification of the precursor cleavage site, the role of AGO1 and AGO7 in the “short” miR390-dependent ta-siARF precursor processing should be investigated in comparison to classical TAS3-like precursors [4, 20].
Recently a novel silencing method utilizing artificial ta-siRNAs was proved to be efficient for the chimeric Arabidopsis thaliana TAS1 and TAS3 genes [31, 32]. This method provides an excellent tool not only for silencing of genes-of- interest but also for investigating functionality of TAS genes and their specific sequence modifications. Application of this method to “short” TAS3-like genes identified in this paper may result in a identification of their possible functions and mapping of cleavage sites in RNA precursors.
Acknowledgments
The authors thank A. C. Timonin for stimulating discussions. This work was supported by Grants 08-04-00846 and 09-04-00707 from the Russian Foundation for Basic Research.
References
- 1.Baulcombe D. RNA silencing in plants. Nature. 2004;431(7006):356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 2.Vazquez F. Arabidopsis endogenous small RNAs: highways and byways. Trends in Plant Science. 2006;11(9):460–468. doi: 10.1016/j.tplants.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes & Development. 2006;20(7):759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 4.Vaucheret H. Plant ARGONAUTES. Trends in Plant Science. 2008;13(7):350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Bonnet E, van de Peer Y, Rouzé P. The small RNA world of plants. New Phytologist. 2006;171(3):451–468. doi: 10.1111/j.1469-8137.2006.01806.x. [DOI] [PubMed] [Google Scholar]
- 6.Laporte P, Merchan F, Amor BB, et al. Riboregulators in plant development. Biochemical Society Transactions. 2007;35(6):1638–1642. doi: 10.1042/BST0351638. [DOI] [PubMed] [Google Scholar]
- 7.Chu CY, Rana TM. Small RNAs: regulators and guardians of the genome. Journal of Cellular Physiology. 2007;213(2):412–419. doi: 10.1002/jcp.21230. [DOI] [PubMed] [Google Scholar]
- 8.Liu B, Chen Z, Song X, et al. Oryza sativa Dicer-like4 reveals a key role for small interfering RNA silencing in plant development. The Plant Cell. 2007;19(9):2705–2718. doi: 10.1105/tpc.107.052209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X. MicroRNA metabolism in plants. Current Topics in Microbiology and Immunology. 2008;320:117–136. doi: 10.1007/978-3-540-75157-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Research. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host and Microbe. 2008;3(6):375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Axtell MJ, Bowman JL. Evolution of plant microRNAs and their targets. Trends in Plant Science. 2008;13(7):343–349. doi: 10.1016/j.tplants.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Mallory AC, Bouché N. MicroRNA-directed regulation: to cleave or not to cleave. Trends in Plant Science. 2008;13(7):359–367. doi: 10.1016/j.tplants.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Ambros V, Chen X. The regulation of genes and genomes by small RNAs. Development. 2007;134(9):1635–1641. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- 15.Liu B, Li P, Li X, et al. Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiology. 2005;139(1):296–305. doi: 10.1104/pp.105.063420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeda A, Iwasaki S, Watanabe T, et al. The mechanism selecting the guide strand from small RNA duplexes is different among Argonaute proteins. Plant & Cell Physiology. 2008;49(4):493–500. doi: 10.1093/pcp/pcn043. [DOI] [PubMed] [Google Scholar]
- 17.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121(2):207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127(3):565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 19.Chen HM, Li YH, Wu SH. Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(9):3318–3323. doi: 10.1073/pnas.0611119104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montgomery TA, Howell MD, Cuperus JT, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133(1):128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 21.Williams L, Carles CC, Osmont KS, Fletcher JC. A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(27):9703–9708. doi: 10.1073/pnas.0504029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Axtell MJ, Bartel DP. Antiquity of microRNAs and their targets in land plants. Plant Cell. 2005;17(6):1658–1673. doi: 10.1105/tpc.105.032185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Axtell MJ, Snyder JA, Bartel DP. Common functions for diverse small RNAs of land plants. Plant Cell. 2007;19(6):1750–1769. doi: 10.1105/tpc.107.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, et al. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell. 2007;19(3):926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chuck G, Candela H, Hake S. Big impacts by small RNAs in plant development. Current Opinion in Plant Biology. 2009;12(1):81–86. doi: 10.1016/j.pbi.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Ha M, Pang M, Agarwal V, Chen ZJ. Interspecies regulation of microRNAs and their targets. Biochimica et Biophysica Acta. 2008;1779(11):735–742. doi: 10.1016/j.bbagrm.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annual Review of Plant Biology. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 28.Li A, Mao L. Evolution of plant microRNA gene families. Cell Research. 2007;17(3):212–218. doi: 10.1038/sj.cr.7310113. [DOI] [PubMed] [Google Scholar]
- 29.Warthmann N, Das S, Lanz C, Weigel D. Comparative analysis of the MIR319a microRNA locus in Arabidopsis and related Brassicaceae. Molecular Biology and Evolution. 2008;25(5):892–902. doi: 10.1093/molbev/msn029. [DOI] [PubMed] [Google Scholar]
- 30.Andachi Y. A novel biochemical method to identify target genes of individual microRNAs: identification of a new Caenorhabditis elegans let-7 target. RNA. 2008;14(11):2440–2451. doi: 10.1261/rna.1139508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montgomery TA, Howell MD, Cuperus JT, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133(1):128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 32.de la Luz Gutiérrez-Nava M, Aukerman MJ, Sakai H, Tingey SV, Williams RW. Artificial trans-acting siRNAs confer consistent and effective gene silencing. Plant Physiology. 2008;147(2):543–551. doi: 10.1104/pp.108.118307. [DOI] [PMC free article] [PubMed] [Google Scholar]